Abstract
Quartz stones are ubiquitous in deserts and are a substrate for hypoliths, microbial colonists of the underside of such stones. These hypoliths thrive where extreme temperature and moisture stress limit the occurrence of higher plant and animal life. Several studies have reported the occurrence of green hypolithic colonization dominated by cyanobacteria. Here, we describe a novel red hypolithic colonization from Yungay, at the hyper-arid core of the Atacama Desert in Chile. Comparative analysis of green and red hypoliths from this site revealed markedly different microbial community structure as revealed by 16S rRNA gene clone libraries. Green hypoliths were dominated by cyanobacteria (Chroococcidiopsis and Nostocales phylotypes), whilst the red hypolith was dominated by a taxonomically diverse group of chloroflexi. Heterotrophic phylotypes common to all hypoliths were affiliated largely to desiccation-tolerant taxa within the Actinobacteria and Deinococci. Alphaproteobacterial phylotypes that affiliated with nitrogen-fixing taxa were unique to green hypoliths, whilst Gemmatimonadetes phylotypes occurred only on red hypolithon. Other heterotrophic phyla recovered with very low frequency were assumed to represent functionally relatively unimportant taxa.
Keywords: Atacama, Chloroflexi, Chroococcidiopsis, Desert, Hyper-arid, Hypolith
Introduction
The Atacama Desert is an ancient coastal arid zone that extends across a large area of coastal Chile in South America (Thomas 1997). The desert is, like most arid regions on Earth, typified by a stony desert pavement terrain. This extends approximately 1,000 km along the Pacific coast of South America. At the hyper-arid core of the Atacama at Yungay (24°06′ S 70°01′ W with 1,051-m elevation), the mean annual rainfall (MAR) is around 2.4 mm making it the driest zone in the desert. Long-term climate data indicate that the hyperarid core of the Atacama Desert at Yungay is the driest non-polar desert location on Earth (Warren-Rhodes et al. 2006; Kuhlman et al. 2008).
Hyper-arid desert pavements are characterized by a lack of higher plants or animals (Thomas 1997), and microorganisms are often the only discernible life. This lends significance to microbial colonization as the predominant form of primary productivity and nitrogen input in such extreme desert regions. A ubiquitous feature of desert pavement microbial ecology has been the occurrence of hypolithic cyanobacterial colonization on quartz rocks both in hot (Schlesinger et al. 2003; Pointing et al. 2007; Warren-Rhodes et al. 2006; Warren-Rhodes et al. 2007) and in cold deserts (Broady 2005; Wood et al. 2008; Pointing et al. 2009; Cowan et al. 2010; Wong et al. 2010a, b). Whilst early studies identified cyanobacteria as the predominant colonist (Cameron and Black 1996; Friedmann and Galun 1974; Berner and Evenari 1978), several recent studies have used DNA fingerprinting techniques to inform community structure at a molecular level (Warren-Rhodes et al. 2006; Wood et al. 2008; Pointing et al. 2007). These have generally lacked a quantitative approach and so beta diversity remains uncharacterized, although such molecular studies indicated that hypolithic communities comprise significantly more complex diversity than previously appreciated. More recently, hypolith community structure has been elucidated using sequence data derived from clone libraries and this has yielded valuable data on beta diversity for cold desert hypoliths (Pointing et al. 2009; Wong et al. 2010a). Whilst clone libraries have well-documented limitations, they remain a widespread tool for inferring community structure but have, as yet, not been applied to Atacama hypoliths. Besides the value in more fully resolving microbial community assembly of ‘typical’ green hypoliths at the arid limit for life, the observation that red pigmentation of a quartz rock supported filamentous microbial colonization created a comparative motive for the current study between green and red hypoliths.
