Abstract
Phenethyl isothiocyanate (PEITC) is a constituent of cruciferous vegetables that has demonstrated cancer preventive activity in a number of cancer models including lung, prostate, and breast cancer. Our objective was to examine the effects of the oral administration of PEITC for 7 days on the hepatic expression of genes important in drug metabolism and toxicity in Sprague Dawley rats. The liver is the major site for the metabolism of various xenobiotics and carcinogens, and determining the effects of PEITC on the gene expression of hepatic enzymes may provide insight into mechanisms underlying the cancer preventive activity of PEITC. Using a microarray containing 282 genes, we observed that PEITC significantly up-regulated UDP-glucuronosyltransferase UGT1A6 and strongly down-regulated nicotinamide N-methyltransferase (NNMT). We also confirmed the down-regulation of NNMT by real-time quantitative RT-PCR. NNMT was recently shown to be elevated in the serum of tumor bearing patients with pancreatic, lung, and colorectal cancer, and may be involved in cell migration. Other genes that were significantly up-regulated were the drug metabolizing enzyme cyp2b15, the anti-apoptotic gene bcl2l2, and the stress regulators Gadd45b, Dnajb9, Dnajb5 and Hspb1. Our results indicate new targets that may be important in the mechanisms of the anticancer effects of PEITC. Of particular significance was the down-regulation of NNMT which may represent a new target for the treatment of a variety of cancers.
INTRODUCTION
Brassica vegetables of the family Cruciferae (e.g. cabbage, watercress, and broccoli) and the genus Raphanus (radishes and daikons) contain isothiocyanates and indoles that have been implicated in the reduction of cancer risk [1]. Intake of broccoli and watercress may reduce the risk for lung cancer through the inhibition of CYP450 enzymes that are responsible for the activation of procarcinogens [2]. In addition to CYP450 inhibition, isothiocyanates are inducers of Phase II metabolic enzymes that play a role in the detoxification of activated carcinogens [3]. Induction of apoptosis also represents an important mechanism by which isothiocyanates exert their anticancer effects [4–5].
Phenethyl isothiocyanate (PEITC) is derived from gluconasturtiin, a glucosinolate of PEITC that occurs naturally in cruciferous vegetables [6]. The action of the enzyme myrosinase, present in cruciferous vegetables, converts gluconasturtiin to PEITC once the vegetable is cut or ingested. Significant plasma concentrations of PEITC can be achieved with dietary consumption; ingestion of a 100 gram dose of watercress results in a maximal plasma concentration of 1 µM in humans. [6–7]. PEITC has been shown to induce several Phase II enzymes in vivo. When 1mmol/kg PEITC was administered to F344 rats orally, induction of NAD(P)H:quinone oxidoreductase and glutathione S-transferase activities were observed in the liver. Additionally, sulfotransferase activity was induced in the nasal mucosa, while UDP glucuronosyl transferase activity was observed in the liver [8]. A diet containing 816 mg/kg PEITC administered to young male rats also significantly induced UDP glucuronosyl transferase activity 1.2 fold and NAD(P)H:quinone oxidoreductase and glutathione S-transferase activities 1.8 and 1.5 fold, respectively [9]. To investigate the mechanisms of action of PEITC as chemopreventive/therapeutic agents and identify other drug metabolizing enzymes that may be targets of PEITC action, we studied the effect of PEITC on gene expression of drug metabolizing enzymes in rat livers. We hypothesized that PEITC alters the gene expression of enzymes that are involved in the metabolism of xenobiotic and endogenous carcinogens. To evaluate this hypothesis, we used an oligomicro array, which has 282 genes that represent Phase I and Phase II enzymes, as well as some drug transporters. Additionally these arrays also contain genes that are involved in apoptosis and cell cycle signaling.
