Abstract
The topology of nuclear receptor (NR) signaling is captured in a systems biological graphical notation. This enables us to identify a number of ‘design’ aspects of the topology of these networks that might appear unnecessarily complex or even functionally paradoxical. In realistic kinetic models of increasing complexity, calculations show how these features correspond to potentially important design principles, e.g.: (i) cytosolic ‘nuclear’ receptor may shuttle signal molecules to the nucleus, (ii) the active export of NRs may ensure that there is sufficient receptor protein to capture ligand at the cytoplasmic membrane, (iii) a three conveyor belts design dissipating GTP-free energy, greatly aids response, (iv) the active export of importins may prevent sequestration of NRs by importins in the nucleus and (v) the unspecific nature of the nuclear pore may ensure signal-flux robustness. In addition, the models developed are suitable for implementation in specific cases of NR-mediated signaling, to predict individual receptor functions and differential sensitivity toward physiological and pharmacological ligands.
Keywords: biochemical network, kinetic model, nuclear receptor, signaling, systems biology
Introduction
The 48 members of the human nuclear receptor (NR) family have been implicated in a diverse range of regulatory functions, such as in development, cellular growth, inflammation and metabolism (El-Sankary et al, 2001, 2002; Phillips et al, 2003; Aouabdi et al, 2006; Carlberg and Dunlop, 2006; Ebert et al, 2006; Cutress et al, 2008). NRs sense lipophilic ligands, with either broad affinity, e.g., fatty acids are sensed by peroxisome proliferator-activated receptors (PPARs), or with high affinity. The latter ligands include (i) steroid hormones, e.g., estradiol (sensed by estrogen receptor (ER)-α and -β), progesterone (progesterone receptor), testosterone (androgen receptor (AR)), cortisol (glucocorticoid receptor (GR)) and aldosterol (mineralocorticoid receptor (MR)), (ii) thyroid hormone (thyroid hormone receptor-α and -β), (iii) retinoic acid (retinoic acid receptor-α, -β and -γ) and (iv) the seco-steroid 1α,25-dihydroxyvitamin D3 (vitamin D receptor (VDR)) (Carlberg and Dunlop, 2006; Ebert et al, 2006; Cutress et al, 2008).
Whereas most other cellular receptors are located in the plasma membrane, NRs derive their family name from the early and paradoxical observation that they are generally located in the nucleus, despite responding to extracellular signals (Fanestil and Edelman, 1966). Hydrophobic, extracellular signal molecules serving as NR ligands are classically envisioned to diffuse through the plasma membrane, the cytosol and gain entry to the nucleus (Gardner, 1975). There they are able to bind to the corresponding NRs, which are already bound to their specific DNA binding site, referred to as response element (RE). We shall refer to this mechanism as the ‘classical’ design of nuclear receptor signaling.
Many studies have since suggested that this is much too simple a picture (reviewed in Cutress et al, 2008; Cao et al, 2009; Levin, 2009a; Bunce and Campbell, 2010). Ligand distribution appears dynamic with some NRs found predominantly in the nucleus (e.g., PXR and PPAR), whereas others are located either in both compartments (e.g., VDR and MR) or mostly in the cytoplasm (e.g., GR and AR). NRs may also reside in cellular organelles and associate with membranes (Levin, 2009b). Moreover, if a given NR is predominantly located in the nucleus, it is not exclusively so and may be relocated outside the nucleus. Recent studies show that ‘nuclear’ NRs, such as PPARs, are shuttled actively between the nucleus and the cytoplasm (Von Knethen et al, 2010). Ligand addition changes receptor location dynamically. For example, the addition of ligand causes a complete shift of GR and AR from the cytoplasm to the nucleus (Pratt et al, 1989; Liu and DeFranco, 2000; Kumar et al, 2004, 2006; Tanaka et al, 2005; Heitzer et al, 2007; Prüfer and Boudreaux, 2007; Ricketson et al, 2007; Cutress et al, 2008).
We have considered a number of key questions of NR function. Is it functionally important for the NR also to be located outside the nucleus? Or is it sufficient for a ligand to diffuse into the nucleus? Subsequent to this question we also considered whether the extranuclear NR location is an inadvertent escape, or leakage, of receptor through the nuclear membrane and detracts from the functionality of the nuclear component? Alternatively, dynamically controlled shuttling out of the nucleus may be functional and represent an aspect of complexity that is evolutionarily advantageous? Finally, do all NRs function in actually the same way with minor variations, or do genuinely distinct mechanisms of signaling exist?
In order to address these issues, we have first asked how complex NR signaling necessarily is by determining a common denominator of the topology of these signaling networks. This reveals a number of additional complexities, as well as properties that at first sight would seem to make these networks ineffective. We subsequently calculate how resolutions of these paradoxes may correspond to newly recognized network design principles that enhance functionalities.
Results
Canonical network topology of endocrine NR signaling
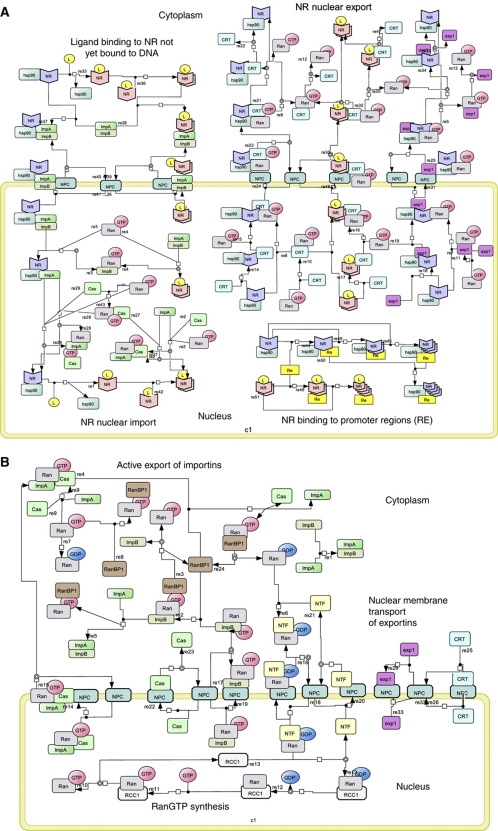
Using the SBML-compliant graphical network notation (SBGN) (Kitano et al, 2005), we constructed a ‘canonical’ endocrine NR signaling network that is rooted in the present literature (Figure 1). The network accounts for NR activation, importin-α and -β binding, nuclear pore complex (NPC)-mediated import, recycling of importins, NR binding to target promoter sequences, exportin-mediated nuclear export of the NR, exportin cycling and free energy-driven Ran recycling. When bound to its ligand, the NR induces transactivation and transrepression of its cognate REs. This topology is ostensibly more complex than that of a hydrophobic signal molecule merely crossing the plasma membrane, moving to the nucleus and binding and activating the NR complexed with its RE. To address to what extent this extra complexity is functional, we undertook the following analysis to reveal that the simplest design does not accurately recapitulate experimentally observed function, and indeed most aspects of this topological complexity serve sophisticated biological function.
Figure 1.