Materials and methods
Micro-environmental data
Environmental data that included long-term mean annual temperature and precipitation were collected. Historical rainfall data were sourced from Dirección General de Aguas (http://www.dga.cl), Dirección Meteorológica de Chile (http://www.meteochile.com or http://docs.lib.noaa.gov/resecue/data_rescue_chile.html) and field measurements (McKay et al. 2003). Nanoclimate data for the hypolithic niche were collected in situ from October 2001–2005, including: (1) air and soil temperature and relative humidity at 1 m above the ground surface, at the ground surface and at ~2–5 mm, 2–5 cm, 10 cm, and 20 cm below the soil surface, using Onset Computer HOBO Pro® dataloggers; (2) presence of liquid water condensed on rock and soil surfaces from dew or fog, using a Campbell 237® leaf wetness moisture sensing grid and/or a Spectrum® moisture sensing grid; and (3) rainfall using an Onset Computer tipping bucket rain gauge. An Onset weather station was also installed, with additional sensors to measure solar flux, photosynthetically active radiation (PAR), barometric pressure, and wind speed and direction. The occurrence of liquid water in the soil sufficient to support photosynthesis was defined as a soil relative humidity of ≥95% (Schlesinger et al. 2003) and on soil or stone surfaces by the Campbell moisture sensing grid as a rise in voltage above the 0.005 V ‘dry’ baseline (or a drop in voltage from the 2.095 V ‘dry’ baseline for the Spectrum sensor). Measurements of photosynthetically active radiation (PAR), UV-A, and UV-B radiation were carried out at solar noon using a Li-Cor LI-I400 datalogger (Li-Cor Inc, Nebraska, USA) for PAR, and a UV UVX radiometer (UVP Inc, California, USA) for UV irradiance. For transmittance studies through rock substrates, the sensor was placed in situ under quartz rocks and edges sealed using plasticine.
Molecular community characterization
Colonized rocks were collected from the hyperarid zone, Yungay area (24°06′ S 70°01′ W with 1,051-m elevation), of the Atacama Desert. The three rocks within the sampling zone at Yungay supporting hypolithic biomass (from the same sampling regime conducted by Warren-Rhodes et al. 2006) were used in the present study. Community DNA recovery was achieved following the protocol for environmental DNA recovery from lithic biofilms described in Warren-Rhodes et al. 2006. The entire colonized surface area of each rock was sampled. 16S rRNA genes were PCR amplified using bacteria-specific primers 8F (AGAGTTTGATCCTGGCTCAG) and 1391R (TGYACWCACCGCCCGTC) (Lane 1991). Archaeal-specific primers 8Fa (TCYSGTTGATCCTGCS) and 1492R (GGTTACCTTGTTACGACTT) (Costello and Schmidt 2006) and eukaryal-specific primers NS1 (GTAGTCATATGCTTGTCTC) and NS4 (CTTCCGTCAATTCCTTTAAG) (White et al. 1990) were also used, but did not give positive amplification. Purified PCR product was used to construct clone libraries for each rock sample (CloningPlus, Qiagen, Valencia, CA, USA). A minimum of 100 clones per library were screened by RFLP (MSP1 and CFO1, Amersham, Bucks, UK) and a total of 104 unique RFLP-defined phylotypes were sequenced (ABI 3730 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) to generate 70 unique 16S rRNA gene sequences (Operational Taxonomic Unit, OTU) of approximately 1,446 bp each. Phylogenetic OTU’s were determined using the computer program DOTUR described in Schloss and Handelsman (2005), with a 97% sequence similarity cutoff. The sufficiency of sampling from each library was estimated using non-parametric rarefaction and OTU Richness determined by Chao1 using Estimate S software (http://viceroy.eeb.uconn.edu/estimateS). Coverage estimates, Shannon’s Diversity Index (Llyod et al. 1968), and Pielou’s Evenness Index (Pielou 1966) were calculated using Primer v6 (Clarke 1993).
Chimera_Check software (Ribosome Database Project, http:/rdp.cme.msu.edu.html) was used to check possible chimeric structures among the sequence data. Approximate phylogenetic affiliations were then determined using BLAST searches of NCBI GenBank database. Multiple alignments were created using ClustalX v.1.81 (Thompson et al. 1997) with reference to sequences selected from the GenBank database. All phylogenetic analysis was performed using both GARLI (Genetic Algorithm for Rapid Likelihood Inference) 0.96 beta (Zwickl 2006) and PAUP* 4.0b10 (Swofford 2001). Maximum likelihood analysis was used to illustrate relationships of the 16S rRNA gene sequences to representative taxa. Bayesian posterior probabilities and bootstrap values of 1,000 replications were calculated and are shown for branch nodes supported by more than 50% of the trees. The phylogenetic differences among communities were quantified using the F st statistic calculated using Arlequin v3.0 (Excoffier and Scheider 2005). Sequence data have been submitted to NCBI GenBank database with accession numbers FJ890991 to FJ891059.