MATERIALS AND METHODS
Materials
Female Sprague Dawley rats were obtained from Harlan (Indianapolis, IN). PEITC and corn oil were obtained from Sigma Aldrich (St. Louis, MO). SV RNA isolation kit was purchased from Promega (Madison, WI). Oligo GEArray® Rat Toxicology & Drug Resistance Microarrays and TrueLabeling-AMP 2.0 kit, and related supplies were obtained from SA Biosciences (Frederick, MD).
Methods
Female Sprague Dawley rats (3–4 animals per group), weighing 170–190 grams were housed in a room with controlled lighting and temperature. The animals were fed with a phytoestrogen free diet (Tekland 2016S) obtained from Harlan. Animals had free access to food and water throughout the study. Rats were acclimated for 1 week before the start of the study. The research protocol for the study was approved by the Institutional Animal Care and Use Committee at the University at Buffalo.
PEITC administration
PEITC (150 µmol/kg) in 0.5 mL corn oil was administered to animals once daily for 7 days. Oral gavage was performed using 20 gauge curved stainless steel gavage needles. Control animals received 0.5 mL corn oil once daily for 7 days. At the end of the 7 days, animals were sacrificed by exsanguination of anesthetized animals. The rat livers were washed in phosphate buffered saline (PBS), snap frozen in liquid nitrogen, and stored at −80C until further processing.
Gene array
RNA was isolated from the livers using the SV RNA isolation system from Promega. Biotinylated cRNA was produced using the TrueLabeling-AMP 2.0 kit from SuperArray. The cRNA was then hybridized on the array membrane under specified conditions, as per the manufacturer’s instructions. After washing, the net intensity of each gene spot on the array was determined using a Kodak Image Station 2000MM.
Data analysis and normalization
A number of sequences such as sod1, ldha, GAPDH and BAS2C were available on the array for normalization of the data. The net intensities of all these genes available for normalization of the array genes were compared for the treated and control groups. We determined that among all of these genes, only BAS2C and GAPDH did not change with treatment. BAS2C is an artificial sequence that does not change between treatments and assesses experimental variation, whereas GAPDH is frequently used as a housekeeping gene. The natural log of the net intensity of each spot was normalized by the average of the log of the net intensities of the housekeeping genes, BAS2C and GAPDH. Genes with significant changes in expression were identified using the unpaired Students t-test, with the level of statistical significance set at p<0.05. Significance Analysis of Microarrays (SAM) was also used to analyze the data, which accounts for errors arising from repeated measurements [10]. While using SAM, the delta value was set such that the false discovery rate for each array was minimized. The false detection rate for comparisons ranged from 0–1%. Results from both tests were compared, and genes that were significant by both tests are reported.
RTQ RT-PCR
From the genes that were shown to be up-regulated, we analyzed NNMT in another group of rats by. Real-time quantitative reverse transcriptase-polymerase chain reaction (RTQ RT-PCR) (n=4 per group). RTQ RT-PCR was performed on Alien RNA (Stratagene, La Jolla, CA) (for normalization) and NNMT using the Stratagene Mx4000TM Multiplex Quantitative PCR System (Stratagene, La Jolla, CA). RTQ RT-PCR reactions were carried out by mixing 4 µl of cDNA, 4 µl of 10× PCR buffer, 2µl of deoxynucleoside triphosphate mix (1mM each dATP, dCTP, dGTP, and dTTP), 2 µl each of 10 µM primer, 0.4 µl reference dye rhodamine-X (1/500 dilution, Molecular Probes, Eugene, OR), 0.4 µl SYBR green I (1/750 dilution, Molecular Probes), 0.25ul of 2 U Taq polymerase (Eppendorf, Westbury, NY), and 25.35 µl H2O and amplified for 40 cycles. The thermal cycling conditions consisted of an initial denaturation step at 95°C for 10 min, then 40 cycles denaturing at 95°C for 30 sec, and an annealing temperature of 57°C for 30 sec and polymerization for 30 sec at 72°C.