(A, B) Network diagram for GR signaling. The NR signaling network is shown in terms of seven modules, in standard SBGN (Kitano et al, 2005). (A) (i) Ligand binding to NR not yet bound to DNA. Core-NR (indicated by the blue wedge shape) has its NLS1 masked by Hsp90, while its NLS2 is accessible. Both its NES1 and its NES2 are exposed. In this state the affinity of NR for DNA is low. Upon binding its ligand, the conformation of the NR is changed, the NR dimerizes, the NLS1 becomes unmasked, the NES2 is masked and the affinity of the NR to DNA is thereby increased. The consequent binding to the DNA (and the engagement of NR in active import into the nucleus, see below) shifts NR from cytoplasm to nucleus when ligand is added (Drouin et al, 1992). (ii) Reversible NR binding to REs: both liganded and core-NRs bind to REs and form tetramers. The DNA binding affinity for NRL is higher than that for core-NR (Garlatti et al, 1994). (iii) NR nuclear import: Both core and NRL bind to importin-α. Core-NR binds to importin-α due to the NLS2, but the NLS1 is occluded by Hsp90 protein. If the NR is liganded, both its NLS1 and its NLS2 are available. This provides higher affinity to importin-α (Pemberton and Paschal, 2005). Binding of the NR alters the conformation of importin-α such that its N terminus becomes accessible to importin-β, which, in turn, can interact with the nucleoporins in the NPC. The NPC allows the importins–cargo complex to pass the nuclear envelope (Sharova, 2002; Tran and Wente, 2006) The importin-β–importin-α–cargo complex binds RanGTP, which is exclusive to the nuclear compartment. Importin-β–importin-α–cargo–RanGTP complex dissociates into an importin-α–cargo and an importin-β–RanGTP moiety. Importin-α–cargo complex associates with RanGTP and CAS, which allows the cargo NR plus hormone to dissociate from the complex. RanGTP favors dissociation of the complexes and hence pushes the balance to dissociation of the cargo complexes in the nucleus, where association may be favored in the absence of RanGTP (i.e., in the cytosol). (iv) NR nuclear export: NR binds to exportin1 (CRM1) via NES1 or to calrecetin (CRT) via NES2. Both exportins bind to RanGTP and the resulting cargo–exportins–RanGTP complex passes through the NPC to the cytoplasm, where free RanGTP is hydrolyzed to RanGDP by RanGAP with the assistance of RanBP1. The lower level of RanGTP in the cytosol, as compared with the nucleus, favors dissociation of the complex into cargo, exportins and RanGTP. (B) (v) Active export of importins: both importin-α–RanGTP–Cas and importin-β–RanGTP complexes can move between nucleus and cytoplasm via the NPC. GAP associates with the NPC on the cytoplasmic side of the nuclear membrane, provoking the hydrolysis of RanGTP to RanGDP in both the complexes (Pemberton and Paschal, 2005), which then dissociate. GTP hydrolysis is assisted by the RanBP1 protein and coupled to the dissociation of RanGDP molecules from importins. (vi) Nuclear membrane transport of exportins: Exp1, CRT and Cas diffuse across the nuclear membrane through the NPC (Pemberton and Paschal, 2005). (vii) RanGTP synthesis: RanGDP is returned into the nucleus in a complex with transport factor NTF2 (Poon and Jans, 2005). The pool of RanGTP is restored when nuclear GEF (containing RCC1 protein and associated with chromatin (Macara, 2001)) replaces GDP with GTP in the Ran molecule (Pemberton and Paschal, 2005). The function of GEF is to provide a 4-step reversible reaction: RCC1 binds RanGDP, GDP is released from the complex, GTP binds to Ran-RCC1 and finally RCC1 splits from Ran-GTP (Riddick and Macara, 2007).
In principle, all reactions depicted in Figure 1 could run in the opposite, and functionally counterproductive, direction. Therefore, we have considered the underlying thermodynamics. NR-shuttling processes are driven by the Gibbs free energy stored in the non-equilibrium ratio of (GTP)/((GDP)·(phosphate)). This ratio is probably the same in the cytoplasmic and nuclear compartments, because of diffusivity of GTP, GDP, phosphate and Mg2+. The unequal distribution of Ran-GTPase-activating protein (GAP) and Ran-guanine nucleotide exchange factor (GEF) activities across the nuclear membrane may sustain higher concentrations of RanGTP (or rather: a higher ratio of RanGTP/RanGDP) in the nucleus than in the cytoplasm. This ratio consequently establishes an exportin and importin gradient that, in turn, drives the nucleo–cytoplasmic distribution of cargo away from thermodynamic equilibrium. None of these three free-energy transduction processes will take place at 100% efficiency, as they all occur out of thermodynamic equilibrium (Westerhoff, 1985). This non-equilibrium pumping system causes importins and exportins to shuttle actively and repeatedly between cytoplasm and nucleus, to induce NR shuttling, and thus to maintain an NR gradient. Analysis of the detailed network (Figure 1) suggests that the effectiveness of nucleo–cytoplasmic shuttling might also depend on a number of additional thermokinetic aspects, including (i) affinity competitions of transport and cargo proteins, (ii) the quality and state of the NLSs and NESs of the cargo proteins, (iii) the saturation of the transport machinery with other cargo and (iv) the non-equilibrium efficiency of the entire process. Consequently, understanding the impact of nucleo–cytoplasmic shuttling on NR signaling is not merely a matter of assessing the quality of the NLSs and NESs of the cargo signaling protein of interest. Rather, the impact depends on the network as a whole, which in turn reflects both the kinetic parameter values of key molecules in the network and network topology. This study focuses on the latter, topological design aspects of the network represented in Figure 1 and in particular on key aspects that we consider to be non-obvious and at times even paradoxical.
Endocrine NR signaling: complex and paradoxical aspects
Figure 1 contains the ‘common denominator’ of the NR signaling networks. This common denominator is surprisingly complex when compared with the classical paradigm of NR signaling. By ‘classical’ we refer to the concept that a given NR resides in the nucleus, attached to a RE waiting for the ligand to bind (design 1 in Figure 2). Though ‘classical’ in our terminology, this concept is not current anymore: more dynamic pictures of this NR signaling abound. Here, however, we shall use this mechanism as the backdrop against which to compare other subsequently proposed mechanisms, including the most current ones. We identify eight aspects of the topology of Figure 1 that are absent from the classical design. Some of these are paradoxical in the sense that on first value, they could be taken to impede rather than enhance signaling, specifically:
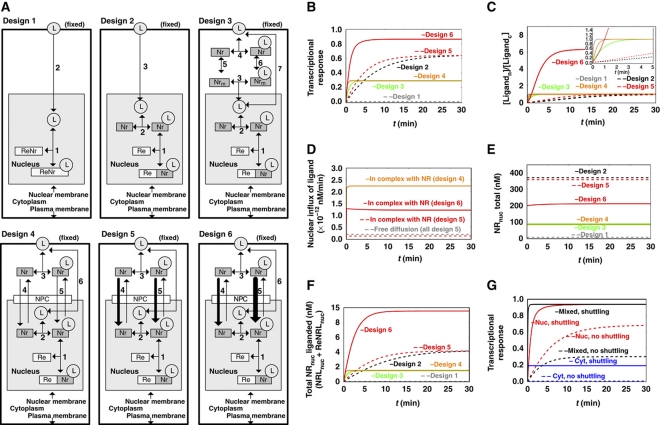
Figure 2.
The expected performance of six different network designs for NR signaling. (A) The six alternative network designs studied: Design 1: Passive diffusion of ligand, which binds directly to the DNA-bound NR. Design 2: NR functioning as NR only, with passive cytoplasmic diffusion of ligand, the NR being in vast excess over RE but confined to the nucleus and helping ligand associate with the RE. Design 3: NR functioning both as NR and as cytosolic shuttling protein. Design 4: NR functioning both as NR and as shuttle from plasma membrane all the way to the DNA, with free NR also shuttling between nucleus and cytoplasm, picking up ligand near the cellular membrane. Designs 5 and 6: active import of the NR, without preferences for import between liganded and core-NRs (design 5), or with core-NR having lower import into the nucleus than NRL (design 6). (B) Transcriptional response to a sudden increase in extracellular ligand (hormone), for the six network designs of (A). The transcriptional response is taken to equal the ratio ReNrL/Retotal, i.e., the fraction of REs attaching ligand-bound NR. The ligand concentration was increased from 0 to 0.005 nM and maintained constant at the latter level. The observation that design 6 is higher than all other designs at long times is robust for parameter changes up to a factor of 3. (C) Time courses of the concentration ratio of nuclear over cytoplasmic ligand for the six network designs. The insert enlarges the early events. (D) Time courses of the nuclear influxes of ligand for the six network designs. Nuclear influx of ligand by free diffusion is equal in all models (gray dashed line). In addition, ligand is imported complexed with the NR (designs 4–6). (E) Time courses of the concentration of the total NR in the nucleus for the six network designs. (F) Time courses of concentration of total ligand-bound receptor in the nucleus (NRLnuc + ReNRLnuc) for the six network designs. (G) Transcriptional responses (taken to equal the ratio ReNrL/Retotal) for different initial localizations of NR. Different initial localizations of the NR were achieved by adjusting the import/export activity ratios of core-NR (nuclear localization—red line; equally distributed between nucleus and cytoplasm—black line; cytoplasmic localization—blue line). The transcriptional response is shown for both high shuttling (solid line) and almost no shuttling (dashed line). Calculations were carried out for a model built according to design 6 (ligand-bound NR having preference for nuclear import). Rate equations and kinetic parameters are given in Supplementary information: Supplementary Table 1 for design 1; Supplementary Table 2 for design 2; Supplementary Table 3 for design 3; Supplementary Table 4 for design 4; Supplementary Table 5 for design 5 and Supplementary Table 6 for design 6. Supplementary Figure 4S shows simulation results for all species in all models. L, ligand (nuclear hormone, e.g., cortisol); Nr, NR (e.g., GR); Re, RE (for model A, NR bound with Re is denoted as ReNr ); NPC, nuclear pore complex. Models are available in JWS Online and can be simulated in its web browser: http://jjj.biochem.sun.ac.za; http://jjj.bio.vu.nl; http://jjj.mib.ac.uk (Snoep and Olivier, 2002; Olivier and Snoep, 2004). Models can be found via the ‘author search’, ‘kolodkin’. Models can be also accessed directly via: http://jjj.bio.vu.nl/webMathematica/Examples/run.jsp?modelName=kolodkinX, with X ranging from 1 to 6 respectively for design 1 to design 6 (at each of the servers listed above). Note: Figure 2D cannot be reproduced with online simulations, which allow determining of the net flux of ligand (as a sum of import and export fluxes) but not the time course of import flux alone. Please contact the authors for more details. Figure 2G can be reproduced by populating design 6 model with parameters from Supplementary Table 6.
