Important interactions controlling the function of CstF-64 in histone RNA processing and general mRNA cleavage/polyadenylation are identified, and an interesting coregulation of CstF-64 and its paralogue CstF-64Tau leads to a model for CstF regulation and its role in modulating poly(A) site choice.
Abstract
Cleavage/polyadenylation of mRNAs and 3′ processing of replication-dependent histone transcripts are both mediated by large complexes that share several protein components. Functional studies of these shared proteins are complicated by the cooperative binding of the individual subunits. For CstF-64, an additional difficulty is that symplekin and CstF-77 bind mutually exclusively to its hinge domain. Here we have identified CstF-64 and symplekin mutants that allowed us to distinguish between these interactions and to elucidate the role of CstF-64 in the two processing reactions. The interaction of CstF-64 with symplekin is limiting for histone RNA 3′ processing but relatively unimportant for cleavage/polyadenylation. In contrast, the nuclear accumulation of CstF-64 depends on its binding to CstF-77 and not to symplekin. Moreover, the CstF-64 paralogue CstF-64Tau can compensate for the loss of CstF-64. As CstF-64Tau has a lower affinity for CstF-77 than CstF-64 and is relatively unstable, it is the minor form. However, it may become up-regulated when the CstF-64 level decreases, which has biological implications for spermatogenesis and probably also for other regulatory events. Thus, the interactions between CstF-64/CstF-64Tau and CstF-77 are important for the maintenance of stoichiometric nuclear levels of the CstF complex components and for their intracellular localization, stability, and function.
INTRODUCTION
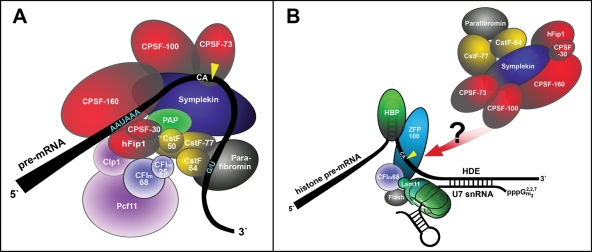
The 3′ ends of eukaryotic mRNAs can be generated through two distinct 3′-end formation pathways. For most mRNAs, the 3′ ends are generated in a process consisting of an endonucleolytic cleavage followed by the addition of a poly(A) tail to the end of the message (Zhao et al., 1999). The exceptions are the replication-dependent histone mRNAs of metazoans and some lower algae for which only an endonucleolytic cleavage is required, leaving a conserved hairpin structure at the end of the mRNA (Müller and Schümperli, 1997; Dominski and Marzluff, 2007). These two processing reactions share several trans-acting factors (Figure 1), that is, the cleavage and polyadenylation specificity factor (CPSF) that contains the active endonuclease CPSF-73, the cleavage stimulation factor subunits CstF-64 and CstF-77, and the scaffolding protein symplekin (Kolev and Steitz, 2005). For histone RNA processing, these proteins form a complex that was previously described as a heat-labile histone processing factor (HLF; Gick et al., 1987) that was found to be cell cycle regulated (Lüscher and Schümperli, 1987). A recent study has revealed yet another shared factor, the 68-kDa subunit of mammalian cleavage factor I (Ruepp et al., 2010). However, CF Im-68 interacts with the U7 snRNP (Ruepp et al., 2010) and does not seem to be part of the HLF (Kolev and Steitz, 2005).
FIGURE 1:
Schematic drawing of known 3′-end processing factors for (A) polyadenylated mRNAs. For reasons of simplicity, the C-terminal domain of RNA polymerase II, poly(A)-binding protein, and factors coupling 3′-end formation to transcription and splicing are not depicted. CF Im and CstF are shown as heterodimer and heterotrimer, instead of heterotetramer and heterohexamer. The arrangement of factors is based on known interactions of individual subunits (Zhao et al., 1999). (B) Replication-dependent histone mRNAs. The U7 snRNA contacts the histone downstream element, and this interaction is stabilized by ZFP100 that bridges the U7 snRNP to HBP/SLBP, bound to the hairpin (Dominski and Marzluff, 2007). The 68-kDa mammalian cleavage factor I subunit CF Im68 (Ruepp et al., 2010) and FLASH (Yang et al., 2009; Bongiorno-Borbone et al., 2010) interact with the U7-specific protein Lsm11 and participate in the reaction. A heat-labile processing factor (HLF) composed of CPSF, symplekin, and the two CstF subunits CstF-64 and CstF-77 (Kolev and Steitz, 2005) is recruited by yet unknown interactions. CPSF-73 is the common endonuclease that performs the endonucleolytic cleavage in both types of reactions (Dominski, 2007). Parafibromin interacts with CstF/CPSF and seems to participate in both reactions (Rozenblatt-Rosen et al., 2009; Farber et al., 2010). The cleavage sites downstream of the conserved CA dinucleotides are indicated by yellow arrowheads.
On poly(A)+ processing substrates, CSPF-160 and CstF-64, as part of the CSPF and CstF subcomplexes, bind to the polyadenylation signal sequence AAUAAA and a GU-rich downstream element, respectively (Zhao et al., 1999). As these elements are not present in replication-dependent histone mRNAs, it will be important to define how these common factors are recruited to histone pre-mRNAs and what their individual functions in the 3′-end processing reaction are.
As part of our own effort to study the function of individual subunits, we have mapped the interaction of symplekin to CstF-64 and vice versa. This has allowed us to generate RNAi-resistant symplekin and CstF-64 mutants that are disturbed in their ability to interact with each other, thereby allowing us to address their isolated function in a depletion background when introduced into HeLa cells by transient expression.
In the course of these experiments, the CstF paralogue CstF-64Tau attracted our attention. We show here that CstF-64Tau can act as a functional component in both types of 3′-end processing reactions and suggest a regulatory network between CstF-77, CstF-64, and its Tau variant.
RESULTS
Determination of CstF-64 interaction site on symplekin
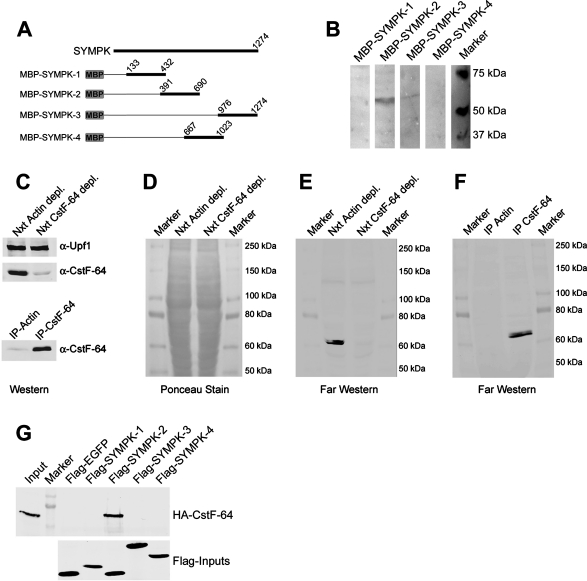
To study the functional importance of CstF-64 in the HLF, we wanted to create symplekin mutants deficient in the interaction with CstF-64. To this end, we first determined which region of symplekin interacts with CstF-64. We produced recombinant maltose-binding protein (MBP) fusions to different parts of symplekin (Figure 2A) with which we probed Far Western blots of HeLa nuclear extract, similar to the method originally used by Manley and coworkers to identify this interaction (Takagaki and Manley, 2000). An interacting band corresponding to a protein of ∼60 kDa was detected only with the symplekin fragment MBP-SYMPK-2 (Figure 2B). To analyze whether this band might represent CstF-64, we depleted nuclear extracts of either β-actin (control depletion) or CstF-64 by immunoprecipitation (Figure 2C). The depleted extracts and the immunoprecipitates were then separated by SDS–PAGE and transferred to nitrocellulose (Figure 2D). The blots were subjected to Far Western analysis by using in vitro–translated Flag-SYMPK-2 fragment as probe and by detecting its binding to the blots with anti-Flag antibody. Like MBP-SYMPK-2 in the previous experiment, the in vitro–translated Flag-SYMPK-2 recognized a band (possibly a doublet) of ∼60 kDa in β-actin–depleted nuclear extract (Figure 2E). Importantly, this band was not present in the CstF-64–depleted extract but rather appeared in the CstF-64 immunoprecipitate (Figure 2F), indicating that it almost certainly represented CstF-64. To further verify whether CstF-64 indeed binds a region between amino acids 391 and 690 in symplekin, we performed MBP pull-downs with in vitro–translated, Flag-tagged versions of all four symplekin fragments and with in vitro–translated, HA-tagged CstF-64 as the binding substrate. Only Flag-SYMPK-2 was able to precipitate HA-CstF-64 (Figure 2G). Taken together, these experiments demonstrate that the interaction site for CstF-64 has to be comprised within residues 391–690 of symplekin.