Results and discussion
Long-term micro-environmental monitoring at the study site carried out by our group indicated that the hypolithic niche beneath quartz rocks received an average of 395(±18) h/year−1 liquid water (Table 1) although of this only 75 h were under conditions that were estimated to support photosynthetic activity (sufficient PAR and temperature) (Warren-Rhodes et al. 2006). This moisture input was derived from fog/dew (68%) and rainfall (32%). We have also conducted radiocarbon dating studies which indicated that Atacama hypoliths may be very long lived and possibly persisted for thousands of years (Warren-Rhodes et al. 2006). Hypolithic colonization can thus be envisaged as a slow, but persistent colonization, and although they clearly tolerate significant environmental stochasticity in terms of water availability and temperature, we have observed that disturbed hypoliths seldom survive, thus suggesting that substrate stability is important.
Table 1.
Summary of beta diversity for green and red hypoliths
| Quartz rock sample | Green 1 (AY1) | Green 2 (AY5) | Red 1 (AY6) |
|---|---|---|---|
| Phylum | Percentage of total phylotypes recovered | ||
| Cyanobacteria | 77 | 85 | 39 |
| Acaryochloris | 1 | ||
| Chroococcidiopsis | 27 | 47 | 29 |
| Nostocales | 48 | 37 | 10 |
| Oscillatoriales | 2 | ||
| Actinobacteria | 5 | 4 | 11 |
| Alphaproteobacteria | 13 | 5 | |
| Bacteroidetes | 2 | 1 | |
| Betaproteobacteria | 2 | ||
| Chloroflexi | 32 | ||
| Deinococci | 3 | ||
| Gammaproteobacteria | 4 | 4 | |
| Gemmatimonadetes | 1 | ||
| Planctomycetes | 1 | ||
| Unknown bacteria | 11 | ||
| Thermobaculum | 6 | ||
Phylotypes were assigned to a given taxonomic group based upon phylogenetic analysis of 16S rRNA gene sequences
The extremely low colonization frequency of quartz rocks at the hyper-arid core of the Atacama Desert mean that locating hypolithic colonized rocks that can be analyzed is highly challenging (i.e. only 3 out of 3,723 stones were colonized at our site, across an area >2,000 m2) (Warren-Rhodes et al. 2006). The study reported here includes all three colonized rocks located within the survey area and therefore represents the total recoverable hypolithic diversity within this area. Despite equal effort in DNA recovery and cloning, the three rocks yielded significantly different rRNA gene-defined communities, and this heterogeneity was supported by microscopy that indicated major phylotypes recovered in each library were also the most commonly encountered morphotypes. The data revealed two major findings; that the two ‘green’ Chroococcidiopsis-dominated hypoliths supported a highly similar diversity although abundance levels varied; and that the ‘red’ hypolith supported chloroflexi rather than cyanobacteria as the dominant taxon plus a markedly different and more speciose heterotrophic component.
The two green hypoliths were the same samples that have been previously analyzed using a DGGE DNA-fingerprinting approach (Warren-Rhodes et al. 2006). Unsurprisingly, our clone libraries revealed more than twice the number of phylotypes than the fingerprinting approach, and also allowed a phylogenetic assignment to near-full length rRNA gene phylotypes. The limitations of the DGGE technique are now well-documented (Jackson and Churchill 1999; Sekiguchi et al. 2001; Kirk et al. 2004; Nocker et al. 2007), and the rationale for the present study was to more comprehensively understand diversity of hypoliths at the driest non-polar desert location on Earth. Our libraries allowed us to more fully define the community composition and generate relatively quantitative beta diversity data (Table 2). For both green hypoliths, the community was dominated by diverse phylotypes within the cyanobacterial genus Chroococcidiopsis and the Nostocales (Fig. 1). The former is a near-ubiquitous colonist of arid deserts worldwide (Bahl et al. 2010), also known to fix nitrogen under certain conditions (Boison et al. 2004). The latter a nitrogen-fixing family that contains several UV-screening pigment-secreting taxa recovered from temperate (Warren-Rhodes et al. 2006) and cold deserts (Pointing et al. 2009; Wong et al. 2010a, b) regions. Production of UV-protective pigments by cyanobacteria, such as scytonemin (Proteau et al. 1993) can be envisaged as an adaptive advantage in high UV desert environments. Both of these cyanobacterial groups are noted for their radiation tolerance (Billi et al. 2000), a proxy for desiccation tolerance.
Table 2.
Summary of diversity data for clone libraries generated from green and red hypoliths
| Sample | No. of positive transformants selected | No. of RFLP-defined phylotypes | No. of OTU (<97% sequence similarity) | Coverage (C) (%) | Chao1 richness ± SD | Shannon’s diversity index (H) | Pielou’s evenness (J) | F st statistic |
|---|---|---|---|---|---|---|---|---|
| Green 1 (AY1) | 108 | 32 | 22 | 90 | 20.8 ± 5.2 | 3.15 | 0.71 | 0.1401 |
| Green 2 (AY5) | 101 | 29 | 19 | 88 | 17.3 ± 5.2 | 2.73 | 0.64 | 0.14164 |
| Red 1 (AY6) | 102 | 43 | 29 | 83 | 32.4 ± 9.7 | 4.04 | 0.83 | 0.13528 |
Fig. 1.