(1) Not all NR molecules are associated with the DNA, potentially limiting the extent of transcription activation. (2) Not all NR molecules reside in the nucleus, which could similarly limit function. (3) NR can move across the nuclear membrane, further leading to possible escape of NR from the proximity of the DNA. (4) Active transport and the corresponding hydrolysis of GTP would waste free energy that is not wasted in the classical design. (5) Why do both an import system (using importins) and an export system (using exportins) exist for NRs? Export systems may lead to redundancy? (6) The possible shuttling of NR from nucleus to cytosol and back could constitute a ‘futile cycle’ wasting even more free energy. (7) Although importins aid the import of NR, it is the export, not the import, of importins that is coupled to GTP hydrolysis. (8) There is a single transport system in the nuclear membrane for all NRs, suggesting fragility due to interferences between different NR and other signaling pathways.
We have examined whether or not the classical design by itself is satisfactory, that is, it is able to recapitulate biological data and function. Subsequently, we investigated which of the eight aspects of topology individually contributes most to the system. We shall now leave the full complexity of Figure 1 behind us and focus on aspects of this complexity, one by one.
The classical, simple interpretation of endocrine NR signaling would not work
In the classical, and simplest, mechanism for endocrine NR-mediated signaling (design 1 in Figure 2A), the dynamics of the transcriptional response were simulated using realistic parameter values. For this and for all other designs, the addition of ligand was modeled as the increase of its fixed concentration in the outer cellular membrane from 0 to 0.005 nM, in which the concentrations are quantified as the aqueous concentrations both extracellular and in the cytosol immediately adjacent to the plasma membrane; we shall assume a rapid equilibration of the ligand between these aqueous phases and the plasma membrane phase. The concentration of ligand in the nucleus (Ln) was treated as aqueous only; i.e., in terms of the total number of molecules divided by the volume of the nucleus. When considering a realistic NR ligand-binding constant in the order of 1 nM, as, e.g., for binding of the cortisol analog dexamethasone to GR (Marissal-Arvy et al, 1999), there was no significant transcriptional response to the exposure of the model to the 0.005 nM of ligand. This is contrary to what might have been expected for this classical model of NR signaling. Indeed, this concentration of ligand led only to an extremely low saturation of the NR with ligand. Only 1 of every 200 NR molecules would have ligand bound and because the number of NR proteins is far lower to the number of potential REs, far <1 out of every 200 REs could be activated. Clearly, this mechanism would not suffice for signaling the presence of ligand at low but realistic concentrations.
The advantage of non-DNA-bound NR protein
We speculated that having excess activated NR proteins contributing to RE activation might result in a stronger transcriptional response. This would deviate from the classical model (and be closer to current views) in that more NR is then not bound to the DNA (Figure 2A, design 2). For some NRs this seems realistic, as there are ∼1000 active REs per cell (e.g., for GR REs (de Kloet et al, 2000; Reddy et al, 2009) and for ER REs (Lin et al, 2007)) versus ∼100 000 NR molecules per cell (e.g., for GR (Nordeen et al, 1989; Van Steensel et al, 1995)). For other NRs the number of NR molecules in the cell may be approximately equal to the number of active REs. Because of the much larger distribution volume for ligand molecules outside the cells, which we assume to be at equilibrium with the free ligand in the plasma membrane, we took the concentration of the latter as fixed (i.e., as unaffected by binding of ligand to NR protein) at either 0 or 0.005 nM (aqueous). Indeed, allowing NR to diffuse freely through the nucleus, a higher concentration of ligand-bound NR in the nucleus was calculated than for the model of the previous section, which had all NR bound to the DNA, at a concentration even higher than that of RE. This occurred even though only a small fraction of NR proteins would have ligand bound. Indeed, the transcriptional response now reached ∼60% of the maximal response (design 2, Figure 2B).
What cytosolic NR could contribute?
Our simulations show that although the design with excess NR led to an increased steady-state response, the response was slow (∼25 min; Figure 2C, design 2), because ligand flowing into the nucleus was sequestered by binding to the excess NR, and initially the flux of ligand across the cytosol was unable to keep up with the demand leading to limitation of activation.
The lipophilic nature of NR ligands causes them to accumulate in membranes due to partition coefficients of well over a thousand (∼2500 in the case of cortisol; Oren et al, 2004). Although this facilitates passage across the plasma membrane, it should make traversing the cytosol to reach the nucleus more difficult. As the distance between plasma membrane and nuclear membrane vastly exceeds the diameter of the plasma membrane, this issue may not be trivial. We propose that in addition to their DNA binding and transcription activation role, NRs serve as ferry boats for NR ligands; ligand binds to the lipophilic NR ligand-binding pocket and is transported from one cellular location to another, which is similar to the transport of fatty acids by fatty acid binding proteins (Weisiger, 2002). We calculated whether, and how, cytosolic shuttling of the NR may realistically enhance signaling by picking up ligand from the cytoplasm near the plasma membrane and releasing it near the nucleus, using realistic kinetic parameters and ditto cytoplasmic and nuclear volumes.
Our design 3 (Figure 2A) had the NR equally distributed between the nucleus and the cytoplasm, without NR being able to traverse the nucleocytoplasmic membrane. Ligand was ferried through the cytosol by the cytoplasmic fraction of the NR, then entered the nucleus and associated with the nuclear NR fraction to initiate a transcriptional response. We modeled the diffusion as a single movement from close to the plasma membrane, where the NR was present at concentration Nrc, to a position close to the nuclear membrane, where the NR had a concentration Nrm. We set the diffusion coefficient for NR equal to 1 × 10−12 m2 s−1. This value was used earlier in the models addressing protein diffusion (Kholodenko et al, 2000a), where the diffusion coefficient of model protein was taken in the order of magnitude of experimentally measured diffusion coefficients of various proteins, e.g., GFP (Dayel et al, 1999). The diffusion coefficient for cortisol was taken to be 6-times higher than the diffusion coefficient of the NR, as estimated from the Stokes–Einstein equation. Although the NR diffuses more slowly than the far smaller ligand molecule, the constant high ligand concentration in the cellular membrane, combined with the concentration of the NR being higher than the concentration of free ligand in the cytosol, allowed for higher concentration of the ligand–NR complex, as compared with the concentration of the free ligand. Consequently, increased fluxes of ligand molecules were carried from the plasma membrane to the nuclear membrane by diffusing NR complexes, and the steady state was reached some 25-times faster than that in design 2 (Figure 2B and C; compare design 3 to design 2).
What shuttling of the NR across the nuclear membrane may contribute?