FIGURE 2:
Mapping of CstF-64 interaction site in symplekin to residues 391–690. (A) Schematic drawing of MBP-symplekin fusion proteins used in (B). Numbers indicate the amino acid residues. (B) Far Western analysis with MBP-symplekin fusions. Nuclear extract was resolved by SDS–PAGE, transferred on PVDF membranes, and incubated with MBP-symplekin fusions. Bound MBP fusions were detected with a monoclonal α-MBP antibody, followed by detection with a species-specific, HRP-coupled secondary antibody. 1/10-s exposure. (C) Immunodepletion of CstF-64 from nuclear extract. Nuclear extract was depleted of either β-actin or CstF-64 by using corresponding rabbit polyclonal antibodies coupled to protein G Sepharose. The amount of CstF-64 in depleted extract as well as in 25% of the immunoprecipitate was assessed by Western blot. The nonsense-mediated decay factor Upf1 was probed as loading control for the nuclear extracts. (D) Ponceau S stain of the membrane used for Far Western analysis in (E). (E, F) Far Western analyses of nuclear extracts immunodepleted of β-actin or CstF-64 (E) and of the corresponding immunoprecipitates (F). The blots were probed with in vitro–translated, Flag-tagged SYMPK-2 symplekin fragment, which was subsequently detected with α-Flag antibody. (G) Coimmunoprecipitation of HA-CstF-64 with in vitro–translated Flag-symplekin fragments. The symplekin fragments were first incubated with α-Flag M2 agarose and subsequently with in vitro–translated, HA-tagged CstF-64. The inputs correspond to 1/10 of the material used in the pull-down.
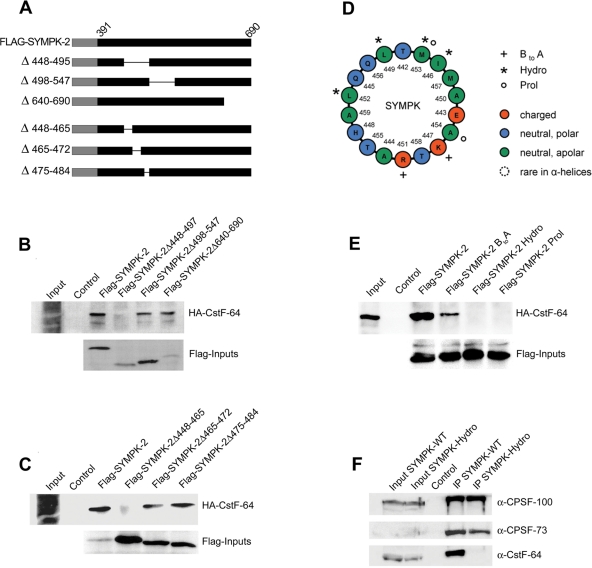
To define the binding site more precisely, in vitro transcription/translation templates corresponding to various deletions of symplekin fragment SYMPK-2 and containing a T7 RNA polymerase promoter and a Flag tag as 5′ extension (Figure 3A) were produced by fusion PCR. After in vitro translation, these fragments were incubated with anti-Flag M2 agarose, followed by the addition of in vitro–translated HA-CstF-64, and the coprecipitated HA-CstF-64 was detected by Western blot. While SYMPK-2Δ498–547 and SYMPK-2Δ640–690 still interacted with CstF-64, the deletion of residues 448–497 abolished the interaction (Figure 3B). In a next set of deletions, SYMPK-2Δ465–472 and SYMPK-2Δ475–484 still interacted with CstF-64, whereas the interaction was lost when residues 448–465 were deleted (Figure 3C). Based on these findings, the downstream border of the interaction domain has to be near residue 465, and the upstream border must lie between residues 391 and 448. We did not refine the mapping any further, as our main goal was to establish interaction mutants.
FIGURE 3:
Further characterization of CstF-64 interaction site in symplekin. (A) Schematic drawing of Flag-SYMPK-2 fusion proteins containing deletions. Numbers and a thin line indicate the deleted amino acid residues. (B–C) Mapping of the interaction with CstF-64 by using internal and C-terminal deletions of the symplekin fragment SYMPK-2. Full-length, Flag-tagged SYMPK-2 (corresponding to residues 391–690) and deletions thereof were translated in vitro, incubated with α-Flag M2 agarose, and then incubated with in vitro–translated HA-CstF-64. After precipitation and Western blot, HA-CstF-64 binding was detected with α-HA-HRP. Inputs of the bait proteins were detected by using α-Flag M2-peroxidase. All inputs correspond to 1/10 of the amount used in the pull-down. This analysis defined the CstF-64 interaction region between symplekin residues 391–448 (upstream) and ∼465 (downstream). (D) Helical wheel projection of an α-helix spanning residues 442–460 whose role was further explored with mutants. The projection was generated with freeware developed by Marcel Turcotte at the University of Ottawa, Ontario, Canada and graphically edited. The last amino acid of the helix, A460, could not be rendered by this graphical procedure. The mutated residues are indicated by plus signs (BtoA), asterisks (Hydro), or small circles (Prol; see main text). (E) Analysis of mutant symplekin fragment SYMPK-2 by the binding assay already used in (B) and (C). (F) Coimmunoprecipitation of 3′-end processing proteins with either full-length, C-terminally Flag-tagged symplekin or the identically tagged symplekin Hydro mutant from HeLa total cell extract. Inputs correspond to 1/10 of the material used for the coimmunoprecipitations. Untransfected cells served as negative control.
We then predicted the three-dimensional structure of this region with the program ESyPred3D (Lambert et al., 2002). This analysis revealed that the deletion of amino acid residues 448–465 disrupts a putative amphipathic α-helix (residues 442–460; Figure 3D). To confirm that this helix is involved in the binding of CstF-64, we exchanged, on one hand, several hydrophobic residues by glycine (SYMPK-2-Hydro: I446G, L449G, L452G, M453G) or broke the helical structure, on the other hand, by introducing a proline (SYMPK-2-Prol: M453P, A454D). To address the question of whether the basic residues in the helix play a role in the interaction with HA-CstF-64, they were exchanged by glutamic acid (SYMPK-2-BtoA: K447E, R451E). All these mutants were analyzed by the same in vitro pull-down procedure as described above. While the exchange of basic residues only reduced the interaction with HA-CstF-64, the exchange of the four hydrophobic residues as well as the introduction of a proline abolished the interaction completely (Figure 3E).
To verify that this interaction also occurs between full-length symplekin and CstF-64 in a total cell extract, we transfected HeLa cells with plasmids encoding either full-length, C-terminally Flag-tagged symplekin or the identically tagged Hydro mutant. While the interactions with CPSF-100 and CPSF-73 remained unchanged, the interaction with CstF-64 was lost upon exchanging the hydrophobic residues (Figure 3F). Therefore, we conclude that CstF-64 binds to this predicted α-helix of symplekin.
Impairment of symplekin/CstF-64 interaction reduces histone RNA 3′-end formation efficiency in vivo
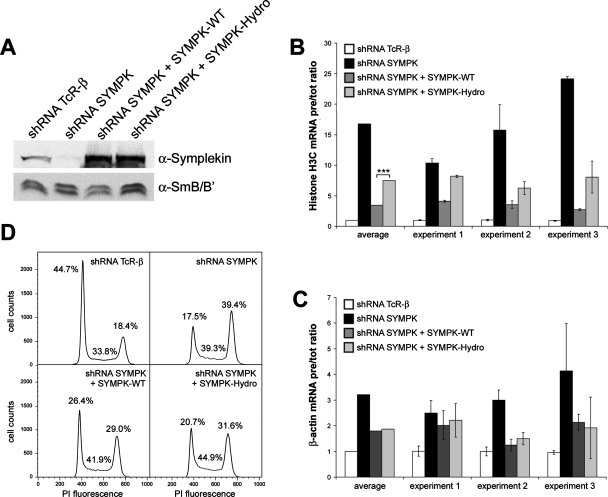
With the help of the Hydro mutant, we were able to address the functional importance of CstF-64 in histone RNA 3′-end processing by symplekin complementation experiments in vivo. HeLa cells were depleted of symplekin by RNA interference and complemented with an empty pcDNA vector or with RNAi-resistant constructs expressing either wild-type symplekin or the symplekin Hydro mutant (Figure 4A). The 3′-end processing efficiency was measured by quantitative PCR in terms of molar ratios between uncleaved histone H3C pre-mRNA and total H3C RNA (i.e., pre-mRNA + mature mRNA; Ruepp et al., 2010). As this is an indirect assay, the values represent an apparent in vivo processing efficiency. The depletion of symplekin reduced this processing efficiency ∼18-fold (Figure 4B). Wild-type symplekin rescued it approximately sixfold (up to a level approximately threefold lower than under TCR-β depletion), but the ability of the Hydro mutant to rescue processing was strongly reduced (reaching an only seven- to eight-fold lower ratio than under TCR-β depletion). For the polyadenylated β-actin mRNA, the apparent in vivo processing efficiency was less strongly affected by symplekin depletion (three- to fourfold reduction; Figure 4C). Interestingly, it could be rescued by both the wild-type symplekin and the Hydro mutant, with no significant difference between the two. To rule out that the difference in histone RNA processing efficiency in the two complementations might be caused by changes in the levels of 3′ processing factors, 17 general and histone-specific 3′-end formation factors were analyzed by Western blotting (Supplemental Figure S1). This revealed at the most very minor differences, the most notable of which was a slight elevation of the cleavage/polyadenylation factors CF Im and CF IIm in response to symplekin depletion. However, this effect was no longer seen in depleted cells complemented with either wild-type or Hydro mutant symplekin. Thus, the observed changes in histone RNA processing efficiency do not appear to be due to secondary effects mediated by other processing factors. Rather, these results imply that the interaction between symplekin and CstF-64 in the HLF is indeed required for efficient histone RNA 3′-end processing but is less important for cleavage/polyadenylation (see Discussion).
