Abstract
We have generated transgenic medaka (teleost, Oryzias latipes), which allow us to monitor germ cells by green fluorescent protein (GFP) fluorescence in live specimens. Two medaka strains, himedaka (orange–red variety) and inbred QurtE, were used. The transgenic lines were achieved by microinjection of a construct containing the putative promoter region and 3′ region of the medaka vasa gene (olvas). The intensity of GFP fluorescence increases dramatically in primordial germ cells (PGCs) located in the ventrolateral region of the posterior intestine around stage 25 (the onset of blood circulation). Whole-mount in situ hybridization and monitoring of ectopically located cells by GFP fluorescence suggested that (i) the increase in zygotic olvas expression occurs after PGC specification and (ii) PGCs can maintain their cell characteristics ectopically after stages 20–25. Around the day of hatching, the QurtE strain clearly exhibits sexual dimorphisms in the number of GFP fluorescent germ cells, a finding consistent with the appearance of leucophores, a sex-specific marker of QurtE. The GFP expression persists throughout the later stages in the mature ovary and testis. Thus, these transgenic medaka represent a live vertebrate model to investigate how germ cells migrate to form sexually dimorphic gonads, as well as a potential assay system for environmental substances that may affect gonad development. The use of a transgenic construct as a selective marker to efficiently isolate germ-line-transmitting founders during embryogenesis is also discussed.
To achieve the ability to transmit genetic information via sexually dimorphic gametes to the next generation, germ cells are processed through many steps during early embryogenesis and gonadogenesis. Genetic epistasy and the molecular mechanisms involved in these steps have been extensively investigated in model invertebrate organisms such as Caenorhabditis elegans and Drosophila melanogaster (for review, see refs. 1 and 2). Despite intensive studies in vertebrates (3, 4), a precise picture of the formation of the germ line and gonads has yet to be developed, largely because of the difficulty in applying modern molecular genetic techniques to analysis of these phenomena in vertebrates.
A short generation time, a variety of strains, a small genome size, and its linkage map render medaka (Teleostei, Oryzias latipes) suitable for molecular genetics to analyze many biological phenomena (e.g., refs. 5–8). Furthermore, techniques to generate transgenic medaka have already been developed (9). We have been carrying out brother–sister mating of the Qurt strain for 12 generations to produce a more homogeneous genetic background (QurtE). This strain lacks most pigments except for leucophores, a Y-linked trait (10). Therefore, QurtE is almost transparent, which allows the viscera and brain to be seen in the living fish. The appearance of leucophores as early as 2 days after fertilization (before the sexually dimorphic gonad forms) allows identification of genetic males and females. Combined with the introduction of the green fluorescent protein (GFP) gene as a germ cell marker, these characteristics make the QurtE an ideal model to elucidate how germ cells behave during gonad formation.
The Drosophila vas gene originally identified by maternal effect screens encodes an RNA helicase (11). This gene affects the posterior localization and translation of the posterior determinants, Nanos and Pumilio, in oocytes and is also known to be a component of pole plasm, a determinant of pole cells (i.e., germ cells) (for review, see ref. 12). The pole plasm in Drosophila was originally identified by ultrastructural analysis as an electron-dense cytoplasmic inclusion. Similar structures in many vertebrate germ cells have also been referred to as germ plasm. Furthermore, there are several reports of exclusive expression of vasa-related genes in vertebrate germ cells (13–24), suggesting a functional similarity of vasa-related genes in the formation of the germ cells. Therefore, the regulatory regions of a vertebrate vasa gene could be used to direct transgene expression exclusively in germ cells.
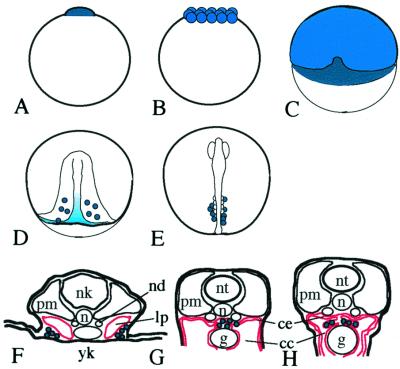
Previously, we isolated the medaka vasa cDNA and gene (olvas, named after Oryzias latipes vasa) and showed its exclusive expression in the germ cells of medaka (24). A brief outline of medaka germ cell migration is shown in Fig. 1. As maternally derived olvas transcripts diminish during late gastrulation, some cells located around the posterior region of embryonic shield seem to retain or increase olvas expression (Fig. 1 A–D). After bilateral alignment along the ventrolateral region of the embryonic body, germ cells migrate to form a clump at the dorsal region of coelomic epithelium with the development of somites, lateral plate mesoderm, and gut (Fig. 1 E–G) and then form two bilateral clusters that develop into the gonad (Fig. 1H).