For the standard curve for NNMT, the PCR product for NNMT was cloned into a pCR® 2.1 TOPO® vector (Invitrogen, Carlsbad, CA) and transformed into One Shot chemically competent Escherichia coli cells (Invitrogen, Carlsbad, CA). Plasmid containing the PCR product sequence was extracted from E. coli cells using a Wizard Plus DNA purification kit (Promega, Madison, WI). The product was resolved by electrophoresis through a 1.2% agarose gel to confirm target size and the presence of a single PCR product. The standard curves using dilution of dSDNA were run in duplicate along with the unknown samples, also in duplicate on the same plate. The reported copy number was estimated from the linear regression of the standard curve on the same plate. Statistical analysis using an unpaired t-test (p<0.05) was performed on the copy numbers, corrected using Ct values from alien RNA runs.
RESULTS
Gene Array
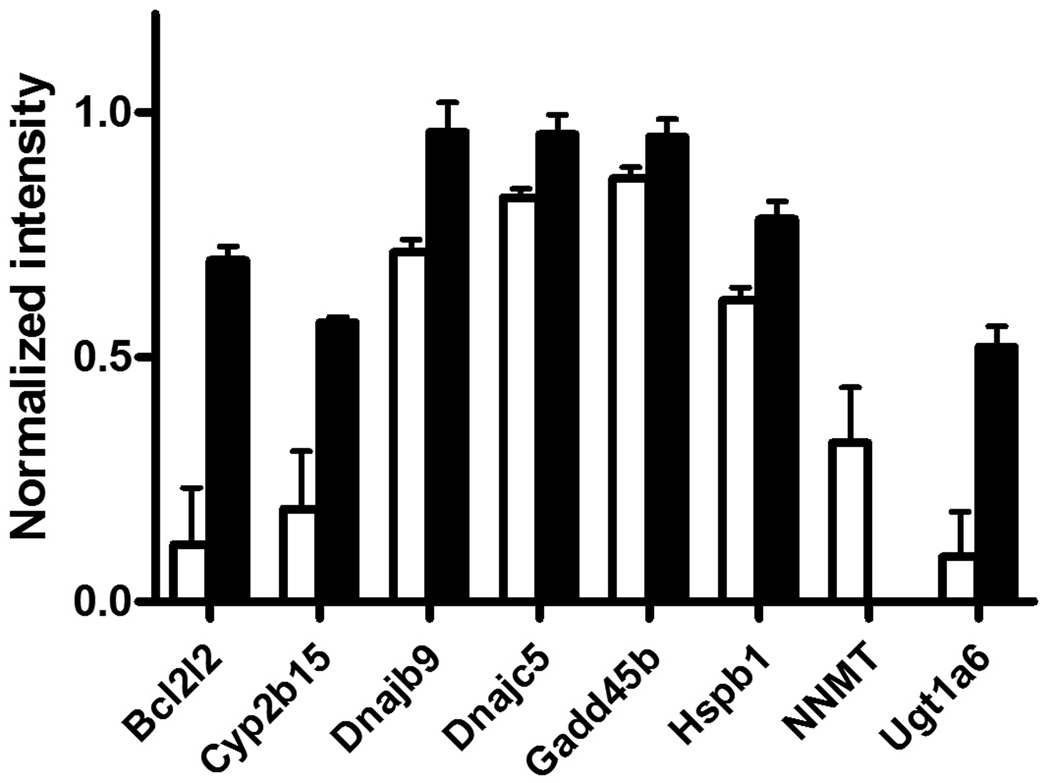
As shown in Table 1, a total of 6 genes, namely, Cyp2b15, ugt1a6, bcl2l2, Dnajb9, Dnajb5, and Hspb1, were significantly up-regulated by PEITC treatment. The variability in the gene array results, as determined in 3–4 animals, is presented in Figure 1. One gene nicotinamide N-methyl transferase (NNMT) was significantly down-regulated. The expression of NNMT in the treated animals was completely abolished in the treated animals, leading to a normalized intensity of zero and therefore, the absence of a closed bar for NNMT in Figure 1.
Table 1.