FIGURE 4:
In vivo complementation of symplekin. (A) Western blot detecting symplekin in cells depleted of TCR-β (negative control), symplekin, or in cells depleted of symplekin and simultaneously expressing Flag-tagged, RNAi-resistant wild-type or Hydro mutant symplekin. (B, C) Apparent in vivo processing efficiencies of histone H3C (B) and β-actin (C) RNA in identically treated cells. The processing efficiencies were calculated as ratios of pre-mRNAs to total RNAs and then normalized with respect to the ratio obtained in cells treated with TCR-β−specific shRNA. The less efficient histone processing complementation obtained with the symplekin Hydro mutant compared to wild-type symplekin is statistically significant (***) with a p value of 0.000027 determined by Student’s t test. (D) Cell cycle analysis by flow cytofluorometry of HeLa cells depleted of symplekin and complemented with wild-type symplekin or the symplekin Hydro mutant. The graphs represent the cell cycle distribution of cells, harvested 5 d posttransfection. The percentages of cells in the different stages of the cell cycle are indicated.
Interestingly, we observed that the depletion of symplekin affects not only histone RNA 3′-end processing but also the cell cycle. Upon depletion of symplekin, cells accumulate in the G2/M phase (Figure 4D). This effect can also be rescued by wild-type symplekin expression and to a lesser extent by expression of the Hydro mutant.
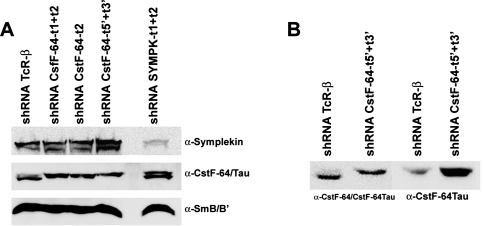
The CstF-64 paralogue CstF-64Tau is expressed in HeLa cells and gets up-regulated upon CstF-64 depletion
To confirm the results gained by these symplekin complementation experiments, we performed CstF-64 depletions by RNA interference. To our surprise, we observed only a moderate effect on histone RNA 3′-end processing and none, contrary to expectation, for polyadenylated mRNAs (see Figure 6, B–D). When we analyzed the efficiency of the depletion by Western blot, we found that the band corresponding to CstF-64 was indeed heavily reduced but that a second band at ∼70 kDa became more intense (Figure 5A).
FIGURE 6:
Codepletion of CstF-64 and CstF-64Tau affects RNA 3′-end processing. HeLa cells were transfected with negative control siRNAs or siRNAs targeting CstF-64, CstF-64Tau, or a combination of the two. (A) Western blot detecting CstF-64 and CstF-64Tau with a rabbit polyclonal anti-CstF-64/CstF-64Tau antibody (H-300) or CstF-64Tau with a CstF-64Tau–specific rabbit polyclonal antibody. SmB/B′ was detected with the monoclonal Y12 antibody. (B–D) Effects of depletion of CstF-64, CstF-64Tau, or both on the apparent in vivo 3′-end processing of β-actin (B), histone H3C (C), or c-myc (D) mRNAs. The levels of the mRNA precursors and of the total mRNAs were measured by qRT-PCR and normalized to the levels measured in the control cells. Shown are the average ratios between the pre- and total mRNA from three biological replicates. The error bars represent the standard deviation among these replicates. (E) Cell cycle analysis by flow cytofluorometry of HeLa cells depleted of CstF-64, CstF-64Tau, or a combination of both in comparison to control cells. The graphs represent the cell cycle distribution of cells, harvested 5 d posttransfection. The percentages of cells in the different stages of the cell cycle are indicated.
FIGURE 5:
Western blots detecting CstF-64 and CstF-64Tau. (A) Western blot revealing CstF-64 and a 70-kDa band corresponding to CstF-64Tau in total extracts of HeLa cells transfected with plasmids encoding shRNAs specific for T-cell receptor β, CstF-64, and symplekin mRNAs. CstF-64 and CstF-64Tau were detected with the C-20 antibody against the C-terminus of CstF-64. Symplekin was detected with a monoclonal anti-symplekin antibody while SmB/B′, which served as loading control, was detected with the monoclonal antibody Y12. (B) Western blot showing CstF-64Tau from the same total extracts of HeLa cells as above transfected with shRNAs specific for T-cell receptor β or CstF-64. CstF-64Tau was detected in the two right-hand lanes with a CstF-64Tau-specific rabbit polyclonal antibody, whereas in the two left-hand lanes the C-20 antibody reacting with both proteins was used.
CstF-64 has a paralogue, CstF-64Tau, which was first found in testes (Wallace et al., 1999; Dass et al., 2001) but appears to be also expressed in a wider variety of cell types and tissues (Shi et al., 2009). As the antibody used for detection (α-CstF-64 C-20, Santa Cruz Biotechnology, Santa Cruz, CA) had been raised against a peptide from the C-terminus of CstF-64 that is identical in CstF-64Tau, we wanted to find out whether this 70-kDa band might correspond to CstF-64Tau and whether its abundance changes in response to the reduction of CstF-64 levels. Thus we performed in vivo depletions with sets of shRNAs targeting different parts of CstF-64 mRNA: either the combination of target 1 and target 2, target 2 alone, or the combination of two targets lying in the 5′- and 3′-untranslated regions (UTRs; see Supplemental Table S1 for description of target sequences). While target 1 has three mismatches compared to CstF-64Tau, target 2 differs in five nucleotides, rendering a possible coreduction rather unlikely. The 5′- and 3′-UTRs of these genes are very different, with no significant homology. Additionally, we performed a depletion of symplekin, which is an important interactor of CstF-64. As can be seen in Figure 5A, cells treated with a control shRNA against T-cell receptor β possess rather low levels of the 70-kDa band. However, this changes upon depletion of CstF-64, where the 70-kDa band becomes equally strong if not stronger than that of CstF-64 in control cells. Moreover, in the symplekin depletion, the ratio of the two bands is also altered, with the 70-kDa band becoming stronger.
To verify whether the 70-kDa band corresponds to CstF-64Tau, we performed a Western blot with a CstF-64Tau-specific antibody (Figure 5B). The experiment showed that the 70-kDa band indeed corresponds to CstF-64Tau and that it increases upon depletion of CstF-64.
The Tau variant can take over the function of CstF-64 in RNA 3′-end processing
Having shown that CstF-64Tau levels are increased upon CstF-64 depletion, we hypothesized that the Tau variant may take over the function of CstF-64. This would be an explanation for the lack of a 3′-end formation deficiency observed upon CstF-64 depletion, as mentioned above. To analyze this possibility, we performed depletions of CstF-64, CstF-64Tau, and a combination of both. The levels of both proteins were strongly reduced by the individual depletions; however, a low amount of CstF-64 was still present after codepletion (Figure 6A). The apparent in vivo 3′-end processing efficiency was measured by quantitative PCR in terms of molar precursor/total mRNA ratios. Interestingly, in the double depletion, but not in either of the individual depletions, we could observe an impairment of the apparent in vivo processing efficiency for the polyadenylated β-actin (2.6-fold) and c-myc (3.3-fold) mRNAs (Figure 6, B and D). In the case of the poly(A)− replication–dependent histone H3C mRNA, the 3′-end processing was affected approximately twofold by the depletion of CstF-64 but more strongly (approximately threefold) by the double depletion of CstF-64 and the Tau variant (Figure 6C). These results demonstrate that CstF-64Tau is able to take over the function of CstF-64 when the latter is depleted, although this functional substitution may be less complete in histone RNA processing than in cleavage/polyadenylation.
When we analyzed the cell cycle distribution of these depleted cells, we observed no significant change after a single depletion of CstF-64 (Figure 6E). In contrast, the depletion of CstF-64Tau increased the number of cells in S and G2/M at the expense of G1 phase cells. This shift was even more pronounced in the double depletion.