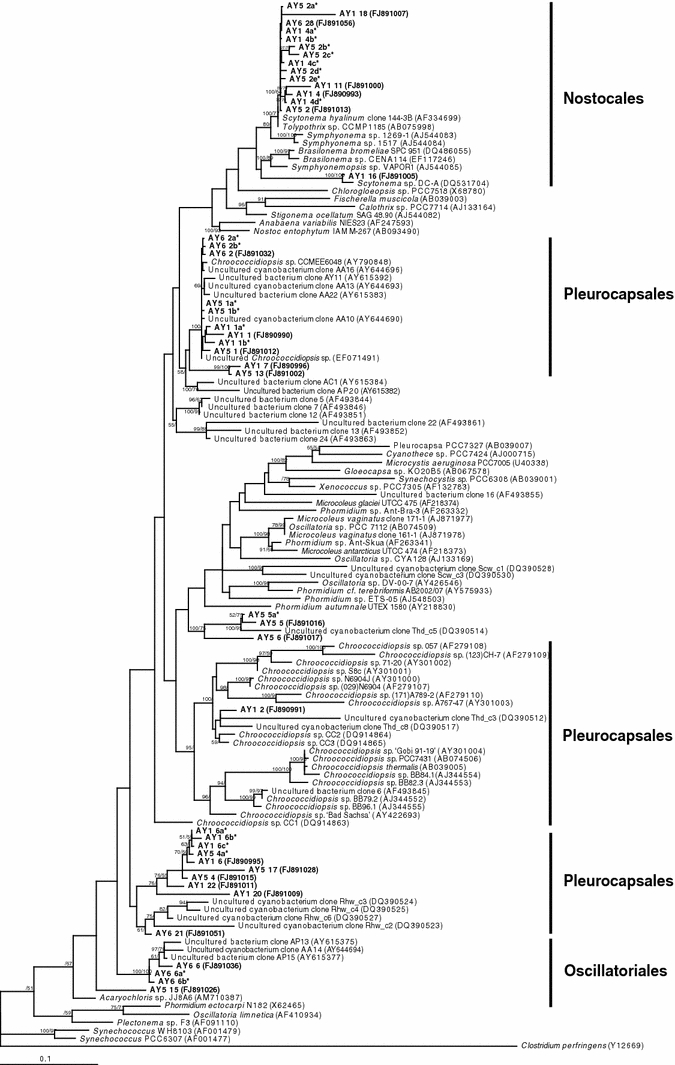
Phylogenetic relationships among cyanobacterial 16S rRNA phylotypes recovered from Atacama hypoliths. Phylotypes recovered during this study are shown in bold type. Phylotypes indicated by an asterisk share >97% sequence similarity with their most closely affiliated phylotype indicated by a NCBI GenBank accession number (in brackets) on the tree. NCBI GenBank accession tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar 0.1 nucleotide changes per position
The second most abundant phylum was the Alphaproteobacteria (Fig. 2). These phylotypes were specific to green hypoliths and whilst phylogenies for this phylum are skewed toward human associates as a result of availability of identified environmental taxa among published sequences, they did also affiliate with the nitrogen fixing genus Agrobacterium. Other studies have revealed that N2-fixing organisms are often the abundant and early colonizers of arid soil environments that give way to other species over time (Yeager et al. 2004; Garcia-Pichel et al. 2001; Walker 1993), presumably as they create nitrogen sufficiency within these otherwise nitrogen-depleted desert soils. Other phylotypes were recovered with very low abundance (Table 1; Fig. 2) and so can be postulated to represent functionally relatively unimportant taxa, although their latency might indicate the capacity of the community to respond to environmental change.
Fig. 2.