If the NR can also traverse the nucleocytoplasmic membrane, the response can be even faster than that in design 3 (design 4, Figure 2B and C). Again, more ligand molecules would be carried from the plasma membrane to the nuclear membrane in NR complexes than in the free form (Figure 2D). However, as also in this model some of the NRs reside in the cytoplasm (which holds close to 80% of the cellular volume), the nuclear NR concentrations are much lower than they would have been had all NRs been confined to the nucleus (Figure 2E). In turn, this causes REs not to be saturated by receptors, resulting in a lowered steady-state transcriptional response (Figure 2B; compare designs 3 and 4 with design 2).
We conclude that if the ligands for NR signaling are highly hydrophobic, their non-chaperoned diffusion through the cytosol would limit the rate at which transcription responds to changes in extracellular signal. The use of a more hydrophilic NR protein as a ‘ferry boat’ (in addition to the use of the latter in transcription activation) may enable the hormone to diffuse faster into the nucleus, but this could occur at the cost of the extent of the transcriptional response, due to a lower concentration of the receptor in the nucleus.
How the active nuclear import of NR may help?
Up to this point we considered the permeation of the NR through the nuclear membrane to be passive, implying an import/export activity ratio of 1. When we took the import/export activity ratio very high (such as in design 5 in Figure 2A), active NR concentrated in the nucleus (Figure 2E), with a positive effect on activation of transcription (Figure 2B, design 5). Consequently, depending on the ratio of import to export activity, design 5 reflects a trade-off between the fast responsiveness of design 4 and the high sensitivity of design 2 (compare the transcriptional response graph in Figure 2B).
Why both importin and exportin are needed; how active import and export of NR can enhance response speed and extent?
In order to maximize responsiveness, core-NR should be concentrated in the cytoplasm, whereas to gain sensitivity, liganded NR (NRL) should be concentrated in the nucleus. This suggests that performance could be improved by making nuclear import and export selective for liganded over unliganded NR (Figure 2A, design 6). Molecularly, this could be based on the facts that liganded and core-NRs have different affinities for transport proteins and that two different types of transport protein (importins and exportins) are involved in NR signaling, one for import and one for export (see Figure 1).
Indeed, retention of core-NR in the cytoplasm (design 6) provides high influx of ligand into the nucleus, especially when complexed with NR (Figure 2D), and also produces the highest concentration of ligand in the nucleus (Figure 2C). Design 6 provides transcriptional responses higher than design 2 and almost as quick a response as established by design 4 (Figure 2B). As shown by Figure 2C, only in design 6 the concentration of ligand becomes higher in the nucleus than in the cytosol; the system functions to pump ligand actively into the nucleus. Interestingly, design 6 has the paradoxical implication that reduced net import of the NR into the nucleus, due to selectivity in the import for liganded-NR over core-NR, can enhance signaling.
NR does not wait in the cytoplasm for the signal, but it is advantageous if it shuttles continuously
Above we have shown that the retention of core-NR in the cytoplasm improves the sensitivity and responsiveness. This result could leave the impression that the higher the fraction of core-NR residing in the cytoplasm when ligand arrives, the higher transcriptional response. In the designs presented in the following, if import and export activities of core-NR were equal (Figure 2G, black line), then unliganded NR was equally distributed between the nucleus and the cytoplasm, a situation denoted as ‘mixed, shuttling’. A high ratio of import to export activities for unliganded NR resulted in a predominant localization of core-NR in the nucleus (Figure 2G, red lines). In comparision, a low ratio resulted in the localization of core-NR in the cytoplasm (Figure 2G, blue lines). For all three cases, the import rate constant of NRL was taken to be 50-times higher than the import rate constant of core-NR resulting in an increase of nuclear concentration of the receptor after the addition of ligand. In order to evaluate the importance of the absolute shuttling rate, the import and export rate constants of both core-NR and NRL were all decreased 10 000-times, keeping constant the import/export ratio (Figure 2; solid lines show high shuttling rate; dashed lines show the decreased shuttling rate).
In all three cases examined, shuttling without changing the initial concentration gradient of NR helped to increase the responsiveness of transcription (Figure 2G, compare solid and dashed lines). Moreover, for high shuttling rates, both the sensitivity and the responsiveness can be very high. In fact, this is even higher when preferences are achieved by decreasing the import of core-NR (compare high shuttling for mixed and nuclear localization of core-NR in Figure 2G with the results for design 6 in Figure 2B). Interestingly, both at very high and at very low import/export ratios of core-NR, the transcriptional responses were slower (Figure 2G). Therefore, we conclude that high shuttling activity is always advantageous, but this advantage can be further increased through an optimal import/export ratio, which results in the optimal subcellular localization of the NR before the onset of the signaling response.
Active export rather than active import of importins is the best strategy to enhance transcription
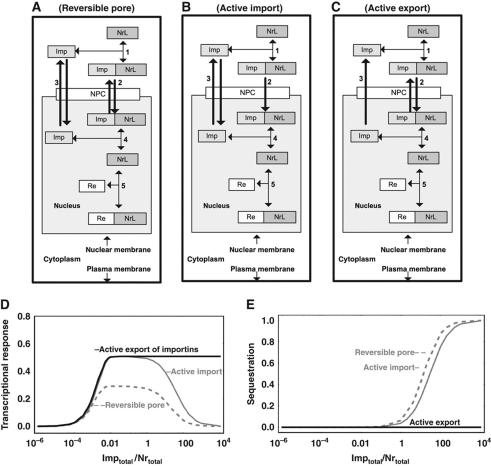
Above we demonstrated that NR shuttling allows for ferrying of ligand that both speeds up and amplifies the transcriptional response, especially if it is accompanied by selective and active import of NRL into the nucleus. Such selectivity requires a protein for the recognition of liganded versus core-NR protein complex. This yields a further aspect of the complexity of NR-mediated signaling, namely, the role of the importin protein that facilitates transport of NRL across the nuclear membrane through the NPC. To establish whether an increased importin concentration would be advantageous, we considered, for the most obvious topology (active import of the importin–NRL complex; Figure 3B), that more importin can help, but may not always do so (the gray line in Figure 3D). At high concentrations importin inhibited the transcriptional response in silico. This occurred when importin concentrations exceeded the total NR concentration (Figure 3E, full gray line). In this case, most of the NR was sequestered by the importin. This phenomenon was not only unique to the active transport mechanism but also occurred when transport was passive (dotted line in Figure 3E).
Figure 3.
Active export of importins rather than active import of the importin–NR complex is advantageous for enhancing the transcriptional response. Three alternative network designs are depicted: (A) Passive facilitated diffusion (reversible pore) of NR across the nuclear membrane; (B) Active nuclear import of importin–NR complex; (C) Active export of importins from the nucleus. (D) Steady-state transcriptional response (ratio ReNrL/Retotal) as function of the total concentration of importins (A–C; note the logarithmic importin concentration axis). The transcriptional response represents the fraction of REs complexed to the NRL. (E) Sequestration (defined as the ratio NrLtotalImptotal/NrLtotal) as function of the total concentration of importins (A–C; note the logarithmic importin concentration axis). Rate equations and kinetic parameters are given in Supplementary Table 7. Supplementary Figure 5S shows simulation results for all species. As compared with Figure 2, the model was simplified by only considering the liganded fraction of the NR (fixed at the level equal to the concentration of NRL in design 4 of Figure 2; in reality, for the case of active transport, the fraction of NRL and consequently the maximal transcriptional response may be larger due to elevated concentration of ligand in the nucleus, in accordance with the design principles discussed above). NrL, liganded NR (e.g., GR); Imp, importins; NrLImp, liganded NR bound with importins; Re, RE for NR on DNA; ReNrL, RE on DNA bound with activated NR; NPC, nuclear pore complex. The model is available in JWS Online and can be simulated in its web browser: http://jjj.biochem.sun.ac.za; http://jjj.bio.vu.nl; http://jjj.mib.ac.uk (Snoep and Olivier, 2002; Olivier and Snoep, 2004). The model can be found via ‘author search’, ‘kolodkin7’. The model can be also accessed directly via: http://jjj.bio.vu.nl/webMathematica/Examples/run.jsp?modelName=kolodkin7 or at any of the other servers listed above. Please note that values of parameters for nuclear import/export rates are set for reversible transport. Simulations of receptor (importin) pump require parameter values from Supplementary Table 7.