Determination of interaction sites for symplekin and CstF-77 on CstF-64
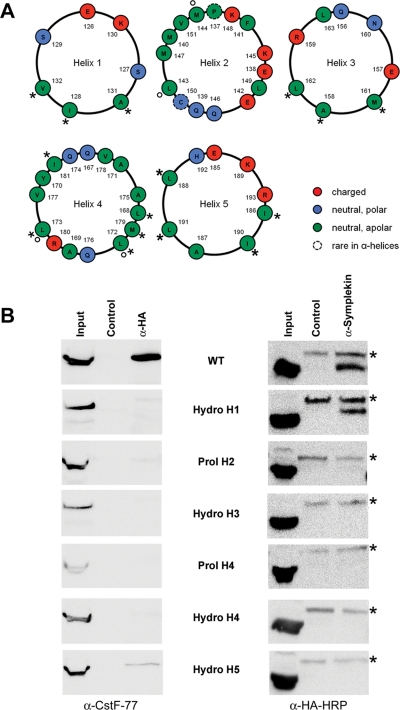
CstF-77 and symplekin are known to bind mutually exclusively to the hinge domain of CstF-64 (Takagaki and Manley, 2000). To be able to study the functional impact of these interactions, we tried to identify CstF-64 mutants that would be affected in the interaction with one, but not the other, of these partners. A bioinformatic analysis of the secondary structure of the hinge domain predicted five putative helices (Supplemental Figure S2), most of them being amphipathic according to the helical wheel projections (Figure 7A). On the basis of this, we hypothesized that symplekin and CstF-77 might contact not exactly the same residues but different combinations of helices 1 to 5. To investigate this possibility, we generated several CstF-64 mutants. For helices 1, 3, 4, and 5, hydrophobic residues were exchanged by glycine (Hydro mutants: Hydro H1: I128G, A131G, V132G; Hydro H3: A158G, M161G, L162G; Hydro H4: L168G, L172G, L173G, M179G, I181G; Hydro H5: I186G, L188G, I190G, L191G), whereas for helices 2 and 4, the chain breaker proline was inserted along with an aspartic acid residue to force the interruption of the helix (Prol mutants: Prol H2: L143P, M144D; Prol H4: L172P, L173D).
FIGURE 7:
Analysis of interaction sites for CstF-77 and symplekin on CstF-64. (A) Helical wheel projections of helices 1–5 of the CstF-64 hinge domain. The projections were generated as described for Figure 3D and graphically edited. The mutated residues are indicated by asterisks (Hydro mutants) or small circles (Prol mutants; see main text). (B) Coimmunoprecipitation of endogenous CstF-77 or symplekin with HA-tagged CstF-64. Total cell extracts from HeLa cells transfected with pcDNA-HA-CstF-64 or mutants thereof were incubated with α-HA or α-symplekin antibodies; α-Mili served as negative control. Endogenous CstF-77 was detected after the α-HA pull-down by using α-CstF-77, followed by the incubation with a species-specific horseradish peroxidase–coupled antibody (left panel). HA-CstF-64 was detected after the α-symplekin pull-down with α-HA-HRP (right panel). The α-HA-HRP antibody interacts unspecifically with a protein in the immunoprecipitates that is marked by an asterisk. Note that the binding to CstF-77 is disrupted upon changing helices 1–4 and reduced for the helix-5 mutant, whereas the binding to symplekin is disrupted upon changing helices 2 to 5, but not helix 1.
To analyze the effects of these mutations, we transfected HeLa cells with HA-tagged CstF-64 or the identically tagged mutants described above and immunoprecipitated total extracts with either anti-HA or anti-symplekin antibodies. We then used anti-CstF-77 or anti-HA antibodies, respectively, to probe Western blots for coimmunoprecipitation of CstF-77 or the HA-tagged CstF-64 variants, respectively. While mutations in helices 1–4 of the hinge domain abolished the interaction with CstF-77, the mutation in helix 5 only reduced the binding (Figure 7B, left column). Interestingly, symplekin binding was abolished by the mutations in helices 2–5 of the CstF-64 hinge domain but was retained with the helix 1 mutant (Figure 7B, right column). The mutually exclusive binding of symplekin and CstF-77 to the hinge domain therefore does not result from binding to the same residues but arises from overlapping binding sites. These findings were also supported by in vitro interaction studies (unpublished data).
Subcellular localization of CstF-64 depends on CstF-77 and not on symplekin
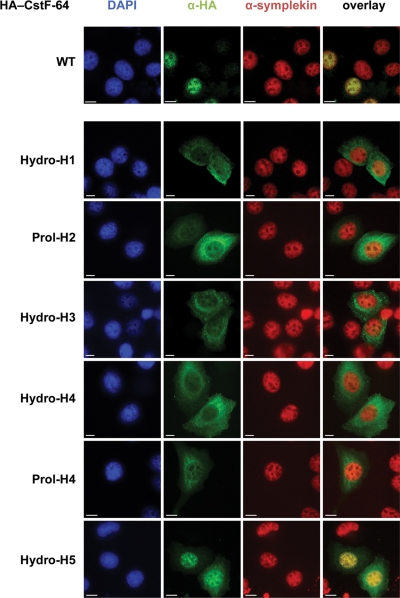
Based on the finding that the hinge domain of CstF-64 interacts with CstF-77 and is essential for nuclear accumulation (Hockert et al., 2010), it was suggested that the nuclear import of CstF-64, which does not contain a nuclear localization signal (NLS) on its own (Takagaki et al., 1992), depends on CstF-77 binding (Hockert et al., 2010). However, as symplekin can compete with CstF-77 for binding to the hinge domain and contains an NLS as well (Takagaki and Manley, 2000), we were interested to clarify which interaction partner is predominantly responsible for CstF-64’s nuclear localization. The two hinge domain mutations in helix 1 (disrupting the interaction with CstF-77 but not symplekin) and in helix 5 (unable to bind symplekin but retaining a residual interaction with CstF-77) appeared to be ideal tools to resolve this question. We performed an immunofluorescence experiment with cells transfected with HA-tagged wild-type CstF-64 and the identically tagged helix mutants. As can be seen in Figure 8, the nuclear accumulation of CstF-64 was lost in all mutants, except for the helix 5 Hydro mutant. For this mutant the CstF-64 staining was predominantly nuclear, but some cytoplasmic staining remained. Importantly, neither symplekin (Figure 8) nor CstF-77 (Supplemental Figure S3A) changed their localization in the mutant-expressing cells. These findings strongly suggest that the interaction with CstF-77 (partly retained in the Hydro 5 mutant) and not the one with symplekin (lost in Hydro 5 but retained in Hydro 1) is responsible for the nuclear import of CstF-64, in agreement with the hypothesis of Hockert et al. (2010). To confirm that indeed CstF-77 and not another yet unidentified factor is required for the nuclear localization of CstF-64, we depleted cells of CstF-77 by RNA interference, which resulted in CstF-64 codepletion and cytoplasmic mislocalization of the remaining protein (Supplemental Figure S3B).
FIGURE 8:
Subcellular localization of CstF-64 depends on CstF-77 and not on symplekin. Cells were grown on coverslips, transfected with pcDNA derivatives encoding either HA-tagged wild-type (WT) CstF-64 or the identically tagged hinge domain mutants indicated on the left. The cells were fixed with Lomant’s reagent 2 d after transfection. CstF-64 was visualized with a rabbit polyclonal α-HA antibody (Y-11) combined with a goat α-rabbit, AlexaFluor 488–labeled secondary antibody. Symplekin was visualized with a mouse monoclonal antibody (Clone 25) in combination with an AlexaFluor 594–labeled, goat α-mouse secondary antibody. DNA staining was achieved by using DAPI-containing mounting medium.
CstF-64 and its paralogue CstF-64Tau are regulated posttranslationally
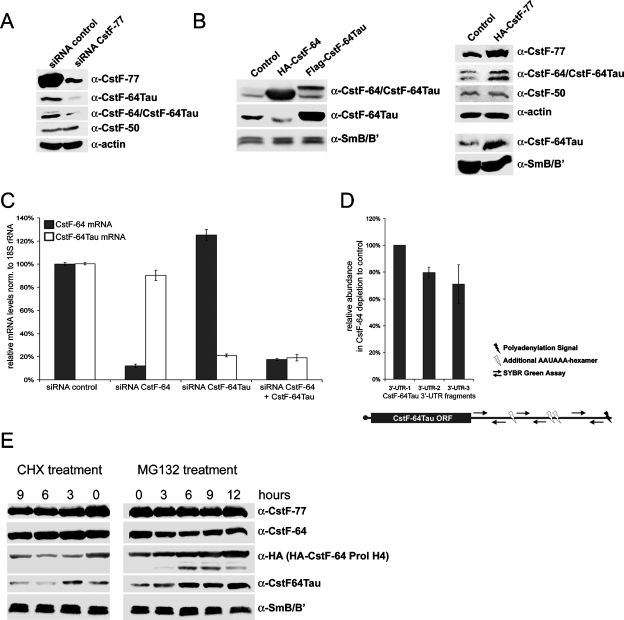
Previous work has already suggested that CstF-64Tau might be regulated at the translational or posttranslational level (Huber et al., 2005). The facts that CstF-64Tau protein levels were increased upon CstF-64 depletion (Figure 5) and that CstF-64 was codepleted upon CstF-77 depletion (Supplemental Figure S3B) strongly suggested that CstF-77 might control the overall levels of CstF-64 and CstF-64Tau and that the two CstF-64 paralogues might mutually control each other’s levels. To corroborate this hypothesis, we depleted cells of CstF-77 by RNA interference and analyzed protein levels by Western blotting. Indeed, the reduction of CstF-77 led to a decrease of CstF-64 and CstF-64Tau protein levels, but not of the level of CstF-50 (Figure 9A). To further challenge our hypothesis, we overexpressed CstF-77, CstF-64, and CstF-64Tau. As expected, CstF-77 overexpression led to an increase of CstF-64 and CstF-64Tau, but again the level of CstF-50 was not altered (Figure 9B). The overexpression of CstF-64 resulted in a decrease in CstF-64Tau levels (Figure 9B), inversely to what we had seen when depleting CstF-64 (Figure 5). Interestingly, CstF-64Tau overexpression did not significantly influence the protein level of CstF-64.