Effect of PEITC on hepatic gene expression in Sprague Dawley rats (n=3-4 per group)
| Role | Gene Name | Ratio (Treatment/Control) | p value (t-test) |
|---|---|---|---|
| Phase metabolism I | Cyp2b15 | 3.01 | 0.047 |
| Phase metabolism II | Ugt1a6 | 5.67 | 0.015 |
| Apoptosis | Bcl2l2 | 6.01 | 0.014 |
| Stress regulator | Dnajb9 | 1.34 | <0.005 |
| Stress regulator | Dnajc5 | 1.16 | 0.01 |
| Stress regulator | Hspb1 | 1.27 | <0.005 |
| Cancer marker | NNMT | 0.00 | <0.005 |
Figure 1.
Effect of PEITC on hepatic gene expression of significantly altered genes in Sprague Dawley rats, as measured using a gene array. Animals (n=3–4 per group) were administered corn oil (white bar, controls) or 150 µmol/kg PEITC (black bar) for 7 days. Data are expressed as net intensity normalized to GAPDH and BAS2C. Error bars represent standard error. All genes represented were statistically different from controls (p<0.05).
A list of the genes present in the gene array can be found in the Supplemental Information.
RTQ RT-PCR
There were no significant differences in the Ct values for Alien RNA amplification, and values were in a narrow range of 8.0 to 9.9. The standard curve for NNMT was Y= 2.99xLog(X)-3.95 (R2=0.999) with the X-axis representing the dilution factor and the Y-axis representing threshold cycle (Ct) number. We were able to confirm our results in an independent group of rats (n=4 per group) and found a 77% down-regulation in NNMT, which was consistent with our gene array results.
DISCUSSION
Isothiocyanates are compounds derived from cruciferous vegetables, such as broccoli, cabbage, and watercress. Based on epidemiological studies, isothiocyanates are widely recommended as preventive agents, and commercially available in herbal supplements [11–13]. The purpose of this study was to evaluate potential mechanisms underlying the effect of PEITC, a component of watercress, as a cancer preventive agent. The liver is the major site of detoxification of endogenous compounds and xenobiotics, including carcinogens. Therefore, to gain insight on the effects of PEITC on metabolism related genes, we examined the effect of the oral administration of PEITC, for 7 days, on hepatic gene expression. The genes we studied were related to drug transport, phase I and phase II metabolism, apoptosis and cell cycle signaling. Based on our previous pharmacokinetic studies [6–7], we selected a dose of 150 µmol/kg dose which yields plasma concentration values similar to those obtained by oral exposure to PEITC from 100 grams of watercress in humans.
A significant novel finding from our gene array experiment was the down-regulation of NNMT by oral PEITC administration, which was further confirmed by RTQ RT-PCR. Nicotinamide N-methyl transferase is a cytosolic enzyme that catalyzes the N-methylation of nicotinamide and other pyridines to form pyridinium ions [14]. Recently the up-regulation of NNMT was implicated in the pathogenesis of neurological diseases, as well as in the increased incidence of certain cancers [15–16]. NNMT has been shown to be a good diagnostic tumor marker in hepatic, thyroid, lung, and colorectal cancer [17–20]. Although the relationship between the increased serum levels of NNMT in cancer is not fully understood, Destalk et al [21] have hypothesized that an increase in NNMT activity leads to a reduction of pyridine nucleotides available for defense against reactive oxygen species. NNMT gene depletion using siRNA in bladder cancer cells resulted in the reduced cellular proliferation and decreased cell migration, indicating that there is a functional role of NNMT in the progression of cancer [22]. In such a scenario, the down regulation of NNMT may play a role in the anticancer activity of PEITC [21]. It is reported that NNMT is regulated by HNF-1 and STAT3 [15, 23]. STAT3 has been shown to be significantly reduced through the administration of benzyl isothiocyanate (BITC), indicating a potential mechanism underlying the effects of isothiocyanates in the down-regulation of NNMT. [24]. NNMT has been shown to be down-regulated by other dietary compounds, as well, although all studies were performed in cancer cell lines. Curcumin treatment for 14 hours, at a concentration of 50 µM, significantly reduced the expression of NNMT in MBA-MB-460 breast cancer cells [15]. In our laboratory, 1 µM PEITC and 1 µM and 10µM indole-3-carbinol treatments, for 72 hours, significantly down-regulated NNMT in MCF7 breast cancer cells. (Leuko, Telang and Morris, unpublished work).