Figure 1.
Schematic representation of the migration of germ cells in medaka embryos (see also ref. 24). Olvas expressing cells or regions are colored blue (dark blue indicates more intense expression). Lateral plate mesoderm and coelomic epithelium are shown in red. (A) 1 cell (stage 2); (B) morula (stage 8); (C) early gastrulation (stage 13); (D) late gastrulation (stage 16); (E) 4 somites (stage 20); (F) stage 25; (G) stage 35 (F–H, cross sections). cc, ceolomic cavity; ce, coelomic epithelium; g, gut; lp, lateral plate mesoderm; nd, nepheric duct; n, notocord; nk, neural keel; nt, neural tube; pm, paraxial mesoderm; yk, yolk.
A similar migration pattern of germ cells was also reported in zebrafish. The maternal vasa product localized to the distal parts of the first cleavage furrow ingresses into the four cells of the zebrafish blastoderm [presumptive primordial germ cells (PGCs)] (20–22). The change of vasa-transcript distribution in the presumptive PGCs at the sphere stage is thought to be one of the earlier processes in germ-line specification that leads to proliferation of PGCs and zygotic expression of vasa gene (22, 25). In medaka, although an asymmetric segregation of vasa product was not reported for medaka, olvas gene is zygotically expressed in the early blastura (A. Shinomiya and S. Hamaguchi, personal communication) followed by PGC proliferation after gastrulation (Fig. 1 C and D), suggesting the existence of the similar specification process as seen in zebrafish. Nevertheless, the molecular mechanisms that establish functional germ cells in gonads, including the process of migration, specification, establishment, and sexual differentiation, remain unclear.
Here, we report the establishment of medaka transgenic lines with GFP fluorescence controlled by the regulatory regions of the olvas gene (olvas-GFP construct) in the germ cells. Expression analysis of GFP shows that PGCs can be maintained ectopically after gastrulation in terms of olvas expression, suggesting that the contact of PGCs with mesoderm or endoderm is important for the maintenance of PGC characteristics. Thus, the transgenic lines provide a useful model to analyze germ cell migration and development from late gastrulation stage to gonadal maturation. Furthermore, this construct eliminates the labor-intensive screening of candidates for germ-line transmitting founders.
Materials and Methods
Strains.
The original Qurt strain [b/b, gu/gu, r/r, X(lf)/Y(+)] was kindly provided by A. Shima and H. Wada of the University of Tokyo. Brother–sister matings of Qurt were carried out for more than 12 successive generations to produce a more homogeneous genetic background (QurtE). Himedaka (orange–red variety) was purchased from the Yatomi Hatchery (Aichi, Japan).
Construction of Olvas-GFP Vector.
The 3′ region of the olvas gene was amplified from a subcloned DNA containing the 3′ genomic portion of olvas gene using primers (T7, 5′-TAATACGACTCACTATAGGG-3′; VI-8, 5′-AGGAGGTGCCGTCATGGCTGGAG-3′). The resulting fragment was double-digested with PstI/EcoRI and blunt-ended by T4 DNA polymerase. This fragment was cloned into a StuI site of pEGFP vector (the enhanced GFP vector; CLONTECH) and designated as pEGFP-3V.
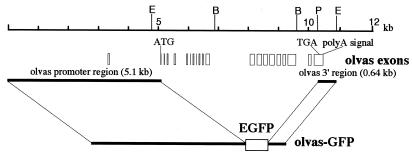
A 5.1-kb genomic fragment encompassing the olvas promoter region was amplified from phage DNA (V5) harboring most of the olvas gene using primers (VP1 M, 5′-CCTCCCAGTCGTCCATATGAATCGTCTGAT-3′; VP3, 5′-AGAGGATCCAATAGAATGAGTAATGGTTCTCTATTTC-3′). The resulting fragment was digested with NdeI, followed by the treatment with T4 DNA polymerase to generate blunt ends and subsequently digested by KpnI. This fragment was cloned into KpnI/blunt-ended NcoI sites of the pEGFP-3V vector to produce the olvas-GFP transgenic vector (see Fig. 2 for details).
Figure 2.
Organization of the olvas gene and structure of the olvas-GFP vector. The upper line indicates the scale that is proportional to the organization of the medaka vasa (olvas) gene shown below. Open boxes denote the exons of the olvas gene. Olvas-GFP is composed of an enhanced GFP gene (EGFP, CLONTECH) flanked by the 5′ region (5.1 kb) including exon 1 and exon 2 and the 3′ region (0.64 kb) of olvas gene. E, EcoRI; B, BamHI; P, PstI.