FIGURE 9:
Regulation of CstF-64 and CstF-64Tau. (A) Western blot detecting CstF subunits and β-actin (loading control) upon CstF-77 depletion by RNA interference. Total extracts from cells transfected with a control siRNA or with siRNAs targeting CstF-77 mRNA were separated by SDS–PAGE and CstF-77, CstF-64Tau, CstF-64/CstF-64Tau, and CstF-50 were detected by Western blot. (B) Western blot detecting CstF subunits and SmB/B′ (loading control) in total extracts from cells overexpressing either HA-CstF-64, Flag-CstF-64Tau, or HA-CstF-77. (C) Relative CstF-64 and CstF-64Tau mRNA levels upon depletion of CstF-64, CstF-64Tau, or a combination of both. The relative mRNA levels were measured by qRT-PCR and normalized to 18S rRNA. (D) Alternative polyadenylation qPCR assay for CstF-64Tau upon CstF-64 depletion. Three different SYBR Green assays were designed. The 3′-UTR-1 assay measures all CstF-64Tau mRNA species, whereas 3′-UTR-2 and 3′-UTR-3 are placed after additional AAUAAA hexamers occurring in the CstF-64Tau 3′-UTR. The relative levels are shown as the ratio of CstF-64Tau mRNA levels in control cells to cells depleted of CstF-64 and were normalized to U6 snRNA. (E) Western blot analysis of protein stabilities from cells treated with the translation inhibitor cycloheximide (CHX) or the proteasomal inhibitor MG132 at different time points. Total cell extracts from cells transfected with HA-CstF-64-Prol H4 were separated by SDS–PAGE. Endogenous CstF-77, CstF-64, CstF-64Tau, and SmB/B′ as well as exogenous HA-CstF-64-Prol H4 were detected by Western blotting.
To analyze at which level this regulation occurs, we determined by quantitative RT-PCR the levels of CstF-64 (CSTF2) and CstF-64Tau (CSTF2T) mRNAs. We found that CstF-64 and CstF-64Tau mRNA levels are not up-regulated upon the depletion of their respective paralogue (Figure 9C). To exclude that a difference in poly(A)-tail position in the CstF-64Tau mRNA upon CstF-64 depletion would lead to a transcript that is translated much more efficiently, we measured CstF-64Tau mRNA levels with two different probes in the 3′-UTR, which would indicate a change in poly(A)-site choice. However, no significant changes could be observed (Figure 9D). This result was also confirmed by Northern blot analysis (Supplemental Figure S4). Furthermore, no change in the localization of the CSTF2T mRNA could be observed by fluorescent in situ hybridization experiments (unpublished data).
These results strongly suggest that CstF-64 and CstF-64Tau levels are controlled at the translational or posttranslational level, in agreement with the conclusion reached by Huber et al. (2005) from their analysis of CstF-64 and CstF-64Tau expression in mouse tissues. Because we observed that, upon CstF-77 depletion, CstF-64 and CstF-64Tau are codepleted (Figure 9A), we addressed the question of whether the interaction with CstF-77 is essential for CstF-64 stability. Therefore, we transfected HeLa cells with the HA-tagged CstF-64-Prol-H4 mutant, followed by incubating the cells with MG132 or cycloheximide (CHX). Because MG132 blocks the proteasome, levels of unstable proteins that are prone to proteasomal degradation will increase over time. Conversely, CHX, which inhibits translation, will lead to a reduction of unstable polypeptides. As can be seen in Figure 9E, the CstF-64 mutant, not able to interact with CstF-77 anymore, is unstable. Its levels increase under MG132 treatment and decrease under CHX treatment, whereas wild-type CstF-64 levels hardly change. Interestingly, wild-type CstF-64Tau is also rather unstable, compared to its paralogue CstF-64 and to CstF-77.
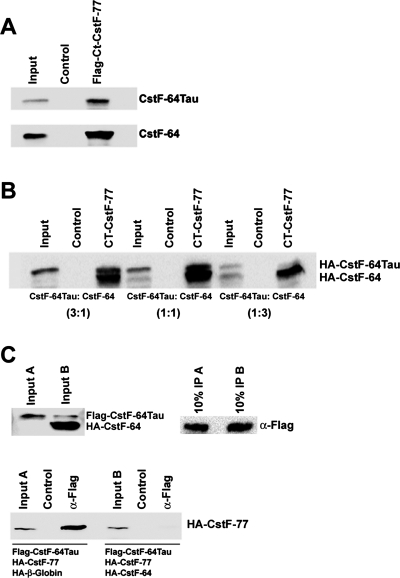
The fact that the interaction with CstF-77 is essential for CstF-64 stability and the results from the CstF-64 depletion and overexpression experiments strongly implicate that CstF-64Tau levels are controlled by the amount of CstF-77 that is not bound by CstF-64 and that therefore remains accessible for CstF-64Tau. This competition model would imply a lower affinity of CstF-64Tau for CstF-77. This possibility was assessed in vitro by coimmunoprecipitations with a Flag-tagged C-terminal fragment of CstF-77 known to interact with CstF-64 (Takagaki and Manley, 2000). To this recombinant protein, we added in vitro–translated CstF-64 and/or CstF-64Tau. When added individually, both proteins were efficiently coprecipitated (Figure 10A). However, when the two proteins were added together in different molar ratios, CstF-64 was preferentially bound, suggesting that it has a higher affinity for the CstF-77 fragment (Figure 10B). We confirmed these results by an inverse setup: Flag-tagged CstF-64Tau was in vitro translated together with HA-tagged CstF-77 and either HA-tagged CstF-64 or HA-tagged β-globin as control. The reaction products were subjected to immunoprecipitation of the Flag-tagged CstF-64Tau, followed by Western blotting to detect coprecipitated CstF-77. While CstF-77 was efficiently coprecipitated by Flag-CstF-64Tau in the presence of β-globin, the coexpression of CstF-64 completely abolished the interaction (Figure 10C). Taken together, these results strongly indicate that CstF-64Tau has a lower binding affinity for CstF-77 than CstF-64.
FIGURE 10:
Interaction of CstF-64 and CstF-64Tau with CstF-77. (A) In vitro pull-down. A recombinant C-terminal Flag-tagged fragment of CstF-77 was incubated with in vitro–translated HA-CstF-64 or HA-CstF-64Tau and with α-Flag-agarose followed by subsequent washes and separation by SDS–PAGE. Agarose beads with BSA served as negative control. Coprecipitated proteins were detected by Western blot with α-HA-HRP. (B) In vitro binding competition. Procedures were as described for (A), except that mixtures of in vitro–translated HA-CstF-64 and HA-CstF-64Tau were used as binding substrates. The ratios indicated in brackets refer to the amount of individual template used in the coupled transcription/translation reaction. (C) Inverse in vitro binding competition. In vitro translation products from reactions A (Flag-CstF-64Tau, HA-CstF-77 [full-length], and HA-β-globin) or B (Flag-CstF-64Tau, HA-CstF-77 [full-length], and HA-CstF-64) were incubated with α-Flag-Agarose to precipitate Flag-CstF-64Tau, followed by washes and separation by SDS–PAGE. In the control reactions, the template for Flag-CstF-64Tau was omitted. The presence of Flag-CstF-64Tau and HA-CstF-64 in the reaction was confirmed by Western blot with 10% of the input samples by using an antibody detecting both proteins simultaneously (C-20 antibody, Santa Cruz). The presence of Flag-CstF-64Tau in the precipitate was confirmed by Western blot with 10% of the precipitate by using α-Flag-HRP. Coprecipitated HA-CstF-77 was detected with α-HA-HRP.
DISCUSSION
Interactions between CstF-64, CstF-77, and symplekin and their role in histone RNA processing, cleavage/polyadenylation, and CstF assembly
To shed light on the individual function of CstF-64 in histone RNA processing and cleavage/polyadenylation and on its assembly and regulation, we have mapped its interaction site on symplekin as well as the symplekin and CstF-77 interaction sites on CstF-64. We could thereby generate an RNAi-resistant symplekin mutant that no longer interacts with CstF but still binds CPSF, and we also produced CstF-64 mutants that retain some interaction with symplekin or CstF-77 but have lost the interaction with the respective other partner.