Another significant finding we observed was an up-regulation of UGT1A6, which is a member of the family of UDP glucuronosyl transferases, a class of phase II enzymes in our gene array. This gene represents a phase II detoxification enzyme that is involved in the attachment of a glucuronide moiety to a compound. Carcinogens, including benzo(a)pyrene, are conjugated by UGT1A6, and increased glucuronidation would result in their increased elimination, and potentially decreased toxicity [25]. Therefore, the up-regulation of this enzyme by PEITC may also represent a cancer preventive mechanism of PEITC. Similar up-regulation of members of the UGT family by isothiocyanates has been previously reported. A 16 hour incubation of liver cancer HepG2 cells with 30 µM sulforaphane caused a 2.8 fold increase in the glucuronidation of bilirubin, suggesting increased activity of UDP glucuronosyl transferase activity and increased elimination of bilirubin [3].
Cyp2b15 was the only Phase I enzyme that was significantly upregulated by PEITC. Little information exists on the modulation of this enzyme by xenobiotics. However, it is reported that in the rat colon, co-incubation with 4mM phenobarbital and 1µM dexamethasone, known constitutive androstane receptor (CAR) and pregnane X receptor (PXR) agonists, caused a 72-fold induction in cyp2b15 mRNA. However, isothiocyanates are known to chemically inhibit a number of CYP450 enzymes, including members of the CYP2B family. While confirmation is needed, it is likely that the increase in gene expression observed in our experiments, may not translate into increased activity as a result of concomitant chemical inhibition.
Another gene that was significantly altered was the anti-apoptotic gene bcl2l2. In our experiments we observed a 6.01 fold up-regulation of bcl2l2. Bcl2l2, also known as Bcl- w, is up-regulated in colon and gastric cancers [26]. In colorectal cancers, Bcl2l2 (bcl-w) was reported to be up-regulated in 92% of adenocarcinomas, compared to 6% adenomas, suggesting a role for the protein in the progression of the cancer [27]. In SNU-16 gastric cancer cells over-expressing Bcl-w, activation of stress activated protein kinase (SAPK) by agents such as etoposide was inhibited, leading to reduced cell death. A 40 µM etoposide treatment (40-hours treatment) produced 20% less cell death in bcl-w overexpressing cells compared to wild type cells, suggesting an anti-apoptotic effect [28]. Overall, these results suggest that the up-regulation of bcl-w by PEITC may represent a deleterious effect in hepatic cells.
Among other genes altered, Gadd45b is involved in hepatic regeneration in mice [29]. Heat shock proteins were also up-regulated in the livers of rats that were treated with PEITC. Heat shock proteins are up-regulated in events of stress, such as increased temperature or cold. Heat shock proteins and related binding proteins play a role in oxidative stress and apoptosis [30]. The consequences of the up-regulation of these genes by PEITC need to be further investigated.
CONCLUSION
PEITC, a dietary agent found in cruciferous vegetables, altered the hepatic gene expression of enzymes that are responsible for the metabolism of carcinogens in vivo. Of the genes altered, the down-regulation of nicotinamide N-methyl transferase was confirmed by RTQ RT-PCR methods. Up-regulation of ugt1a6 represents another significant effect of oral administration of PEITC, as this may contribute to increased elimination of carcinogens. Other significant changes included the up-regulation of drug metabolizing enzymes ugt1a6 and cyp2b15 and the anti-apoptotic gene bcl2l2. The down-regulation of NNMT by PEITC is a novel finding that may represent an important mechanism for the cancer preventive effects of dietary PEITC.