Microinjection.
Fertilized eggs from himedaka (orange–red variety) and QurtE were collected within 30 min after spawning. Attaching filaments were removed with fine forceps. Microinjection was carried out according to Kinoshita et al. (26). The injected eggs were incubated at 25°C and checked for GFP fluorescence by a fluorescent binocular microscope (Leica or Olympus). Embryos or larva with the cells showing GFP fluorescence in the dorsal region above posterior gut (founder candidates) were separated from those without GFP fluorescence. Both groups were raised to maturity in separate tanks at 25°C.
Selection of the Transgenic Lines.
Sexually mature medaka raised from injected eggs were pair-mated to nontransgenic medaka to obtain F1 embryos. Embryos were screened to identify germ-line-transmitting founders (F0) by both the acquisition of GFP fluorescence and PCR using primers for GFP. The offspring (F1) from each of the identified founders were raised to maturity and crossed with nontransgenic medaka to confirm that the embryos (F2) showing GFP fluorescent PGCs had a segregation ratio of approximately 50%.
Whole-Mount in Situ Hybridization.
Whole-mount in situ hybridization was performed basically according to the methods of Shinomiya et al. (24).
Results
Construction of GFP-Expressing Vector Driven by Medaka Vasa (olvas) Regulatory Regions.
Olvas cDNAs were originally cloned from a medaka ovarian cDNA library (24). Sequence analysis revealed that the 5′ untranslated portion (at least 79 nts) contains exon 1. A single gene often accomplishes tissue- or stage-specific expression by usage of different promoters with alternatively spliced 5′ untranslated exons. As the exon 1 we identified was not translated, alternative exons with different promoters may be present in the olvas gene. The original 5.1-kb putative promoter fragment, therefore, might not contain all of the differentially regulated promoters. To identify potential alternatively spliced exons, we isolated the 5′ portions of olvas cDNAs from testes and embryos by reverse transcription–PCR and determined the nucleotide sequences. All sequences matched either exon 1 or 2 from the ovarian olvas cDNA, confirming that the previously identified promoter(s) upstream of exon 1 function in the testis, embryos, and ovary (data not shown).
We generated a construct (olvas-GFP) by inserting a 5.1-kb putative promoter fragment (including exon 1 and intron 1) and a 0.64-kb 3′ region into the 5′ and 3′ multiple cloning sites of the pEGFP vector, respectively. The olvas-GFP was designed to translate the GFP polypeptide from a putative ATG start codon in exon 2 of the olvas gene. The 3′ region contains the entire 3′ untranslated region of the olvas transcript and its 3′ flanking genomic portion (Fig. 2).
Generation of Stable Transgenic Lines Expressing GFP.
The olvas-GFP construct was microinjected into 1- or 2-cell stage embryos of either himedaka or QurtE. One day after injection, there were no GFP-positive embryos observed. Three to six days after injection, some embryos contained cells showing GFP fluorescence. Some of these cells were found in the dorso-caudal region of the intestine, where the gonad is expected to form. The number of GFP-positive cells varied from one to more than 10. The frequency of germ-line-transmitting founders is summarized in Table 1. GFP fluorescence was observed in the dorsal region of the intestine in 11% (QurtE) and 9% (himedaka) of the injected embryos (Table 1). Positive embryos were raised to sexual maturity and mated to either himedaka or QurtE to confirm germ-line transmission. Of GFP-positive mature candidates, 54% and 16% showed germ-line transmission in himedaka and QurtE, respectively (Table 1). Only two of 39 mature medaka from GFP-negative embryos (5%) were germ-line-transmitting founders (himedaka). These results demonstrate that the olvas-GFP construct can be used as a reliable selective marker to identify the candidates for germ-line-transmitting founders from the injected embryos. Nine himedaka lines and three QurtE lines were established from GFP-positive founders (F0). Chimerism of germ cells in F0 founders ranged from 5% to 67% as judged by the frequency of appearance of GFP-positive germ cells in F1 embryos. All F2 offspring showing GFP fluorescence represented approximately 50% of offspring from an F1-transgenic/nontransgenic cross. These results demonstrate that olvas-GFP is stably transmitted to offspring according to Mendelian genetics and that transgenic lines with olvas-GFP have been successfully established.
Table 1.
Results of microinjection of olvas-GFP construct into two medaka strains
| Strain | No. of injected eggs | No. of hatching | GFP positives | Sexual maturity | No. of germline transmitters | Transmitters from GFP positives |
|---|---|---|---|---|---|---|
| Himedaka | 410 | 145 (35%) | 37 (9%) | 24 | 13 (4%) | 54% |
| QurtE | 470 | 198 (42%) | 50 (11%) | 19 | 3 (1%) | 16% |
Detection of GFP Fluorescence Exclusively in Germ Cells in Transgenic Lines.