The fact that the symplekin Hydro mutant still interacts with CPSF-100 and CPSF-73 suggests that it has retained some of its functions, even though it has lost the ability to bind CstF-64 in vitro and in vivo (Figure 3). This implies that CPSF and CstF bind to symplekin independently of each other.
By expressing the RNAi-resistant symplekin Hydro mutant in a symplekin-depletion background, we could observe that histone RNA processing and cleavage/polyadenylation differ in their dependence on the CstF-64/symplekin interaction: While the processing of β-actin mRNA in vivo was equally well complemented by wild-type and mutant symplekin (Figure 4C), the loss of the CstF-64/symplekin interaction clearly impaired the complementation of histone RNA processing (Figure 4B). This was not due to a depletion of histone processing factors (Supplemental Figure S1). However, a possible reason for this difference could be the absence of CstF-50 from the histone HLF (Kolev and Steitz, 2005). This subunit, in cleavage/polyadenylation, may strengthen the cohesion of the CstF complex and its interaction with the other processing factors and thus reduce the dependence on the symplekin/CstF-64 interaction. Alternatively, the interactions of CstF-64 and CPSF-160 with the GU-rich downstream element and the AAUAAA hexamer, respectively, may stabilize the poly(A)+ processing complex and thereby render the CstF-64/symplekin interaction less important. In contrast, for the processing of replication-dependent histone pre-mRNAs, the HLF has to be assembled without CstF-50, and the RNA interactions of CstF-64 and CPSF-160 are replaced by (yet unknown) interactions with the histone-specific components of the processing machinery, that is, the U7 snRNP, the hairpin- or stem-loop-binding protein (HBP/SLBP), and/or the 100-kDa zinc finger protein (ZFP100; reviewed in Dominski and Marzluff, 2007). It should, however, be mentioned that other explanations are also possible and that the two proposed ones are not mutually exclusive.
By using the CstF-64 mutants that selectively bind either symplekin or CstF-77, we could show that the subcellular localization of CstF-64 depends on CstF-77 and not on symplekin (Figure 8). The interaction sites for these two proteins were known to be located in the hinge domain of CstF-64 (Takagaki and Manley, 2000). Now, the analysis of our mutants demonstrates that these binding sites overlap but are not identical. Presumably, during CstF assembly the interaction with CstF-77 prevails, but there may be a structural rearrangement when the cleavage/polyadenylation complex or the HLF assemble.
Similar to symplekin depletion, the depletion of CstF-64 also affected the apparent in vivo processing efficiencies of histone and cleavage/polyadenylation substrates (Figure 6). However, both CstF-64 and CstF-64Tau had to be depleted to produce an effect on cleavage/polyadenylation, whereas histone 3′-end processing was already affected by CstF-64 depletion alone but was more strongly inhibited when both CstF-64 and its paralogue were codepleted. This probably means that CstF-64Tau can less efficiently substitute for CstF-64 in histone RNA processing than in cleavage/polyadenylation. It is also interesting to speculate about what could be the function of CstF-64 in histone RNA processing since its RNA-binding activity to GU-rich sequences is presumably not required (although this possibility has not been investigated). The most likely explanation that will have to be tested experimentally is that CstF-64, like symplekin and perhaps other HLF components, could be necessary for the integrity of the HLF complex. A difference in HLF integrity could also be a reason why the depletion of CstF-64/CstF-64Tau had a quantitatively smaller impact on histone RNA processing than the depletion of symplekin. Perhaps a “residual HLF” lacking CstF components, but containing symplekin and CPSF, is still partly functional, whereas symplekin depletion completely disintegrates the HLF.
Interestingly, the depletions of both symplekin and CstF-64 had an effect on the cell cycle distribution. Such cell cycle effects can be expected for factors involved in histone RNA 3′ processing, as histones are required to package the newly synthesized DNA during S phase. However, the expected effect would be a cell cycle arrest during S phase or at the G1/S boundary. This is the case for depletions of HBP/SLBP (S phase arrest; Wagner et al., 2005; Sullivan et al., 2009), ZFP100, and the U7 snRNP-specific proteins Lsm10 and Lsm11 (G1 arrest; Wagner and Marzluff, 2006; Ruepp et al., 2010). Based on these latter results, it was even proposed that mammalian cells may possess a histone checkpoint in G1 (Marzluff et al., 2008). In contrast, the components of the cleavage/polyadenylation machinery are less likely to cause a cell cycle phenotype upon their depletion. An exception is a report showing an accumulation of cells in G0/G1 after a strong CstF-64 reduction in the chicken DT40 cell line (Takagaki and Manley, 1998).
In our hands, symplekin depletion caused a relative increase of cells in G2/M that could be partly rescued by complementation with wild-type symplekin and less efficiently by the Hydro mutant (Figure 4D). This unusual cell cycle phenotype is most likely not related to histone RNA processing, when one considers the depletion phenotypes of the histone-specific processing factors mentioned above. It is even uncertain whether the shift in cell cycle distribution is due to an RNA processing deficiency at all, as symplekin also affects the transcription of certain cell cycle–related genes (Kavanagh et al., 2006; Buchert et al., 2010) and localizes to cytoplasmic plaques of tight junctions in some cell types (Keon et al., 1996).
The phenotype observed in our single and double depletions of CstF-64 and CstF-64Tau in HeLa cells (increase in S and G2/M populations; Figure 6E) differs from the one observed in chicken DT40 cells (G0/G1 accumulation; Takagaki and Manley, 1998). We note that in our experiments the shift was primarily due to CstF-64Tau depletion and only reinforced by CstF-64 codepletion. As the chicken CstF-64 gene is located on chromosome 4, the Tau paralogue is not required to maintain mRNA 3′-end formation in haploid germ cells and is therefore absent. If one considers that CstF-64 and CstF-64Tau prefer slightly different RNA sequences (Monarez et al., 2007), that CstF is probably hexameric (see below) allowing for homo- and heterodimers of different CstF-64 isoforms and paralogues, and that the downstream GU-rich elements differ widely in sequence, it is quite possible that some mRNAs are more affected than others by a reduction of CstF-64 proteins. Such a differential reduction of certain mRNAs could then cause the different cell cycle phenotypes in human and chicken cells. A precedent for such an effect is USAF, whose depletion leads to mis-splicing of several cell cycle regulatory genes and to M phase arrest (Pacheco et al., 2006).
CstF-64/CstF-64Tau and the regulation of CstF
Our work has revealed an interesting regulation involving CstF-64, CstF-64Tau, and CstF-77. The CstF-64 paralogue, CstF-64Tau, was first identified in testes, where it takes over the function of CstF-64 (whose gene is located on the X chromosome) during spermatogenesis (Wallace et al., 1999; Dass et al., 2001). However, CstF-64Tau was also identified in 3′-end processing complexes from HeLa cells (Shi et al., 2009), suggesting that it might play additional roles in other tissues. This is corroborated by our finding that CstF-64Tau can functionally replace CstF-64 in 3′-end formation of poly(A)+ mRNA in HeLa cells (Figure 6, B and C). For replication-dependent histone mRNAs, already the depletion of CstF-64 resulted in a processing defect, but codepletion of CstF-64 and CstF-64Tau affected histone processing even more (Figure 6D). One explanation for this subtle difference could be that CstF-64 interacts more strongly with the HLF than the Tau variant in agreement with its higher affinity for CstF-77 (Figure 10; see below). On the other hand, we cannot formally exclude the possibility that this effect is indirect, as CstF-64 depletion might affect the 3′-end formation of an mRNA for a factor required for histone mRNA processing.
This, together with the other findings reported in this article, suggests a model for CstF regulation. In this model, CstF-77 dictates the expression level and nuclear accumulation of CstF-64 (and CstF-64 Tau). Should the CstF-77 level become low, CstF-64 would remain cytoplasmic and be subject to rapid degradation. This guarantees the maintenance of stoichiometric levels of the two factors, and it prevents that CstF-64 on its own, not integrated into CstF, binds to pre-mRNAs. The fact that the level of CstF-50 is not coregulated might be related to its additional involvement in a DNA damage response (Kleiman and Manley, 2001; Mirkin et al., 2008; Cevher et al., 2010). However, it is also possible that there is no need for a regulation of CstF-50 because it has no RNA-binding affinity.
Furthermore, the abundance of CstF-64 with respect to the availability of CstF-77 regulates the expression of CstF-64Tau. This is most important in male germ cells during spermatogenesis when CstF-64 disappears due to X-chromosome inactivation. Then CstF-64Tau no longer has to compete with CstF-64 for CstF-77 and is stabilized by its interaction with CstF-77. Such a mechanism might even play a role in other cells and tissues when the level of CstF-64 fluctuates. One example could be the switch from membrane-bound to secreted-form immunoglobulin M that occurs during differentiation of B lymphocytes and which is controlled by a reduction of CstF-64 (Takagaki et al., 1996; Takagaki and Manley, 1998). Another phenomenon in which the coregulation between CstF-64 and CstF-64Tau might play a role is during the cell cycle, as CstF-64 fluctuates during the G0 to S phase transition (Martincic et al., 1998).