Supplementary Material
ACKNOWLEDGEMENTS
Financial Support from National Institutes of Health grant NIH CA121404. UT was supported by a fellowship from Daiichi Sankyo, Inc.
REFERENCES
- 1.Zhao BSA, S A, Lee EJ, Poh WT, Teh M, Eng P, Wang YT, Tan WC, Yu MC, Lee HP. Dietary isothiocyanates, glutathione S-transferase -M1, -T1 polymorphisms and lung cancer risk among Chinese women in Singapore. Cancer Epidemiol Biomarkers Prevention. 2001;10:1063–1067. [PubMed] [Google Scholar]
- 2.Hecht SS, Carmella SG, Murphy SE. Effects of watercress consumption on urinary metabolites of nicotine in smokers. Cancer Epidemiol Biomarkers Prev. 1999;8:907–913. [PubMed] [Google Scholar]
- 3.Basten GP, Bao Y, Williamson G. Sulforaphane and its glutathione conjugate but not sulforaphane nitrile induce UDP-glucuronosyl transferase (UGT1A1) and glutathione transferase (GSTA1) in cultured cells. Carcinogenesis. 2002;23:1399–1404. doi: 10.1093/carcin/23.8.1399. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Tang L, Gonzalez V. Selected isothiocyanates rapidly induce growth inhibition of cancer cells. Molecular cancer therapeutics. 2003;2:1045–1052. [PubMed] [Google Scholar]
- 5.Izzotti A, Larghero P, Cartiglia C, Longobardi M, et al. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate, and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010 doi: 10.1093/carcin/bgq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Y, Morris ME. Determination of phenethyl isothiocyanate in human plasma and urine by ammonia derivatization and liquid chromatography-tandem mass spectrometry. Analytical biochemistry. 2003;323:39–47. doi: 10.1016/j.ab.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Ji Y, Kuo Y, Morris ME. Pharmacokinetics of dietary phenethyl isothiocyanate in rats. Pharmaceutical research. 2005;22:1658–1666. doi: 10.1007/s11095-005-7097-z. [DOI] [PubMed] [Google Scholar]
- 8.Guo Z, Smith TJ, Wang E, Sadrieh N, et al. Effects of phenethyl isothiocyanate, a carcinogenesis inhibitor, on xenobiotic-metabolizing enzymes and nitrosamine metabolism in rats. Carcinogenesis. 1992;13:2205–2210. doi: 10.1093/carcin/13.12.2205. [DOI] [PubMed] [Google Scholar]
- 9.Dingley KH, Ubick EA, Chiarappa-Zucca ML, Nowell S, et al. Effect of dietary constituents with chemopreventive potential on adduct formation of a low dose of the heterocyclic amines PhIP and IQ and phase II hepatic enzymes. Nutrition and cancer. 2003;46:212–221. doi: 10.1207/S15327914NC4602_15. [DOI] [PubMed] [Google Scholar]
- 10.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambrosone CB, M S, Freudenheim JL, Marshall JR, Zhang Y, Shields PG. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. Journal of Nutrition. 2004;134:1134–1138. doi: 10.1093/jn/134.5.1134. [DOI] [PubMed] [Google Scholar]
- 12.Fowke JH, C F, Jin F, Qi D, Cai Q, Conaway C, Cheng JR, Shu XO, Gao YT, Zheng W. Urinary isothiocyanate levels, brassica, and human breast cancer. Cancer Research. 2003;63:3980–3986. [PubMed] [Google Scholar]
- 13.Giovannucci E, Rimm EB, Liu Y, Stampfer MJ, Willett WC. A prospective study of cruciferous vegetables and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:1403–1409. [PubMed] [Google Scholar]
- 14.Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem. 1994;269:14835–14840. [PubMed] [Google Scholar]
- 15.Tomida M, Ohtake H, Yokota T, Kobayashi Y, Kurosumi M. Stat3 up-regulates expression of nicotinamide N-methyltransferase in human cancer cells. Journal of cancer research and clinical oncology. 2008;134:551–559. doi: 10.1007/s00432-007-0318-6. [DOI] [PubMed] [Google Scholar]