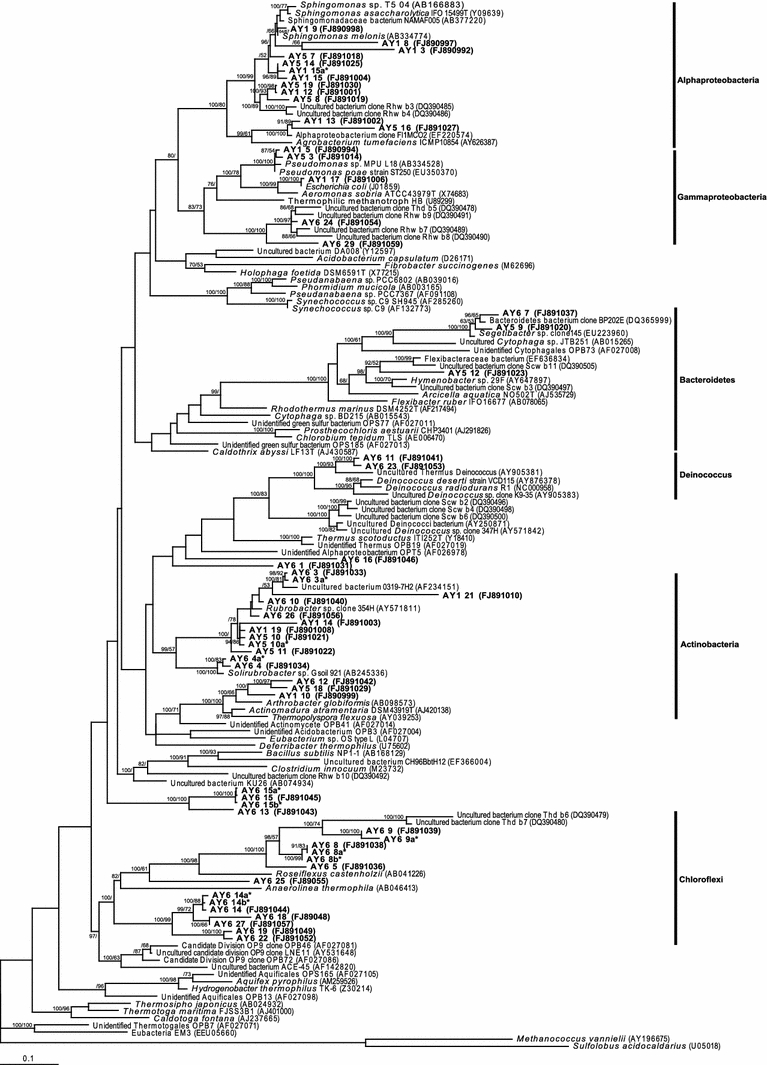
Phylogenetic relationships among bacterial 16S rRNA phylotypes recovered from Atacama hypoliths. Phylotypes recovered during this study are shown in bold type. Phylotypes indicated by an asterisk share >97% sequence similarity with their most closely affiliated phylotype indicated by a NCBI GenBank accession number (in brackets) on the tree. NCBI GenBank accession tree topologies are supported by Bayesian posterior probabilities (first number) and bootstrap values for 1,000 replications (second number). Scale bar 0.1 nucleotide changes per position
The red hypolith represented an opportunity to describe a novel hypolithic colonization, often previously assumed to represent an abiotic red/brown patina on rocks. Our study revealed surprisingly different community structure between red and green hypoliths (Table 1). The most striking feature was the dominance by a collection of phylogenetically relatively diverse chloroflexi (Fig. 2). The chloroflexi have traditionally been associated with hot spring environments (Jing et al. 2005; Lau et al. 2006, 2008, 2009; Lacap et al. 2007), but recently have emerged as colonists of hot (Pointing et al. 2007) and cold (Wong et al. 2010a) desert hypoliths, in addition to cold tundra soils (Costello and Schmidt 2006) and endoliths (Wong et al. 2010b). As a phylum they may therefore display a general adaptation to arid environments. The phylotypes affiliated with the genera Roseiflexus, Anaerolinea and also formed a deep-branching cluster of uncharacterized Chlofoflexi (Fig. 2), possibly indicating a novel group of green non-sulfur bacteria. The cyanobacterial genus Chroococcidiopsis was the second most abundant group, whilst the Nostocales comprised only 10% of the community. Interestingly, no other putative nitrogen fixing taxa were indicated from the phylogenies, and this may reflect a higher degree of nitrogen sufficiency within this micro-niche, or generally more abundant nutrients since the chloroflexi are heterotrophically capable and may have out-competed the cyanobacteria under such conditions. The difference is unlikely a function of moisture availability since the sampled area experiences very similar water input and residence times (Warren-Rhodes et al. 2006).