Importantly, the covariation between CstF-64 and CstF-64Tau may have an influence on the recognition of GU-rich downstream elements. Because CstF-77 can form a dimer (reviewed in Mandel et al., 2008), the CstF complex may be a hexamer at least during some step(s) of pre-mRNA processing. Such a hexamer could therefore contain homo- as well as heterodimers of the RNA-binding CstF-64 subunit. As, moreover, CstF-64 and CstF-64Tau differ somewhat in their sequence preference for RNA binding (Monarez et al., 2007), this opens the possibility for differential and regulated recognition of polyadenylation sites. This possibility may also be exploited in the nervous system, where alternatively spliced variants of CstF-64 have been identified (Shankarling et al., 2009).
In conclusion, this regulation of CstF may ensure several features that are important for a multisubunit complex geared to deal with multiple RNA targets in various biological systems: 1) to ensure the proper assembly of the individual subunits in stoichiometric amounts; 2) to prevent possible deleterious effects of free CstF-64 subunits binding to RNA; and 3) to allow for regulation of alternative cleavage/polyadenylation in different tissues and functional states. Whether some of this regulation also applies to histone RNA 3′-end formation is an open question, but at least our experiments show that CstF-64Tau can function in this process, albeit not quite as well as CstF-64 (Figure 6C).
MATERIALS AND METHODS
Plasmids, antibodies, and oligonucleotides
Descriptions of plasmids and antibodies are presented in the Supplemental Materials and Methods. Plasmids encoding shRNAs and siRNA oligonucleotides used for in vivo depletions are shown in Supplemental Tables S1 and S2, respectively. Sequences of real-time quantitative RT-PCR probes and primers are indicated in Supplemental Tables S3 and S4.
Cell culture and transfection
HeLa cells were grown in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Bio-Concept, Salem, NH), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C in a moist atmosphere containing 5% CO2. For plasmid transfection, they were grown to 60–70% confluence in cell culture dishes of appropriate sizes. For transfection, plasmids were complexed with DreamFect (OZ Biosciences, Marseille, France) and equally distributed over the cells.
RNA interference was achieved either by pSUPuro-based depletion (Brummelkamp et al., 2002), followed by selection of transiently transfected cells with 1.5 μg/ml puromycin, or by transfection of siRNAs with Lullaby (OZ Biosciences) according to the manufacturer’s instructions.
MBP fusion protein purification and MBP pull-downs
To study protein–protein interactions in vitro, MBP fusion proteins were expressed in Escherichia coli BL21 (DE3) LysS or BL21 (DE3) RIPL transformed with pMalc-derived plasmids encoding MBP-MS2 (negative control) or MBP fused in frame to various symplekin fragments. After purification over amylose beads (New England Biolabs, Ipswich, MA), the proteins were analyzed by SDS–PAGE.
Amylose beads were coupled with equimolar amounts of either MBP-MS2, MBP-SYMPK-1, MBP-SYMPK-2, MBP-SYMPK-3, or MBP-SYMPK-4 in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) supplemented with 0.1% NP-40 at 4°C for 1.5 h with gentle agitation on a wheel and subsequently incubated with RNase-treated HA-CstF-64 obtained by coupled in vitro transcription/translation in rabbit reticulocyte lysate (TNT T7 Quick kit, Promega, Madison, WI) for 2 h with gentle agitation. Subsequently, beads were washed with 0.1% NP-40 in PBS, then input and bound fraction were analyzed by SDS–PAGE and detected by incubation with anti-HA-HRP (Roche, Mannheim, Germany) and the enhanced chemiluminescence (ECL) method (GE Healthcare, Waukesha, WI).
Far Western analysis
Nuclear extract, generously provided by Reinhard Lührmann, was resolved by SDS–PAGE and electrophoretically transferred to a polyvinylidene fluoride (PVDF) membrane (Roche Applied Science, Rotkreuz, Switzerland). All subsequent steps were performed at 4°C unless noted differently. After transfer, the blots were rinsed in TBST (10 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20) and incubated for 16 h in blocking/renaturation buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2% bovine serum albumin [BSA]). For Far Western analysis, the blots were cut into 9 × 2-cm strips and incubated for 16 h with the affinity-purified MBP fusion proteins (0.5–20 μg/ml) in overlay buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EGTA, 1% NP-40, 0.5% BSA, 0.25% gelatin). The blots were washed with washing buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20, 0.2% gelatin) three times for 5 min at room temperature and then incubated for 1 h in TBST containing 1% BSA. Bound MBP fusion proteins were revealed by sequential incubation with a rabbit primary anti-MBP polyclonal antiserum (1:1000, New England Biolabs) and a secondary goat anti-rabbit antibody coupled to horseradish peroxidase (1:3125, Promega), and signals were detected by the ECL method.
For the depletion of CstF-64 and β-actin from nuclear extract, Protein G Sepharose beads (40-μl bed volume) were incubated head over tail in IIP-150/NP-40 (50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 0.1% NP-40) with 60 μg of rabbit polyclonal antibodies directed against CstF-64 or β-actin at 4°C overnight. After incubation, the beads were washed twice with IIP-150/NP-40. Residual wash buffer was removed from the beads, followed by the addition of 250 μl HeLa nuclear extract and incubation for 4 h head over tail at 4°C. The suspension was centrifuged, and supernatant (depleted nuclear extract) was recovered. The beads were washed three times with IIP-150/NP-40, followed by removal of residual wash buffer and subsequent boiling in SDS-loading buffer. The amount of CstF-64 in depleted extract and 25% of the immunoprecipitate was assessed by Western blot with goat polyclonal α-CstF-64 and donkey α-goat IRDye 800CW.
For Far Western blotting, the depleted nuclear extracts and immunoprecipitates were loaded on an SDS-polyacrylamide gel, followed by transfer onto nitrocellulose membranes. The Far Western blots were then performed as mentioned above, apart from the use of in vitro–translated Flag-SYMPK-2 as bait, followed by the detection of bound Flag-SYMPK-2 with a primary mouse a-Flag antibody (OctA probe) and a secondary goat α-mouse antibody coupled to IRDye 800CW. Signals were detected using the LI-COR Odyssey Infrared Imaging System (Lincoln, NE).
Interaction mapping by in vitro pull-downs
To characterize the binding of CstF-64 to symplekin and vice versa, several deletion mutants were generated by fusion PCR. Briefly, a first PCR added a FLAG-epitope tag to the N-terminus of the symplekin fragment SYMPK-2. Internal deletions were then produced by amplifying the pieces upstream and downstream of the desired deletion end points separately with internal primers designed to create an overlap. The final fusion PCR then joined these two fragments and introduced a T7 RNA polymerase promoter upstream of the FLAG tag. The PCR reactions were performed in a final volume of 50 μl by using either PCR FastStart Master Mix (Roche) or 1× Herculase II buffer, 300 nM dNTPs, 1 unit Herculase II (Stratagene, La Jolla, CA), 400 nM of the primers, and 1 ng of HA-pcDNA symplekin. The first PCR consisted of a denaturation at 9°C for 2 min, followed by 25–30 cycles of denaturing for 1 min at 95°C, annealing for 30 s at 52°C, elongation at 72°C for 45 s, and ending with a final elongation step of 5 min. For the fusion of the two amplicons, 5 μl of each primary PCR was used as template, and the procedure consisted of a denaturation at 95°C for 2 min, followed by 35 cycles of denaturing for 1 min at 95°C, annealing for 30 s at 52°C, elongation at 72°C for 45 s, and ending with a final elongation step of 5 min.
The PCR products were purified over GFX PCR purification columns (GE Healthcare), and then they served as templates for in vitro translation by the TNT Quick Coupled Transcription/Translation System (Promega), following the manufacturer’s instructions.
To generate CstF-64 mutants, pcDNA-HA-CstF-64 was subjected to QuikChange mutagenesis (Stratagene) according to the manufacturer’s instructions, and in vitro translation was performed by using the generated plasmids.
The in vitro–translated, Flag-tagged symplekin fragments or the CstF-77 fragment was incubated for 1.5 h at room temperature with a 40-μl bed volume of anti-Flag M2 agarose (Sigma-Aldrich, Buchs, Switzerland) in PBS containing 0.1% NP-40 (PBS/NP-40), washed once with PBS/NP-40, and incubated with in vitro–translated, HA-tagged CstF-64 or CstF-64 mutants for 1 h at 4°C. After four washes with PBS/NP-40, the beads were transferred to a new tube in order to reduce background, followed by completely removing the supernatant with a 27¾-gauge needle, boiling in SDS-loading buffer, and loading on an SDS-polyacrylamide gel, followed by Western blotting. The bound HA-CstF-64 was detected with anti-HA-HRP (Roche), and the Flag-tagged proteins were detected with anti-Flag M2 peroxidase (Sigma).