- 16.Aoyama K, Matsubara K, Kondo M, Murakawa Y, et al. Nicotinamide-N-methyltransferase is higher in the lumbar cerebrospinal fluid of patients with Parkinson's disease. Neurosci Lett. 2001;298:78–80. doi: 10.1016/s0304-3940(00)01723-7. [DOI] [PubMed] [Google Scholar]
- 17.Tomida M, Mikami I, Takeuchi S, Nishimura H, Akiyama H. Serum levels of nicotinamide N-methyltransferase in patients with lung cancer. Journal of cancer research and clinical oncology. 2009 doi: 10.1007/s00432-009-0563-y. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Hong SJ, Lim EK, Yu YS, et al. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J Exp Clin Cancer Res. 2009;28:20. doi: 10.1186/1756-9966-28-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sartini D, Santarelli A, Rossi V, Goteri G, et al. Nicotinamide N-methyltransferase upregulation inversely correlates with lymph node metastasis in oral squamous cell carcinoma. Mol Med. 2007;13:415–421. doi: 10.2119/2007-00035.Sartini. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roessler M, Rollinger W, Palme S, Hagmann ML, et al. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res. 2005;11:6550–6557. doi: 10.1158/1078-0432.CCR-05-0983. [DOI] [PubMed] [Google Scholar]
- 21.Dostalek M, Hardy KD, Milne GL, Morrow JD, et al. Development of oxidative stress by cytochrome P450 induction in rodents is selective for barbiturates and related to loss of pyridine nucleotide-dependent protective systems. J Biol Chem. 2008;283:17147–17157. doi: 10.1074/jbc.M802447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27:6679–6689. doi: 10.1038/onc.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Capezzone M, Xu X, Hershman JM. Activation of nicotinamide N-methyltransferase gene promoter by hepatocyte nuclear factor-1beta in human papillary thyroid cancer cells. Mol Endocrinol. 2005;19:527–539. doi: 10.1210/me.2004-0215. [DOI] [PubMed] [Google Scholar]
- 24.Sahu RP, Srivastava SK. The role of STAT-3 in the induction of apoptosis in pancreatic cancer cells by benzyl isothiocyanate. Journal of the National Cancer Institute. 2009;101:176–193. doi: 10.1093/jnci/djn470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock KW, Kohle C. UDP-glucuronosyltransferase 1A6: structural, functional, and regulatory aspects. Methods in enzymology. 2005;400:57–75. doi: 10.1016/S0076-6879(05)00004-2. [DOI] [PubMed] [Google Scholar]
- 26.Bae IH, Park MJ, Yoon SH, Kang SW, et al. Bcl-w promotes gastric cancer cell invasion by inducing matrix metalloproteinase-2 expression via phosphoinositide 3-kinase, Akt, and Sp1. Cancer Res. 2006;66:4991–4995. doi: 10.1158/0008-5472.CAN-05-4254. [DOI] [PubMed] [Google Scholar]
- 27.Wilson JW, Nostro MC, Balzi M, Faraoni P, et al. Bcl-w expression in colorectal adenocarcinoma. Br J Cancer. 2000;82:178–185. doi: 10.1054/bjoc.1999.0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HW, Lee SS, Lee SJ, Um HD. Bcl-w is expressed in a majority of infiltrative gastric adenocarcinomas and suppresses the cancer cell death by blocking stress-activated protein kinase/c-Jun NH2-terminal kinase activation. Cancer Res. 2003;63:1093–1100. [PubMed] [Google Scholar]
- 29.Papa S, Zazzeroni F, Fu YX, Bubici C, et al. Gadd45beta promotes hepatocyte survival during liver regeneration in mice by modulating JNK signaling. The Journal of clinical investigation. 2008;118:1911–1923. doi: 10.1172/JCI33913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beere HM. "The stress of dying": the role of heat shock proteins in the regulation of apoptosis. J Cell Sci. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.