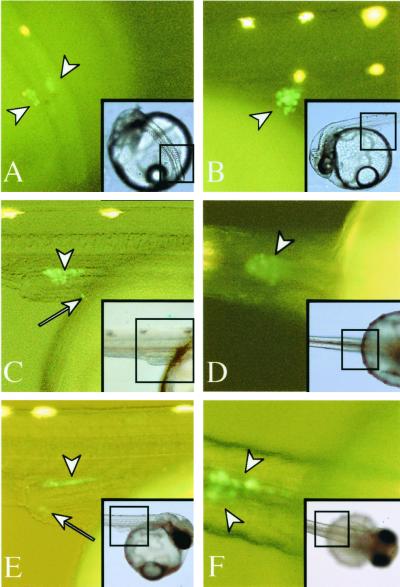
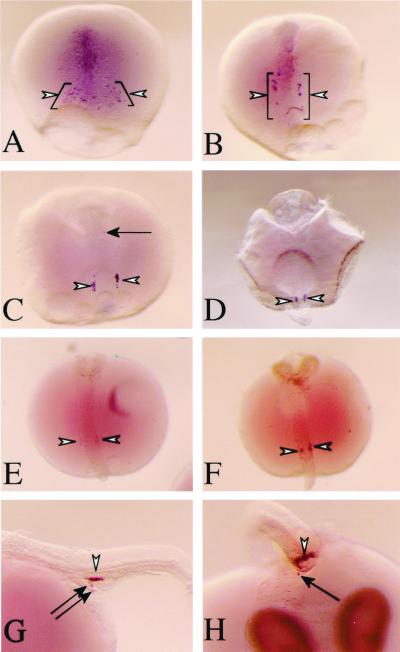
In a very few cases, GFP fluorescence was first weakly detected in cells aligned along the posterior region of the embryonic body (stage 20, 4-somite stage). This fluorescence was undetectable in most embryos (data not shown). By stage 25 (the onset of blood circulation/21-somite stage), GFP fluorescence became intense in all embryos. Cells with GFP fluorescence on the surface of yolk mass were recognized as two clusters lateral to the embryonic body (Fig. 3A). The GFP fluorescent cells migrated dorsally from the surface of yolk mass (Fig. 3B) and formed a single clump at the dorsal region of the intestine by stage 30 (Fig. 3 C and D) (30 somites). This movement was consistent with previous reports on the migration of PGCs with the development of lateral mesoderm (24, 35) (Fig. 1 F and G). By stage 32, the clump began to extend along the anteroposterior axis above the intestine. Lateral views show that this clump is composed of a monolayer of large GFP-positive cells. Within 24 h, the clump moved bilaterally from the midline to form two flattened clusters in the area of the gonads (Fig. 3 E and F). The clusters continued to extend along the anteroposterior axis as somitogenesis proceeded during late embryogenesis and thickened to several layers of germ cells by the day of hatching (Fig. 4 A and B).
Figure 3.
GFP fluorescent germ cells. Each fluorescent image was magnified from the area encompassed by the black square in the corresponding Inset. Insets show an overview of each embryo in the bright field. White arrowheads indicate cells showing GFP fluorescence. GFP fluorescent cells that mismigrated into ectopic locations are indicated by arrows. Yellow spots without arrows are autofluorescence of either leucophores or yolk. Dorsolateral (A) and lateral (B) views of stage 25. Two clumps of GFP-positive cells are clearly seen in the ventrolateral region of the intestine. Lateral (C) and ventral (D) views of stage 30. Both clumps have migrated dorsally and formed a single clump on the intestine. (E) Lateral view of stage 32. The clump situated on the intestine extends along the anteroposterior axis. (F) Ventral view of stage 33. The clump has redivided into bilateral clusters. Most cells located on the intestine are observed by GFP fluorescence, whereas some cells (C and E, arrows) remain on the surface of the yolk or in the lateral region of the intestine.
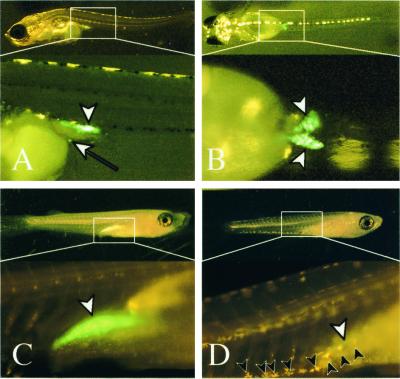
Figure 4.