We measured light transmittance (PAR, UVA, and UVB) through all rocks from the point of soil surface to the depth of colonization, with no marked differences between green and red colonized rocks. We therefore discount a selective effect from light transmittance. Whilst low abundances of commonly encountered arid soil heterotrophic bacteria (Yeager et al. 2004; Garcia-Pichel et al. 2001; Walker 1993) from the phyla Actinobacteria and Bacteriodetes were shared between green and red hypoliths, the red hypolith also supported additional unique heterotrophic phylotypes (Table 2; Fig 2). The actinobacterial phylotypes affiliated with the genus Rubrobacter, a noted radiation tolerant desert bacterium (Ferreira et al. 1999). This included the Deinococci, which affiliated with those recovered from arid soils in other hot deserts (Rainey et al. 2005; Chanal et al. 2006; Pointing et al. 2007). The Deinococci are noted for their desiccation and radiation tolerance (De Groot et al. 2005)) although they appear to be a feature of hot rather than cold desert hypoliths. The remaining red hypolith phylotypes affiliated with the Gemmatimonadetes, which are emerging as a common taxon in hypoliths from hot (Pointing et al. 2007) and cold (Pointing et al. 2009) deserts, plus unidentified beta proteobacteria and other unidentified phyla. The greater diversity and abundance of heterotrophic phylotypes may indicate that the chloroflexi-dominated hypolith represents a community structured under more nutrient sufficient conditions.
Statistical analyses of the phylogenetic data (Table 2) indicated that whilst the red hypolith community was most biodiverse, all three hypoliths displayed significantly different diversity and evenness indices (Table 2). The phylogenetically informed community structure was demonstrated as significantly different between all three communities (Table 2). This is interesting, since given the high coverage of our libraries (due to the relatively low diversity of hypoliths); this indicates that hypolith communities can be highly heterogenous within a given climatic biome. This is likely due to unmeasured differences in the microclimate beneath each quartz rock.
The Atacama Desert supports several distinct microbial oases, including quartz hypoliths, halite endoliths, and microbial mats in hypersaline waters (Weirzchos et al. 2009). It is noteworthy that similar cyanobacterial taxa occur in all of these niches and so they may act as local reservoirs for ‘extreme’ colonists in a variety of challenging niches. It will be interesting to establish in future work the extent to which common cyanobacterial taxa are involved in community assembly within each niche, and whether non-cyanobacterial taxa, such as the chloroflexi are also widespread in these niches.
Acknowledgments
The authors acknowledge support extended by NASA’s Astrobiology Science and Technology for Exploring Planets (ASTEP) program, and the Hong Kong Research Grants Council (Grant number HKU7733/08M HKU7763/10).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bahl J, Lau MCY, Smith GJD, Vijaykrishna D, Cary SC et al (2010) Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nature Commun (in press) [DOI] [PMC free article] [PubMed]
- Berner T, Evenari M. The influence of temperature and light penetration on the abundance of the hypolithic algae in the Negev desert of Israel. Oecologia. 1978;33:255–260. doi: 10.1007/BF00344852. [DOI] [PubMed] [Google Scholar]
- Billi D, Friedmann IE, Hofer KG, Grilli-Caiola M, Ocampo-Friedmann R. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl Environ Microbiol. 2000;66:1489–1492. doi: 10.1128/AEM.66.4.1489-1492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison G, Mergel A, Jolkver H, Bothe H. Bacterial life and dinitrogen fixation at a gypsum rock. Appl Environ Microbiol. 2004;70:7070–7077. doi: 10.1128/AEM.70.12.7070-7077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broady PA. The distribution of terrestrial and hydro-terrestrial algal associations at three contrasting locations in southern Victoria Land, Antarctica. Algalogical Stud. 2005;118:95–112. doi: 10.1127/1864-1318/2006/0118-0095. [DOI] [Google Scholar]
- Cameron RA, Black GB (1966) Desert algae: soil crusts and diaphanous substrata as algal habitats. Technical Report 32-971. National Aeronautics and Space Administration (NASA), Jet Propulsion Laboratory, California Institute of Technology, Pasadena, California, USA