The thermodynamic driving force of GTP hydrolysis could also be coupled to the export of free importins (Figure 3C). Paradoxically, the latter topology, i.e., active nuclear export of importins (Figure 3C), turned out to be the most advantageous design: also at high importin concentrations, it supported a high transcriptional response (black line in Figure 3D). The importin concentration in the cytosol was now much higher than the importin concentration in the nucleus, and therefore the importin concentration gradient itself drove the import of NR, while the NR sequestration in the nucleus by nuclear importin was small.
All calculations shown in Figure 3 are based on the assumption that NR degradation is much slower than the signaling process and, consequently, does not affect signaling. It has been demonstrated that ubiquitination of NRs, targeting them for proteasomal degradation, occurs when NRs are bound to their REs (Liu and DeFranco, 2000). When we incorporated NR degradation in our models, we continued to find that active nuclear export of importins benefited the transcriptional response (Supplementary Figure S1).
Flux through the NPC may be robust even if all pathways run through the same pore
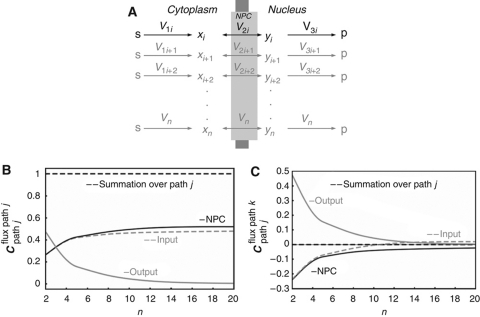
All large proteins that are subject to nucleocytoplasmic transport, as well as all RNAs, pass through the NPC (Tran and Wente, 2006). The design of the NPC as a single unspecific pore appears to impose a limitation, as all transported macromolecules, irrespective of function, would compete for transport. One might then expect that the transport pathways depend strongly on each other's activity. Functionally unrelated molecular networks could, therefore, exhibit inadvertent crosstalk.
We used metabolic control analysis (Burns et al, 1985) and enzyme kinetics to examine this issue in a generic model, with a single NPC (Figure 4A). As reported in more detail in the Supplementary information, the nuclear import rate of each particular signaling protein (xi, with yi representing its imported form) through the NPC was described by a reversible kinetic equation incorporating competitive inhibition by other cargo from both sides of the nuclear membrane. When many other cargo proteins compete for transport and their individual concentrations become small relative to the total cargo concentration, several simplifications apply. First, the nuclear import rate (vi) becomes proportional to the cytosolic concentration of the corresponding cargo (xi) and the corresponding elasticity coefficient, i.e., the log-log dependence of rate on concentration (Burns et al, 1985), approaches 1, whenever the gradient of substance i across the nuclear membrane is sizeable (i.e., xi≫yi):
Figure 4.
Control properties of NPC transport in the case of excess competing processes. (A) NPC transport model. Note that (i) the import and export of proteins, i.e., x's and y's, compete for the transport (all reactions are dependent on substrates and products), (ii) the pathways do not exchange mass flow between each other but only regulatory influences (competitive inhibition); this system displays an hierarchical design (Kahn and Westerhoff, 1991) and (iii) the cycling of importins etc. between nucleus and cytosol is not taken into account. (B, C) Numerical illustration of control design of NPC as function of the number of reactions through the pore. (B) Control of the flux through path j by (gray dashed line) the input reaction of path j, by (black solid line) the NPC, and by (gray solid line) the exit reaction of path j. The sum of the aforementioned flux control coefficients is given by the black dashed line; (C) control of the flux through path k by the input reaction of path j (gray dashed line), by the NPC (black solid line), and by the exit reaction of path j (gray solid line) and the summation of the three corresponding control coefficients (dashed black line). Rate equations and kinetic parameters are given in Supplementary Table 8. Supplementary Figure 6S shows simulation results for all species.
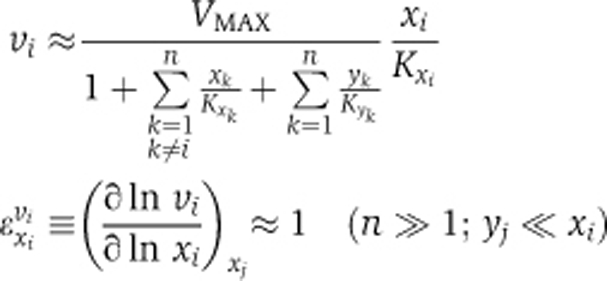 |
VMAX and the Kxk are constant rate characteristics of transport of species k.
Second, the rate becomes independent of the concentration of any specific other cargo molecule, including the already imported forms, causing all cross-elasticity coefficients to become zero:
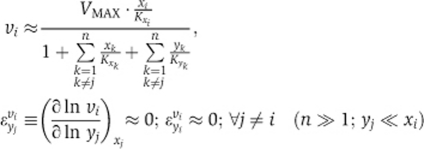 |
Consequently, all pathways should become independent of each other. When the same pore is used for the transport of different cargoes, the concentration level of each cargo is far below the concentration that by itself would challenge the carrying capacity of the transport system. By analogy, at rush hour, two roads leading into a roundabout influence each other's traffic intensely, if they are the only two, but exert relatively little influence, if there are 10 other roads feeding into the roundabout.
Numerical simulations confirmed this scenario, whereby when the number of NR pathways (n) exceeded 6, ‘cross-control’ of the flux of one NR by other receptors was close to zero (Figure 4C), leaving most control over the transport of an NR to its own pathway (Figure 4B). Paradoxically, the high promiscuity of the NPC prevents crosstalk between different NR pathways.
Discussion
Only a few main categories of signal transduction govern gene expression activity in response to extracellular signals. The distinction is largely in the physico-chemical properties of the signal molecule. For example, extracellular signals carried by hydrophilic molecules, such as epidermal growth factor, bind receptors in the plasma membrane. In this category, no signaling molecule is transported across the membrane, but a signal is, through changes in the state of a transmembrane receptor. This leads to the increased local concentration (Kholodenko et al, 2000b) just below the plasma membrane of a single protein, alters the state of other membrane-anchored molecules, such as RAS, and indirectly the states of components of a MAP kinase cascade. A phosphorylated protein at the end of such a cascade then binds to a gene-locus control region and activates transcription. In this type of signal transduction, no molecule needs to move all the way from the outside of the cell to the chromatin.
In a second category, and the subject of this study, the extracellular signal is a hydrophobic molecule, thereby able to cross the plasma membrane by itself. That hydrophobic molecule then moves even further to the nucleus and binds to a NR, which then activates transcription. In this category not just a signal but also a signal molecule moves all the way from outside the cell to the targeted genomic region.
It would seem that in this second category of signal transduction, only a signal-activated transcription factor would need to be involved. That transcription factor would be the only protein ‘receiving the signal’. In this scenario, this ‘receptor’ could indeed be only located in the nucleus and await the hydrophobic signaling molecule to arrive. The design of this category seems to excel by simplicity, which would be welcome in our attempts to comprehend cell function.
In this study, we tested whether this category of signal transduction actually follows this simplest design. We constructed the common denominator network for NR signaling. We found that even this common denominator was nowhere near as simple as this design. We then identified eight aspects of the network topology where reality appears to be more complex and we found reasons why all eight topologies were supportive of the function of signal transduction. A first and general conclusion of our study is that most, if not all, aspects were somewhat important; all contributed at least somewhat to signaling. The importance of each of the different aspects may become clear when applying these models to specific and individual NRs; then a more or less subtle balance of the various topological contributions may emerge. In this manner, we have generated a valuable tool kit for the NR research community.
The current assessment used generic mathematical models to identify potential functions of these topological features. The first, for the classical design 1 of Figure 2, showed significant disadvantages of exclusively nuclear localization of the NR: when all NR was constitutively bound to the DNA, the transcriptional response was very low, perhaps paradoxically so. A high concentration of free NR in the nucleus improved sensitivity, but made the responsiveness slow (∼25 min). Forward rate constants for all association reactions in our models were chosen as diffusion limited. In reality their values could be lower; this would slow down the response even further. On the contrary, experimental measurements indicate faster formation of the GR–RE complex (∼10 min). Moreover, this time is mostly composed of a slow lag-phase, after which the formation of GR–RE complex is very fast (∼1 min; Stavreva et al, 2009). Consequently, the slow responsiveness that exclusively nuclear NR localization would entail is not only disadvantageous, but also less realistic.