GFP fluorescent germ cells after hatching. Lateral (A), dorsal (B, Upper), and ventral (B, Lower) views at day of hatching (himedaka). An ectopically located germ cell on the yolk surface cells can be observed by GFP fluorescence (white arrow in A). Female (C) and male (D) 3 weeks after hatching (Qurt E). Leucophores are seen as yellow pigments on genetic males (black arrowheads in D), whereas females (C) have no leucophores. Three weeks after hatching (C and D), sexual dimorphisms of gonads with the fluorescence within the gonads are apparent. The female gonad (white arrowhead in C, ovary) contains numerous oocytes, whereas germ cells have not proliferated well in the testis, which can be seen as a green streak (white arrowhead in D). gonad (white arrowheads).
Leucophores appear at stage 25 only in males of the QurtE strain, allowing discrimination of genetic male and female embryos before gonads form (10). The number of GFP-positive cells increases in genetic females around the day of hatching, whereas the cell number in genetic males appears constant. The difference in cell number becomes more apparent 2 days after hatching (data not shown). Fig. 4 (C and D) shows GFP fluorescence in ovary and testis of QurtE strain 3 weeks after hatching, clearly demonstrating the sexual dimorphisms of gonad visible by GFP fluorescence.
Expression profiles of GFP fluorescence during embryogenesis strongly suggest that GFP is expressed in PGCs during gastrulation and in germ cells during late embryogenesis (compare Figs. 1 and 3). A previous study on the expression of olvas transcripts during medaka embryogenesis demonstrated that maternal olvas transcripts in most cells diminished during early gastrulation. A limited number of cells around the posterior region of the embryonic shield (presumptive PGCs), however, retained or acquired olvas transcripts and moved toward the embryonic body as gastrulation proceeded (Figs. 1 D and E and 5 A and B) (24). We found that the acquisition of GFP fluorescence in PGCs (stage 25) was preceded by the earliest detection of olvas transcripts in the presumptive PGCs. Therefore, we performed whole-mount in situ hybridization using olvas and GFP cDNAs as probes to determine whether GFP transcripts were also present before stage 25. In some cases, we detected weak signals of GFP transcripts around stage 20 (data not shown). In most embryos, however, GFP transcripts were not evident until around stage 25, reflecting the increase in GFP fluorescence in PGCs. After stage 25, the expression profile of GFP transcripts correlated with that of olvas transcripts (Fig. 5 C–F). Cells acquiring GFP fluorescence were typically rounder and larger than other cells, which is characteristic of PGCs. The results of in situ hybridization and the pattern of cells acquiring GFP fluorescence demonstrate that GFP expression occurs specifically in the germ lines.
Figure 5.
Detection of endogenous olvas (A–D, G, and H) and GFP (E and F) transcripts by whole-mount in situ hybridization (white arrowheads). (A) Stage 16, olvas-expressing cells decrease in number and converge on the embryonic shield. Stages 18 (B) and 21 (C), olvas-expressing cells align bilaterally along the trunk. (D) Stage 27, two clumps form at the ventrolateral region of the intestine. Stages 21 (E) and 27 (F) are also shown. Black arrows in C, G (stage 32, lateral view), and H (stage 32, a view from the tail) indicate cells expressing endogenous olvas transcripts in ectopic locations.
Maternal Expression of GFP Fluorescence in Transgenic Lines.
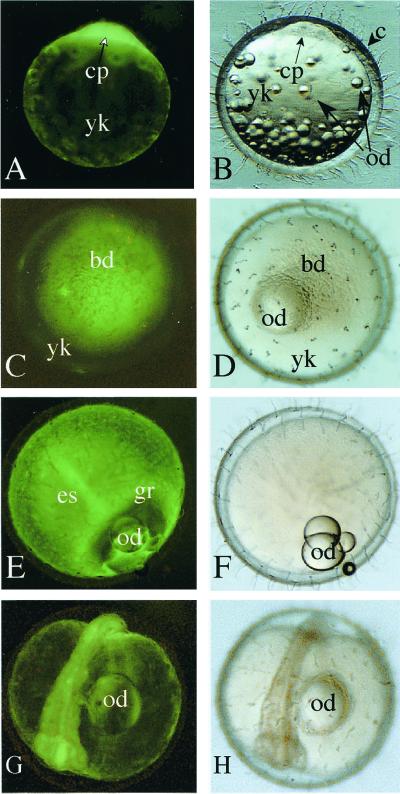
Previously, olvas transcripts were found to be maternally provided during oogenesis (24). Therefore, we mated olvas-GFP transgenic females and nontransgenic males to observe maternal expression patterns of GFP fluorescence. In fertilized eggs, GFP fluorescence in the cytoplasm of a single cell was observed (Fig. 6 A and B). As cell division proceeded to the gastrulation stage, blastoderms contained a mixture of cells with either intense or weak GFP fluorescence (Fig. 6 C and D). After late gastrulation stage, GFP fluorescence became more intense at the germ ring and embryonic shield (Fig. 6 E and F). In contrast to the rapid attenuation of maternally derived olvas transcripts (24), however, the intensity of GFP fluorescence decreases its intensity very slowly during embryogenesis (Fig. 6 G and H). GFP fluorescence persisted until the day of hatching. Some transgenic lines retained very intense GFP fluorescence in the yolk surface cells after hatching. GFP fluorescence in germ cells was more intense than that in somatic cells, allowing identification of germ cells on a weakly fluorescent somatic cells. Whole-mount in situ hybridization using the embryos from transgenic females failed to detect GFP transcripts in somatic cells (data not shown). Therefore, it is likely that GFP fluorescence in the somatic cells is derived from a maternally supplied, stable GFP polypeptide.