Coimmunoprecipitations from HeLa total cell extract
HeLa cells were transfected with either pcDNA6-HA-symplekin-Flag or pcDNA6-HA-symplekin-Flag Hydro. Two days posttransfection, cells were harvested, washed with PBS, and resuspended in hypotonic lysis buffer (10 mM Tris, pH 7.8, 10 mM NaCl, 2 mM EDTA, 0.5% Triton-X-100) containing 125 μg/ml RNase A and supplemented with 1× complete EDTA-free protease inhibitor (Roche). The cells were lysed for 10 min on ice, followed by the addition of NaCl to 150 mM. The extract was cleared by centrifugation at 4°C, with 16,100 × g for 15 min. The cleared extract was then incubated head over tail for 2 h at 4°C with 60 μl anti-Flag M2 agarose, followed by five washes with NET-2 HD (50 mM Tris, pH 7.8, 150 mM NaCl, 0.1% NP-40), boiling of the beads in SDS loading buffer, and SDS–PAGE and Western blot. Coimmunoprecipitated proteins were then visualized by using goat anti-CstF-64 (C-20, Santa Cruz), rabbit anti-CstF-50 (Laboratory of Walter Keller, Basel, Switzerland), rabbit anti-CPSF-73 (009, Laboratory of Walter Keller), and rabbit anti-CPSF-100 (Laboratory of Walter Keller) with the corresponding species-specific, horseradish peroxidase (HRP)–coupled secondary antibody.
Immunofluorescence
The 2 × 105 cells were seeded onto coverslips in 6-well plates. For immunostaining, the cells were washed with PBS, fixed for 30 min at 37°C in DSP-IF buffer (PBS containing 2 mM MgCl2, 10% glycerol, 0.5 mM dithiobis-(succinimidyl) propionate; DSP, Pierce, Rockford, IL), and subsequently washed five times with the same buffer without DSP. Cells were either first stored at 4°C in 1× PBS, 0.2 M glycine, and 0.04% sodium azide or directly permeabilized for 20 min with PBS containing 0.1% Triton X-100 and 0.2 M glycine. To block unspecific binding of the primary antibodies, the cells were incubated for 30 min with Image-iT FX signal enhancer (Invitrogen) or with 10% BSA in PBS, followed by incubation with the primary antibodies diluted in 10% BSA in PBS, either for 2 h at room temperature or overnight at 4°C. After fives washes with PBS-0.05% NP-40, secondary antibodies were added and incubated for 1 h at room temperature. All antibodies were used at the dilutions indicated in the Supplemental Materials and Methods. The coverslips were mounted in Mowiol 4–88 reagent (Calbiochem, Darmstadt, Germany). Images were collected on a Leica TCS SP2 AOBS laser scanning confocal microscope equipped with a HCX PL APOlbd.BL 63.0 1.2W objective (Leica Microsystems, Exton, PA).
Cell cycle analysis
HeLa cells were trypsinized, washed with PBS, and fixed by dropwise addition to ice-cold 70% ethanol. Before staining with propidium iodide (PI), cells were washed twice with PBS, resuspended in PI-staining solution (0.1% Triton X-100 in PBS, 0.2 mg/ml RNase A, 0.2 mg/ml PI), and incubated for 1 h at room temperature in the dark. Cell cycle profiles were recorded by cytofluorometry (Beckton Dickinson BD FACSCalibur, San Jose, CA), and data were evaluated with FlowJo software (Tree Star, Ashland, OR).
Reverse transcription and quantitative real-time PCR
Usually 400–1000 ng RNA was reverse transcribed in 50 μl StrataScript 6.0 RT buffer in the presence of 4 mM of all four deoxynucleoside triphosphates, 300 ng random hexamers, 40 U RNasIn (Promega), and 1 μl StrataScript 6.0 reverse transcriptase (Stratagene). For real-time PCR, reverse-transcribed material corresponding to 40 ng RNA was amplified in 25 μl MasterMix Plus for probe Assay ROX (Eurogentec, Seraing, Belgium) with MESA GREEN qPCR MasterMix Plus for SYBR Assay ROX (Eurogentec) primers (Supplemental Table S3) or with TaqMan primers and probes (Supplemental Table S4), and real-time data were acquired with an ABI Prism SDS 7000 (Applied Biosystems, Carlsbad, CA) and analyzed with the associated software and Microsoft Excel according to Pfaffl (2001), taking amplification efficiencies into account for TaqMan assays or by using the comparative CT method (2−ΔΔCT method) for SYBR Green assays. The apparent in vivo 3′-end formation efficiency is expressed in molar ratios of precursor over total RNA. The TaqMan assays measure total H3C, β-actin, and c-myc mRNAs with primer/probe sets spanning the respective translation initiation codons. The corresponding pre-mRNAs are measured with primer/probe sets spanning the 3′ processing sites. The histone H3C assay has been described previously (Ruepp et al., 2010).
Supplementary Material
Acknowledgments
We thank W. Keller and G. Martin (Biozentrum Basel) for generous gifts of clones and antibodies, C.C. MacDonald and R.S. Pillai for antibodies, R. Lührmann (MPI Göttingen) for HeLa nuclear extracts, O. Mühlemann (University of Bern) for LI-COR secondary antibodies, and S. Barabino (University of Milano Bicocca) for very helpful suggestions and discussions. This work was supported by the Canton Bern and Swiss National Science Foundation grant 3100A0-120064 to D.S.
Abbreviations used:
- BSA
bovine serum albumin
- CHX
cycloheximide
- CPSF
cleavage and polyadenylation specificity factor
- ECL
enhanced chemiluminescence
- HBP/SLBP
hairpin- or stem-loop-binding protein
- HLF
heat-labile processing factor
- HRP
horseradish peroxidase
- MBP
maltose-binding protein
- NLS
nuclear localization signal
- PBS
phosphate-buffered saline
- PI
propidium iodide
- PVDF
polyvinylidine fluoride
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0543) on November 30, 2010.
REFERENCES
- Bongiorno-Borbone L, De CA, Barcaroli D, Knight RA, Di IC, Melino G, De Laurenzi V. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene. 2010;29:802–810. doi: 10.1038/onc.2009.388. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buchert M, et al. Symplekin promotes tumorigenicity by up-regulating claudin-2 expression. Proc Natl Acad Sci USA. 2010;107:2628–2633. doi: 10.1073/pnas.0903747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE. Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. EMBO J. 2010;29:1674–1687. doi: 10.1038/emboj.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J Biol Chem. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber LJ, Kort EJ, Wang P, Chen J, Teh BT. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mRNA. Mol Carcinog. 2010;49:215–223. doi: 10.1002/mc.20591. [DOI] [PubMed] [Google Scholar]
- Gick O, Krämer A, Vasserot A, Birnstiel ML. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockert JA, Yeh HJ, MacDonald CC. The hinge domain of the cleavage stimulation factor protein CstF-64 is essential for CstF-77 interaction, nuclear localization, and polyadenylation. J Biol Chem. 2010;285:695–704. doi: 10.1074/jbc.M109.061705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber Z, Monarez RR, Dass B, MacDonald CC. The mRNA encoding tauCstF-64 is expressed ubiquitously in mouse tissues. Ann NY Acad Sci. 2005;1061:163–172. doi: 10.1196/annals.1336.017. [DOI] [PubMed] [Google Scholar]
- Kavanagh E, Buchert M, Tsapara A, Choquet A, Balda MS, Hollande F, Matter K. Functional interaction between the ZO-1-interacting transcription factor ZONAB/DbpA and the RNA processing factor symplekin. J Cell Sci. 2006;119:5098–5105. doi: 10.1242/jcs.03297. [DOI] [PubMed] [Google Scholar]
- Keon BH, Schafer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- Lüscher B, Schümperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze MT, Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc Natl Acad Sci USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin N, Fonseca D, Mohammed S, Cevher MA, Manley JL, Kleiman FE. The 3′ processing factor CstF functions in the DNA repair response. Nucleic Acids Res. 2008;36:1792–1804. doi: 10.1093/nar/gkn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monarez RR, MacDonald CC, Dass B. Polyadenylation proteins CstF-64 and τCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J. 2007;401:651–658. doi: 10.1042/BJ20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Schümperli D. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin Cell Dev Biol. 1997;8:567–576. doi: 10.1006/scdb.1997.0182. [DOI] [PubMed] [Google Scholar]
- Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell. 2006;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp MD, Vivarelli S, Pillai R, Kleinschmidt N, Azzouz TN, Barabino SM, Schümperli D. The 68 kDa subunit of mammalian cleavage factor I interacts with the U7 small nuclear ribonucleoprotein and participates in 3′ end processing of animal histone mRNAs. Nucl Acids Res. 2010 doi: 10.1093/nar/gkq613. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarling GS, Coates PW, Dass B, MacDonald CC. A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol Biol. 2009;10(22) doi: 10.1186/1471-2199-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA. 2009;15:459–472. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, MacDonald CC, Shenk T, Manley JL. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci USA. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Berkow A, Marzluff WF. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem Soc Trans. 2005;33:471–473. doi: 10.1042/BST0330471. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Marzluff WF. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol Cell Biol. 2006;26:6702–6712. doi: 10.1128/MCB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci USA. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.