Our modeling next indicated a considerable increase of the rate of response when the NR was allowed to enter (and leave) the cytoplasm, but at the cost of sensitivity. We predicted the effect of additional cytoplasmic localization of NRs to be substantial yet subtle: highly hydrophobic NR ligands moved mostly in association with their specific NR. In our model, NR bound ligand in the proximity to the cellular membrane, where we considered the aqueous concentration of ligand to be low, as it should be mostly in the bordering hydrophilic phase. Inside the membrane, the concentration of the hormone should be many times higher (Oren et al, 2004). Hence, a scenario could be envisaged, whereby the NR might contribute more to ligand movement if it could directly collect the ligand from the membrane. In fact, the latter scenario is quite realistic. Recently, it was shown that 5–10% of total cellular ER is found at the plasma membrane (Levin, 2009b), where it may interact with GPR30 and induce rapid signaling through, e.g., p38-β MAP kinase. The scenario that liganded ER may leave the membrane surface was not considered. There is experimental evidence suggesting that liganded ER may leave the plasma membrane and head for the nucleus. For example, fluorescence microscopy experiments in ROS cells (Spona et al, 1980; Ong et al, 2004) showed that before addition of estrogen, ERα-RFP was distributed over the nucleus and cytoplasm, but after addition of estrogen, all receptor shifted to the nucleus. This should also have depleted any pool close to the plasma membrane.
The transport system could be considered to involve three conveyor belts, including one that would consist of importin cycling and another one exportin cycling. RanGTP binds competitively with NR to importin with the effect that the outward driving force of RanGTP (outward because of the activity of RanGEF in the nucleus and RanGAP in the cytosol) makes the importin belt transport the NR more inward than outward. Conversely, RanGTP binds positively cooperatively with NR to exportin, causing the RanGTPase driving force to make the exportin conveyor belt export NR. Together, the exportin and importin conveyor belts serve for a rapid cycling of NR, which should ensure a rapid response of transcription to changes in signal concentration in the cytosol, without amplifying the signal intensity. If the importin cycle were more active with NRL as cargo, and/or the exportin cycle more active with core-NR, an additional signal amplification effect should arise (design 6). Importin and exportin conveyor belts together then drive the cycling of the third conveyor belt, consisting of the receptor that brings ligand into the nucleus. Consequently, apart from its classical role in transcription activation, the NR may be also used as a ‘smart’ ferry boat: coming into the nucleus with ligand and leaving the nucleus when empty. As both binding of ligand to the receptor and binding of ligand-bound receptor to DNA are reversible stochastic processes (Voss et al, 2009), a single ligand-bound receptor in the nucleus may either bind directly to DNA or may loose the ligand; then the core-receptor may either bind a new ligand molecule or may be exported out of the nucleus. The probability of each event would depend on the relative magnitudes of the relevant parameters. The ‘ferry boat’ is ‘smart’ because (i) it likes to have ligand on board when it ‘sails’ into the nucleus, but not when it ‘sails’ out, and (ii) when it dwells in the nucleus it likes to bond to the DNA and activate transcription.
An important outcome of preferential import of ligand-bound receptor and export of core-receptor (design 6) is that it would be the only design where the addition of ligand would result in the observable shift of NR intracellular localization. For realistic parameters, e.g., of GR signaling, an addition of 0.1 nM of DEX should increase the total GR concentration in the nucleus from 15 to 30% (Supplementary Figure S2B). An addition of 1 nM of DEX should increase the fraction of total nuclear GR even further, up to 70% (Supplementary Figure S2C). These model predictions are consistent with the results of single dose experiments described before in the literature (Kumar et al, 2004; Charmandari et al, 2005, 2007) and experimentally confirmed (Supplementary Figure S3).
Our analysis proved that, paradoxically, the transport of all cargo through the same NPC makes the transport of any particular cargo robust with respect to perturbations in the availability of any other cargo. Only when the transport of any individual cargo is greatly increased, does a competition effect at the transport level become significant. The design of having many different signaling and bulk transport routes share the same mechanism, may be a way to reduce competition until the situation arises, where the energetic capacity of the system as a whole would be compromised. The emergent flux independence due to the utilization of a single NPC for many transport systems should have general implications. The effect has indeed been observed experimentally: single-molecule video microscopy indicated that nuclear import dynamics are mainly determined by cargo–NR–pore interactions and are robust to other cell processes and other transported molecules (Dange et al, 2008). This is not to say that there are no other designs that would avoid competition. Clearly, giving all transported species their own transporter operating far below its Vmax should also make their transports independent of one another and provide the ability to increase it when needed. However, this would require higher totals of transport proteins, at a higher synthetic burden to the cell. The single pore mechanism seems an attractive design alternative.
Our calculations predict that there is an optimal ratio of nuclear to cytoplasmic fractions of the NR that depends on the specific properties of the ligand and on the transcription activation requirements. This may help to explain the observation that different NRs have different predominant intracellular localizations. For instance, the VDR is present both in the nucleus and cytoplasm, and after the addition of ligand its nuclear/cytoplasmic ratio increases only slightly (Racz and Barsony, 1999; Menezes et al, 2008), but GR is concentrated in the cytoplasm before ligand addition and shifts into the nucleus upon addition of ligand (Prüfer and Boudreaux, 2007; Ricketson et al, 2007). This issue warrants further study, which will require interaction between modeling and quantitative experimentation, and again the tool kit generated in the current study may provide a welcome resource with which to test and analyze these predictions.
Because the earliest (‘classical’) paradigms of NR signaling had the NR attached to its RE, ‘waiting’ for its ligand (Brink et al, 1992; Van Steensel et al, 1995), pathology related to NR signaling was attributed mostly to the concentration of ligand, the expression level and the integrity of the NR. At present, it is well known that the NR is not always attached to chromatin and that its intracellular localization is important for signaling. Clinical data support the latter paradigm. For instance, alterations of the nucleocytoplasmic ratio of the VDR are correlated with the progression of lung cancer (Menezes et al, 2008). Our analysis of design principles shows that the efficiency of signaling may depend not only on the intracellular localization of the NR, as set by the import/export activity ratio, but also on the absolute rate of nucleocytoplasmic shuttling. This rate emerges from the whole network of GTPase-dependent reactions involved in nucleocytoplasmic transport. It suggests new etiologies, as well as new potential drug targets.
This study readdressed the significant complexity of NR signaling, in a novel way. Whereas the diversity of these networks is accepted generally, it is rarely discussed which topological aspects are important for which aspects of biological function. Assessing this importance was the novel contribution of the current study by using mathematical models based on realistic physical, chemical and biological data. We have not been able to address all complex aspects of NR signaling. For example, it is well established that many ligands for NRs are also substrates for metabolism by the target genes of said NRs; hence, 1,25D3 activation of the VDR results in increased CYP24/24-hydroxylase expression, which is responsible for 1,25D3 degradation. We have also not considered the full mechanism of transcription activation downstream of the formation of the NR–ligand–DNA complex; NR dimerization, co-repressors and co-activators complexation and chromatin modulation. Whereas we acknowledge the importance of these processes, and appreciate that they may impact upon the total network response, it is important to focus on a clearly defined network module, allowing a focussed examination of the design principles underlying nucleocytoplasmic shuttling (Figure 1). We discussed eight aspects of this part of signaling together, rather than just one or two: multiple mechanistic aspects turned out to be important at the same time; i.e., the complexity of Figure 1 may really have offered selective advantage in evolution.
The models that we have produced (and are available to the reader) were relatively generic. Yet further testing may also help verify which design principles are most functional in actual signaling pathways. For this, actual parameter values will need to be inserted, which can result in an important integration between more modeling and more experimentation. The design principles we have identified may well be in more general use and may also be important for yet other signal-transduction pathways, such as SMAD signaling (Nicolas et al, 2004; Dupont et al, 2009).
In conclusion, in this study we have shown that complex networks of biochemical and signaling reactions can harbor subtle design principles that can be understood rationally in terms of simplified models. Of course, these predictions should be substantiated in experimental studies of specific cases of NR signaling that in turn may reveal additional design aspects.