Figure 6.
Maternal expression of GFP fluorescence during embryogenesis. (A, C, E, and G) Fluorescent images. (B, D, F, and H) Bright field images. (A and B) One-cell stage, lateral view. The cytoplasm of a single cell shows GFP fluorescence. (C and D) Late morula (stages 11 and 12, animal pole view). (E and F) Gastrulation (stage 16, lateral view). (G and H) Otic vesicle formation stage (stage 21, five somites). bd, blastoderm; c, chorion; cp, cytoplasm; es, embryonic shield; gr, germ ring; od, oil droplet; yk, yolk.
Discussion
We have successfully established transgenic lines that enable us to observe and monitor germ cells by GFP fluorescence in live medaka. We used two strains, himedaka and QurtE, to generate transgenic lines. QurtE transgenic lines (olvas-QurtE) provide great advantages for investigating how germ cells migrate to form sexually dimorphic gonads. QurtE is almost transparent (10), allowing easy observation of GFP fluorescence in the live specimens. Genetic males are easily distinguished from genetic females by the appearance of leucophores before the gonads form during embryogenesis. Twelve generations of brother–sister matings of QurtE provide a more homogenous genetic background.
GFP fluorescence was detected only in the germ line after stage 25 in transgenic lines. F1 transgenics with hemizygous olvas-GFP transgene(s) produced transgenic offspring (F2) with a segregation ratio of approximately 50% after mating to nontransgenic medaka. Analysis of GFP fluorescent cells during embryogenesis and the results of whole-mount in situ hybridization indicate that GFP gene is expressed exclusively in germ cells and that the olvas-GFP construct contains the regulatory region(s) that can drive zygotic and maternal expression of the endogenous olvas gene. These results demonstrate that medaka lines with the olvas-GFP transgene in the germ line were established.
Our results also demonstrate that this construct could be used as a selective marker to isolate the candidates for founders with germ-line integration of any other transgene. As shown in previous studies, injected fish are mosaics for expression and integration of the transgene (9, 27–30). Consequently, all injected embryos (Table 1) needed to be raised to adults for extraction of genomic DNA for PCR or Southern blot hybridization to select F0 germ-line-transmitting founders. GFP fluorescent germ cells, however, are excellent indicators of F0 founders (Table 1). The ease of detecting the fluorescence eliminates the labor-intensive maintenance and analysis of all possible candidates. Selection of F0 founders by this method increases 5–10 times efficiency (Table 1).
Olvas transcripts were first detected in a small number of cells at stage 16 (24), preceding the earliest detection of GFP fluorescence driven by the transgene by approximately 1 day (stage 20). The detection of GFP fluorescence requires a cellular concentration of 100 nM GFP chromophore and posttranscriptional modification to form the GFP chromophore (CLONTECH). These criteria may account for the time lag of 1 day; however, other reports show time lags ranging from 0.7 to 5 h (e.g., refs. 31–33).
Consequently, other explanations may be more relevant. The mouse vasa homolog (Mvh) is not detected in EG cells established from migrating PGCs. Mvh protein, however, can be induced by contact with somatic cell lines derived from the gonad, raising the possibility that the interaction between PGCs and gonadal somatic cells is important for Mvh protein expression (34). This hypothesis is also supported by the fact that Mvh is first detected in germ cells that have entered the urogenital ridge (15). At stage 25, medaka PGCs, which are located in the ventrolateral region of the trunk, have contact with the mesoderm and endodermal layers (35). This contact at stage 20–25 might trigger the dramatic increase in zygotic expression of the olvas gene and olvas-GFP transgene.