- Chanal A, Chapon V, Benzerara K, Barakat M, Christen R, Achouak W, et al. The desert of Tatouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ Microbiol. 2006;8:514–525. doi: 10.1111/j.1462-2920.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- Costello EK, Schmidt SK. Microbial diversity in alpine tundra wet meadow soil: novel chloroflexi from a cold, water-saturated environment. Environ Microbiol. 2006;8:1471–1486. doi: 10.1111/j.1462-2920.2006.01041.x. [DOI] [PubMed] [Google Scholar]
- Cowan DA, Pointing SB, Stevens MI, Cary SC, Stomeo F, Tuffin IM (2010) Distribution and abiotic influences on hypolithic communities in an Antarctic Dry Valley. Polar Biol. doi:10.1007/s00300-010-0872-2 [e-pub ahead of print]
- De Groot A, Chapon V, Servant P, Christen R, Saux MFL, Sommer S, Heulin T. Deinococcus deserti sp. nov., a gamma-radiation-tolerant bacterium isolated from the Sahara Desert. Int J Syst Evol Microbiol. 2005;55:2441–2446. doi: 10.1099/ijs.0.63717-0. [DOI] [PubMed] [Google Scholar]
- Excoffier LGL, Scheider S. Arlequin v3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Ferreira AC, Nobre MF, Moore E, Rainey FA, Battista JR, da Costa MS. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles. 1999;3:235–238. doi: 10.1007/s007920050121. [DOI] [PubMed] [Google Scholar]
- Friedmann EI, Galun M. Desert algae, lichens, and fungi. In: Brown GW, editor. Desert biology. New York, New York, USA: Academic Press; 1974. pp. 165–212. [Google Scholar]
- Garcia-Pichel F, Lopez-Cortez A, Nubel U. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microbiol. 2001;67:1902–1910. doi: 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CR, Churchill PF. Analysis of microbial communities by denaturing gradient gel electrophoresis: applications and limitations. Recent Res Dev Microbiol. 1999;3:81–91. [Google Scholar]
- Jing H, Aitchison JC, Lacap DC, Peerapornpisal Y, Sompong U, Pointing SB. Community phylogenetic analysis of moderately thermophilic cyanobacterial mats from China, the Philippines, and Thailand. Extremophiles. 2005;9:325–332. doi: 10.1007/s00792-005-0456-1. [DOI] [PubMed] [Google Scholar]
- Kirk JL, Beaudette LA, Hart M, Moutoglis P, Klironomos JN, Lee H, Trevors JT. Methods of studying soil microbial diversity. J Microbiol Methods. 2004;58:169–188. doi: 10.1016/j.mimet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Kuhlman KR, Venkat P, La Duc MT, Kuhlman GM, McKay CP. Evidence of a microbial community associated with rock varnish at Yungay, Atacama Desert, Chile. J Geophys Res. 2008;113:G04022. doi: 10.1029/2007JG000677. [DOI] [Google Scholar]
- Lacap DC, Barraquio W, Pointing SB. Thermophilic microbial mats in a tropical geothermal location display pronounced seasonal changes but appear resilient to stochastic disturbance. Environ Microbiol. 2007;9:3065–3076. doi: 10.1111/j.1462-2920.2007.01417.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, UK: Wiley; 1991. pp. 115–148. [Google Scholar]
- Lau CY, Jing H, Aitchison JC, Pointing SB. Highly diverse community structure in a remote central Tibetan geothermal spring does not display monotonic variation to thermal stress. FEMS Microbiol Ecol. 2006;57:80–91. doi: 10.1111/j.1574-6941.2006.00104.x. [DOI] [PubMed] [Google Scholar]
- Lau CY, Aitchison JA, Pointing SB. Early colonization of thermal niches in a silica-depositing geothermal spring in central Tibet. Geobiol. 2008;6:136–146. doi: 10.1111/j.1472-4669.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- Lau CY, Aitchison JA, Pointing SB. Bacterial community composition in thermophilic microbial mats from 5 hot springs in central Tibet. Extremophiles. 2009;13:139–149. doi: 10.1007/s00792-008-0205-3. [DOI] [PubMed] [Google Scholar]
- Llyod H, Zar JH, Karr JR. On the calculation of information-theoretical measures of diversity. Am Mid Nat. 1968;79:257–272. doi: 10.2307/2423177. [DOI] [Google Scholar]
- McKay CP, Friedmann EI, Gomez-Silva B, Cacere L, Andersen D, Landheim R. Temperature and moisture conditions in the extreme arid regions of the Atacama Desert: four years of observations including the El Nino of 1997–1998. Astrobiology. 2003;3:393–406. doi: 10.1089/153110703769016460. [DOI] [PubMed] [Google Scholar]