Materials and methods
The SBGN graphical network notation for the ‘canonical’ endocrine NR signaling network (Figure 1) has been constructed using CellDesigner version 4.1 beta. The CellDesigner SBML-compliant free package has been downloaded from Kitano et al (2005). Our mathematical models did not address this network as a whole, but parts thereof. For Figures 2 and 3 these simplified models have been built with the following assumptions.
We have simplified the formation of the ligand–NR–RE complex in designs 2–6 (Figure 2) by considering it as only one process: binding of ligand to NR and binding of the ligand–NR complex to RE. Possible occurrence of NR–RE complex (binding of unliganded NR to RE followed by binding of ligand) has been omitted. This is the only simplification possible when one needs to accommodate NR diffusing independently of the DNA.
Instead of considering importin-α and -β as separate complexes, the single importin-α–importin-β complex has been noted as a single importin protein. This simplification is warranted as described in the detailed scheme in Figure 1: importin-α first binds to importin-β and subsequently the complex binds cargo. This sequence of events is supported by the observation that importin-α contains an N-terminal autoinhibitory domain that blocks the NLS binding site. Binding of importin-β unmasks this autoinhibitory blockage and allows importin-α to bind cargo proteins with high affinity (Catimel et al, 2001; Riddick and Macara, 2005).
We did not consider the NPC complexes explicitly for the models presented in Figures 2 and 3. Taking into account the large number of NPCs (∼2000 per cell) and their relatively homogeneous distribution, we modeled the overall transport of cargo through the area of a membrane.
The translocation of importins across the nuclear membrane has been considered as a single reaction, reversible and symmetrical. In reality it is a complex biochemical network of reactions, in which importins interact with many other proteins, such as RanGTP, adaptor proteins, Hsp90 and filaments of the NPC (Figure 1). The translocation through nuclear pores is always reversible (Kopito and Elbaum, 2007). Ultimately, the direction of transport is determined by the nucleocytoplasmic gradient of RanGTP. This gradient is maintained by the exclusively cytoplasmic hydrolysis of RanGTP stimulated by RanGAP, which is associated with the cytoplasmic side of the NPC complex, and the exclusively nuclear regeneration of RanGTP by GEF associated with chromatin. The steady state of the RanGTP gradient is the net result of the transport of many cargo molecules and of the distinct localization and efficiency of GAP and GEF. NRs make up only a small fraction of cargo involved in the global process. Consequently, they do not much affect transport of other cargoes, including other NRs. If there is excess RanGAP and RanGEF activity and excess GTP, RanGTP gradients can be considered as externally fixed and can be presented in terms of kinetic parameters of a single reaction. Our calculations reflect these assumptions by keeping the ratio of forward to reverse rate constants for active transport constant (e.g., Figure 2, design 5). We did not keep this ratio at the very high level corresponding to thermodynamic equilibrium, but at a ratio of 100, acknowledging the non-equilibrium nature of the process (Westerhoff, 1985).
NR was considered to bind ligand in the proximity to the cellular membrane, where the concentration of ligand is low, as it should be in a hydrophilic phase. In fact, inside the membrane the concentration of the hormone should be many times higher (Oren et al, 2004) and NR might contribute even more to the movement of ligand if it could directly collect the ligand from the membrane (this possibility is considered in the Discussion section).
Realistic cytoplasmic and nuclear compartment volumes, 1.55 and 0.45 pL, respectively (Riddick and Macara, 2007), have been used in all models. The total concentrations of RE and NR were set to realistic values, i.e., 1.7 × 10−12 nmoles (1000 molecules) per cell for the RE (De Kloet et al, 2000) and 1.7 × 10−10 nmoles (100 000 molecules) per cell for NRs (Nordeen et al, 1989; Van Steensel et al, 1995). The rate constants for complex formation of hormone with NR were chosen as diffusion limited (values for GR and cortisol analog dexamethasone, kassociation=1 nM−1 s−1 and Kd=1 nM (Marissal-Arvy et al, 1999)). Rate constants for complex formation of the ligand-bound NR to the RE were chosen to be diffusion limited as well (values for GR and cortisol kassociation=1 nM−1 s−1 and Kd=1 nM (Drouin et al, 1992); the diffusion coefficient for NR was taken equal to 1 × 10−12 m2 s−1. This value was used earlier in the models addressing protein diffusion (Kholodenko et al, 2000a), where the diffusion coefficient of model protein was taken in the order of magnitude of experimentally measured diffusion coefficients of various proteins, e.g., GFP (Dayel et al, 1999). The diffusion coefficient for cortisol was assumed to be 6-times higher than this, as estimated from the Stokes–Einstein equation and the relative sizes. The external concentration of free ligand was taken to change abruptly from 0 to 0.005 nM. The model used for Figure 4 is illustrative in nature and has been built neither taking into account the differences in nuclear and cytoplasmic volumes nor the physiological parameter ranges.
Balance equations, rate equations and kinetic parameters for all models are presented in the Supplementary information. For all models, the ODEs have been solved numerically using the Mathematica6 commercial package. All models are also available in cps format for simulation in COPASI. In addition, the models are made available in JWS Online and can be simulated in a web browser: http://jjj.biochem.sun.ac.za; http://jjj.bio.vu.nl; http://jjj.mib.ac.uk (Snoep and Olivier, 2002; Olivier and Snoep, 2004).
Models can be found using the regular menu, for instance, via author search ‘kolodkin’. Models can be also accessed directly via: ~/webMathematica/Examples/run.jsp?modelName=kolodkinX, with ∼ either http://jjj.bio.vu.nl, http://jjj.mib.ac.uk or http://jjj.biochem.sun.ac.za and X ranging from 1 to 8 for the respective models. For instance: http://jjj.bio.vu.nl/webMathematica/Examples/run.jsp?modelName=kolodkin1 yields the model for Figure 2 design 1.
We examined the robustness of the conclusions of this paper by varying parameter values and checking whether the conclusions persisted. Our conclusions were mostly robust for up to fivefold changes in parameter values, but the precise details are given below.
Figure 2 (Supplementary Table 11): Design 6 is the most advantageous. This conclusion was not affected by at least fivefold perturbation of any single parameter in the model. The only exception was related to the rate of nuclear import of NRL. If active nuclear import of NRL in design 6 is decreased more than threefold, then the advantages of design 6 as compared with design 2 almost disappear. This fits well in the context of the main messages of our manuscript. Indeed, an advantageous feature of the design 6 is exactly the active import of ligand into the nucleus achieved by preferential nuclear import of the NRL.
Figure 3 (Supplementary Table 12): Active export of importins prevents sequestration of the receptor in the nucleus by importins. This conclusion was not affected by l0-fold perturbation of any single parameter in the model.
Figure 4 (Supplementary Table 13): Flux through the NPC may be robust even if all pathways run through the same pore. This conclusion was not affected by l0-fold perturbation of any single parameter in the model.