The signals of GFP transcripts detected by whole-mount in situ hybridization were much weaker than those of olvas transcripts. Therefore, a third possibility is that the olvas regulatory regions used in the olvas-GFP construct lack some enhancer-like element(s). Similarly, a positional effect of the transgene in the medaka genome may also lower expression levels.
GFP fluorescent cells were often mislocated around the trunk and/or on the surface of the yolk (Fig. 3 C and E). In rare cases, they were found in the head region or in a tail somite. Ectopically located cells began appearing after stage 25, and ectopic GFP fluorescence persisted after hatching for at least a couple of days (Fig. 4A; white arrow). These ectopically located cells also expressed olvas transcripts (Figs. 5 C, G, and H; black arrows), indicating that ectopically located PGCs, as identified by GFP fluorescence, maintain their characteristics with respect to olvas expression.
Assuming that the olvas-GFP construct contains most of the endogenous regulatory elements of the olvas gene, there appears to be two steps to establish PGCs in medaka. First, a small number of cells are specified as PGCs before olvas transcripts are restricted to cells around the embryonic shield at around stage 16 (24). Zygotic expression of the olvas gene is first detected around stage 12 (A. Shinomiya and S. Hamaguchi, personal communication) and its expression is kept very low between stages 16 and 20. Consequently, this process could be a permissive step similar to the mechanisms that establish germ cells in C. elegans and Drosophila. In the second step, olvas transcripts are up-regulated in PGCs at the time when PGCs come in contact with mesodermal cells. The up-regulation is easily detected by GFP fluorescence resulting from olvas-GFP transgene expression and may be associated with the capability of PGCs to locate in ectopic regions. Previous histological observations showed some interaction of PGCs with mesodermal cells during these stages (35). In addition, nuage-like structures of PGCs, which are similar to the electron-dense structures observed in germ cells of many vertebrates, simultaneously change their morphology from strand-like to amorphous fibrous upon contact with mesoderm (21, 25, 36). Thus, the contact of PGCs with the mesoderm seems to be critical to establishing the germ line by triggering increased zygotic expression of olvas, conferring maintenance of PGC characteristics and providing positional cues similar to those reported in zebrafish (37).
In conclusion, we have established transgenic lines that express GFP exclusively in germ cells that can be visualized in a living vertebrate. These lines offer a useful model for analyzing gonadal development. The apparent sexual dimorphism of the gonads indicated by GFP fluorescence can also provide a useful indicator of the effects of environmental substances on gonadal development. Finally, the transgenic vector, olvas-GFP, allows easy identification of germ-line-transmitting founders.
Acknowledgments
We are grateful to Professor H. Bern and Dr. C. Morrey for critical reading of the manuscript, Ms. N. Sumikawa and T. Takagi for fish maintenance, and Ms. C. Sugita for technical assistance. Qurt medaka were generously provided by Drs. A. Shima and H. Wada of the University of Tokyo. We also appreciate Professor K. Morohashi's constant encouragement to generate transgenics. This work was supported in part by Grants-in-Aid for Scientific Research of Priority Area (11236210 to M.T., 11236203 to M.K.) and for research for the future from the Japan Society for the Promotion of Science (JSPS-RFTF 96L100401), by funds from the Bio Design Program from the Ministry of Agriculture, Forestry, and Fisheries, Japan, and by Japan Society for the Promotion of Science Research Fellowships for Young Scientists (10-8720 to D.K.).