- Nocker A, Burr M, Camper AK. Genotypic microbial community profiling: a critical technical review. Microbial Ecol. 2007;54:276–289. doi: 10.1007/s00248-006-9199-5. [DOI] [PubMed] [Google Scholar]
- Pielou EC. The measurement of diversity in different types of biological collections. J Theor Biol. 1966;13:131–144. doi: 10.1016/0022-5193(66)90013-0. [DOI] [Google Scholar]
- Pointing SB, Warren-Rhodes KA, Lacap DC, Rhodes KL, McKay CP. Hypolithic community shifts occur as a result of liquid water availability along environmental gradients in China’s hot and cold hyperarid deserts. Environ Microbiol. 2007;9:414–424. doi: 10.1111/j.1462-2920.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- Pointing SB, Chan Y, Lacap DC, Lau CY, Jurgens J, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proteau PJ, Gerwick WH, Garcia-Pichel F, Castenholz R. The structure of scytonemin, an ultraviolet sunscreen pigment from the sheaths of cyanobacteria. Cell Mol Life Sci. 1993;49:825–829. doi: 10.1007/BF01923559. [DOI] [PubMed] [Google Scholar]
- Rainey FA, Ray K, Ferriera M, Gatz BZ, Fernanda Nobre M, Bagaley D, et al. Extensive diversity of ionizing-radiation-tolerant bacteria recovered from Sonoran Desert soil and description on nine new species of the genus Deinococcus obtained from a single soil sample. Appl Environ Microbiol. 2005;71:5225–5235. doi: 10.1128/AEM.71.9.5225-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger WH, Pippin J, Wallenstein M, Hofmockel K, Klepeis D, Hahall B. Community composition and photosynthesis by photoautotrophs under quartz pebbles, southern Mojave Desert. Ecology. 2003;84:3222–3231. doi: 10.1890/02-0549. [DOI] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi H, Tomioka N, Nakahara T, Uchiyama H. A single band does not always represent single bacterial strains in denaturing gradient gel electrophoresis analysis. Biotechnol Lett. 2001;23:1205–1208. doi: 10.1023/A:1010517117046. [DOI] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b10. Sunderland: Sinauer Associates; 2001. [Google Scholar]
- Thomas DSG. Arid zones: their nature and extent. In: Thomas DSG, editor. Arid zone geomorphology. 2. Chichester, UK: Wiley; 1997. pp. 3–12. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal–Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LR. Nitrogen fixers and species replacements in primary succession. In: Miles J, Walton DWH, editors. Primary succession on land. Oxford, UK: Blackwell Scientific; 1993. pp. 249–272. [Google Scholar]
- Warren-Rhodes KA, Rhodes KL, Pointing SB, Ewing SA, Lacap DC, Gómez-Silva B, et al. Hypolithic cyanobacteria, dry limit of photosynthesis and microbial ecology in the hyperarid Atacama Desert. Chile Microb Ecol. 2006;52:389–398. doi: 10.1007/s00248-006-9055-7. [DOI] [PubMed] [Google Scholar]
- Warren-Rhodes KA, Rhodes KL, Boyle LN, Pointing SB, Chen Y, et al. Cyanobacterial ecology across environmental gradients and spatial scales in China’s hot and cold deserts. FEMS Microbiol Ecol. 2007;61:470–482. doi: 10.1111/j.1574-6941.2007.00351.x. [DOI] [PubMed] [Google Scholar]
- Weirzchos J, de los Rios A, Davila AF, Camara B, Valea S, et al. Primary producers in extreme arid environment of the Atacama Desert: where, how and when? Geochim Cosmochim Acta (Suppl) 2009;73:A1439. [Google Scholar]
- White TJ, Burns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Wong FKY, Lacap DC, Lau MCY, Aitchison JC, Cowan DA, Pointing SB. Hypolithic microbial community of quartz pavement in the high-altitude tundra of central Tibet. Microbial Ecol. 2010;60:730–739. doi: 10.1007/s00248-010-9653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong FKY, Lau MCY, Lacap DC, Aitchison JC, Cowan DA, Pointing SB. Endolithic microbial colonization of limestone in a high-altitude arid environment. Microbial Ecol. 2010;59:689–699. doi: 10.1007/s00248-009-9607-8. [DOI] [PubMed] [Google Scholar]
- Wood SA, Rueckert A, Cowan DA, Cary SC. Sources of edaphic cyanobacterial diversity in the Dry Valleys of Eastern Antarctica. ISME J. 2008;2:308–320. doi: 10.1038/ismej.2007.104. [DOI] [PubMed] [Google Scholar]
- Yeager CM, Kornosky JL, Housman DC, Grote EE, Belnap J, Kuske CR. Diazotrophic community structure and function in two successional stages of biological soil crusts from the Colorado Plateau and Chihuahuan Desert. Appl Environ Microbiol. 2004;70:973–983. doi: 10.1128/AEM.70.2.973-983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin, USA