Abbreviations
- PGC
primordial germ cell
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Saffman E E, Lasko P. Cell Mol Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wylie C C. Cell. 1999;96:165–170. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- 3.McLaren A. Genes Dev. 1999;13:373–376. doi: 10.1101/gad.13.4.373. [DOI] [PubMed] [Google Scholar]
- 4.Swain A, Lovell-Badge R. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- 5.Wada H, Naruse K, Shimada A, Shima A. Mol Mar Biol Biotechnol. 1995;4:269–274. [PubMed] [Google Scholar]
- 6.Hori H, Watanabe I K. Fish Biol J Medaka. 1995;7:71. [Google Scholar]
- 7.Wittbrodt J, Meyer A, Schartl M. BioEssays. 1998;20:511–515. [Google Scholar]
- 8.Ohtsuka M, Makino S, Yoda K, Wada H, Naruse K, Mitani H, Shima A, Ozato K, Kimura M, Inoko H. Genome Res. 1999;9:1277–1287. doi: 10.1101/gr.9.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozato K, Wakamatsu Y, Inoue K. Mol Mar Biol Biotechnol. 1992;1:346–354. [PubMed] [Google Scholar]
- 10.Wada H, Shimada A, Fukamachi S, Naruse K, Shima A. Zool Sci. 1998;15:123–126. doi: 10.2108/zsj.15.123. [DOI] [PubMed] [Google Scholar]
- 11.Hay B, Ackerman L, Barbel S, Jan L Y, Jan Y N. Cell. 1988;55:577–587. [Google Scholar]
- 12.Rongo C, Lehman R. Trends Genet. 1996;12:102–109. doi: 10.1016/0168-9525(96)81421-1. [DOI] [PubMed] [Google Scholar]
- 13.Castrillon D H, Quade B J, Wang T Y, Quigley C, Crum C P. Proc Natl Acad Sci USA. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. . (First Published August 1, 2000; 10.1073/pnas.160274797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe M, Itoh K, Abe K, Akizawa T, Ikenishi K, Furusawa M. Dev Growth Differ. 1992;34:223–231. doi: 10.1111/j.1440-169X.1992.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Proc Natl Acad Sci USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komiya T, Itoh K, Ikenishi K, Furusawa M. Dev Biol. 1994;162:354–363. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- 17.Forristall C, Pondel M, Chen L, King M L. Development (Cambridge, UK) 1995;121:210–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- 18.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Development (Cambridge, UK) 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 19.Olsen L C, Aasland R, Fjose A. Mech Dev. 1997;66:95–105. doi: 10.1016/s0925-4773(97)00099-3. [DOI] [PubMed] [Google Scholar]
- 20.Yoon C, Kawakami K, Hopkins N. Development (Cambridge, UK) 1997;124:3157–3166. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 21.Braat A K, Zandbergen T, van de Water S, Goos H J T, Zivkovic D. Dev Dyn. 1999;216:153–167. doi: 10.1002/(SICI)1097-0177(199910)216:2<153::AID-DVDY6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Braat A K, van de Water S, Goos H, Bogerd J, Zivkovic D. Mech Dev. 2000;95:271–274. doi: 10.1016/s0925-4773(00)00344-0. [DOI] [PubMed] [Google Scholar]
- 23.Yoshizaki G, Sakatani S, Tominaga H, Takeuchi T. Mol Reprod Dev. 2000;55:364–371. doi: 10.1002/(SICI)1098-2795(200004)55:4<364::AID-MRD2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Shinomiya A, Tanaka M, Kobayashi T, Nagahama Y, Hamaguchi S. Dev Growth Differ. 2000;42:317–326. doi: 10.1046/j.1440-169x.2000.00521.x. [DOI] [PubMed] [Google Scholar]
- 25.Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. J Cell Biol. 2000;149:875–888. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita M, Toyohara H, Sakaguchi M, Inoue K, Yamashita S, Satake M, Wakamatsu Y, Ozato K. Aquaculture. 1996;143:267–276. [Google Scholar]
- 27.Stuart G W, McMurray J V, Westerfield M. Development (Cambridge, UK) 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- 28.Stuart G W, Vielkind J R, McMurray J V, Westerfield M. Development (Cambridge, UK) 1990;109:577–584. doi: 10.1242/dev.109.3.577. [DOI] [PubMed] [Google Scholar]
- 29.Culp P, Nusslein-Volhald C, Nancy N. Proc Natl Acad Sci USA. 1991;88:7953–7957. doi: 10.1073/pnas.88.18.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler C, Vielkind J R, Schartl M. Mol Gen Genet. 1991;226:129–140. doi: 10.1007/BF00273596. [DOI] [PubMed] [Google Scholar]
- 31.Davis I, Girdham C H, O'Farrell P H. Dev Biol. 1995;170:726–729. doi: 10.1006/dbio.1995.1251. [DOI] [PubMed] [Google Scholar]
- 32.Timmons L, Becker J, Barthmaier P, Fyrberg C, Shearn A, Fyrberg E. Dev Genet. 1997;20:338–347. doi: 10.1002/(SICI)1520-6408(1997)20:4<338::AID-DVG5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Hazelrigg T, Liu N, Hong Y, Wang S. Dev Biol. 1998;199:245–249. doi: 10.1006/dbio.1998.8922. [DOI] [PubMed] [Google Scholar]
- 34.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 35.Hamaguchi S. Cell Tissue Res. 1982;227:139–151. doi: 10.1007/BF00206337. [DOI] [PubMed] [Google Scholar]
- 36.Hamaguchi S. Cell Tissue Res. 1985;240:669–673. [Google Scholar]
- 37.Weidinger G, Wolke U, Koprunner M, Klinger M, Raz E. Development (Cambridge, UK) 1999;126:5295–5307. doi: 10.1242/dev.126.23.5295. [DOI] [PubMed] [Google Scholar]