Supplementary Material
Acknowledgments
We thank various EC framework programs (notably, the Marie Curie research training network NucSys, BioSim, NISB, EC-MOAN, YSBN, UNICELLSYS), NWO-FALW, NWO-ZON (contract grant number: 91206069), the BBSRC (BBC0082191 [MCISB] and ERASysBio) and the EPSRC (DTC), for support of some of this work (see also www.systembiology.net/support). MJC acknowledges support from the NCI Cancer Center Support Grant to the Roswell Park Cancer Institute (CA016056).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aouabdi S, Gibson G, Plant N (2006) Transcriptional regulation of the PXR gene: identification and characterization of a functional peroxisome proliferator-activated receptor alpha binding site within the proximal promoter of PXR. Drug Metab Disp 34: 138–144 [DOI] [PubMed] [Google Scholar]
- Brink M, Humbel BM, De Kloet ER, Van Driel R (1992) The unliganded glucocorticoid receptor is localized in the nucleus, not in the cytoplasm. Endocrinol 130: 3575–3581 [DOI] [PubMed] [Google Scholar]
- Bunce CM, Campbell MJ (eds.) (2010) Nuclear receptors, Proteins and Cell Regulation 8, 455-457. DOI 10.1007/978-90-481-3303-1, Springer Science Business Media BV
- Burns JA, Cornish-Bowden A, Groen AK, Heinrich R, Kacser H, Porteous JW, Rapoport SM, Rapoport TA, Stucki JW, Tager JM, Wanders RJA, Westerhoff HV (1985) Control analysis of metabolic systems. Trends Biochem Sci 10: 16 [Google Scholar]
- Cao HJ, Lin HY, Luidens MK, Davis FB, Davis PJ (2009) Cytoplasm-to-nucleus shuttling of thyroid hormone receptor-1 (Tr1) is directed from a plasma membrane integrin receptor by thyroid hormone. Endocr Res 34: 31–42 [DOI] [PubMed] [Google Scholar]
- Carlberg C, Dunlop TW (2006) An integrated biological approach to nuclear receptor signaling in physiological control and disease. Crit Rev Eukar Gene Expr 16: 1–22 [DOI] [PubMed] [Google Scholar]
- Catimel B, Teh T, Fontes MRM, Jennings IG, Jans DA, Howlett GJ, Nice EC, Kobe B (2001) Biophysical characterization of interactions involving importin-alpha during nuclear import. J Biol Chem 276: 34189–34198 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Kino T, Ichijo T, Jubiz W, Mejia L, Zachman K, Chrousos GP (2007) A novel point mutation in helix 11 of the ligand-binding domain of the human glucocorticoid receptor gene causing generalized glucocorticoid resistance. J Clin Endocrinol Metab 92: 3986–3990 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Raji A, Kino T, Ichijo T, Tiulpakov A, Zachman K, Chrousos GP (2005) A novel point mutation in the ligand-binding domain (LBD) of the human glucocorticoid receptor (hGR) causing generalized glucocorticoid resistance: the importance of the C terminus of hGR LBD in conferring transactivational activity. J Clin Endocrinol Metab 90: 3696–3705 [DOI] [PubMed] [Google Scholar]
- Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE (2008) Structural basis for the nuclear import of the human androgen receptor. J Cell Sci 121: 957–968 [DOI] [PubMed] [Google Scholar]
- Dange T, Grunwald D, Grunwald A, Peters R, Kubitscheck U (2008) Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J Cell Biol 183: 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Hom EFY, Verkman AS (1999) Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys J 76: 2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kloet ER, Meijer OC, Vreugdenhil E, Joels M (2000) The Yin and Yang of nuclear receptors: symposium on nuclear receptors in brain, Oegstgeest, The Netherlands, 13-14 April 2000. Trends Endocrinol Metab 11: 245–248 [DOI] [PubMed] [Google Scholar]
- Drouin J, Sun YL, Tremblay S, Lavender P, Schmidt TJ, Delean A, Nemer M (1992) Homodimer formation is rate-limiting for high-affinity DNA-binding by glucocorticoid receptor. Mol Endocrinol 6: 1299–1309 [DOI] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136: 123–135 [DOI] [PubMed] [Google Scholar]
- Ebert R, Schutze N, Adamski J, Jakob F (2006) Vitamin D signaling is modulated on multiple levels in health and disease. Mol Cell Endocrinol 248: 149–159 [DOI] [PubMed] [Google Scholar]
- El-Sankary W, Bombail V, Gibson GG, Plant N (2002) Glucocorticoid-mediated induction of CYP3A4 is decreased by disruption of a protein: DNA interaction distinct from the pregnane X receptor response element. Drug Metab Disp 30: 1029–1034 [DOI] [PubMed] [Google Scholar]
- El-Sankary W, Gibson GG, Ayrton A, Plant N (2001) Use of a reporter gene assay to predict and rank the potency and efficacy of CYP3A4 inducers. Drug Metab Disposit 29: 1499–1504 [PubMed] [Google Scholar]
- Fanestil DD, Edelman IS (1966) Characteristics of renal nuclear receptors for aldosterone. Proc Natl Acad Sci USA 56: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RS (1975) Nuclear thyroid-hormone receptors—evidence for association with nucleolar chromatin. Biochem Biophys Res Commun 67: 625–633 [DOI] [PubMed] [Google Scholar]
- Garlatti M, Daheshia M, Slater E, Bouguet J, Hanoune J, Beato M, Barouki R (1994) A functional glucocorticoid-responsive unit composed of 2 overlapping inactive receptor-binding sites—evidence for formation of a receptor tetramer. Mol Cell Biol 14: 8007–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer MD, Wolf IM, Sanchez ER, Witchel SF, DeFranco DB (2007) Glucocorticoid receptor physiology. Rev Endocr Metab Dis 8: 321–330 [DOI] [PubMed] [Google Scholar]
- Kahn D, Westerhoff HV (1991) Control-theory of regulatory cascades. J Theo Biol 153: 255–285 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Brown GC, Hoek JB (2000a) Diffusion control of protein phosphorylation in signal transduction pathways. Biochem J 350: 901–907 [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN, Hoek JB, Westerhoff HV (2000b) Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol 10: 173–178 [DOI] [PubMed] [Google Scholar]
- Kitano H, Funahashi A, Matsuoka Y, Oda K (2005) Using process diagrams for the graphical representation of biological networks. Nat Biotech 23: 961–966 [DOI] [PubMed] [Google Scholar]
- Kopito RB, Elbaum M (2007) Reversibility in nucleocytoplasmic transport. Proc Nat Acad Sci USA 104: 12743–12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Chaturvedi NK, Nishi M, Kawata M, Tyagi RK (2004) Shuttling components of nuclear import machinery involved in nuclear translocation of steroid receptors exit nucleus via exportin-1/CRM-1 independent pathway. Biochim Biophys Acta-Mol Cell Res 1691: 73–77 [DOI] [PubMed] [Google Scholar]
- Kumar S, Saradhi M, Chaturvedi NK, Tyagi RK (2006) Intracellular localization and nucleocytoplasmic trafficking of steroid receptors: an overview. Mol Cell Endocrinol 246: 147–156 [DOI] [PubMed] [Google Scholar]
- Levin ER (2009a) Membrane oestrogen receptor alpha signalling to cell functions. J Physiol 587: 5019–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ER (2009b) Plasma membrane estrogen receptors. Trends Endocrinol Metab 20: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y et al. (2007) Whole-genome cartography of estrogen receptor alpha binding sites. PloS Gen 3: 867–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JM, DeFranco DB (2000) Protracted nuclear export of glucocorticoid receptor limits its turnover and does not require the exportin 1/CRM1-directed nuclear export pathway. Molecul Endocrinol 14: 40–51 [DOI] [PubMed] [Google Scholar]
- Macara IG (2001) Transport into and out of the nucleus. Microbiol Mol Biol Rev 65: 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissal-Arvy N, Mormede P, Sarrieau A (1999) Strain differences in corticosteroid receptor efficiencies and regulation in Brown Norway and Fischer 344 rats. J Neuroendocrinol 11: 267–273 [DOI] [PubMed] [Google Scholar]
- Menezes RJ, Cheney RT, Husain A, Tretiakova M, Loewen G, Johnson CS, Jayaprakash V, Moysich KB, Salgia R, Reid ME (2008) Vitamin D receptor expression in normal, premalignant, and malignant human lung tissue. Cancer Epidemiol Biom Preven 17: 1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas FJ, De Bosscher K, Schmierer B, Hill CS (2004) Analysis of Smad nucleocytoplasmic shuttling in living cells. J Cell Sci 117: 4113–4125 [DOI] [PubMed] [Google Scholar]
- Nordeen SK, Kuhnel B, Lawlerheavner J, Barber DA, Edwards DP (1989) A quantitative comparison of dual control of a hormone response element by progestins and glucocorticoids in the same cell-line. Mol Endocrinol 3: 1270–1278 [DOI] [PubMed] [Google Scholar]
- Olivier BG, Snoep JL (2004) Web-based kinetic modelling using JWS Online. Bioinformatics 20: 2143–2144 [DOI] [PubMed] [Google Scholar]
- Ong DB, Colley SM, Norman MR, Kitazawa S, Tobias JH (2004) Transcriptional regulation of a BMP-6 promoter by estrogen receptor alpha. J Bone Miner Res 19: 447–454 [DOI] [PubMed] [Google Scholar]
- Oren I, Fleishman SJ, Kessel A, Ben-Tal N (2004) Free diffusion of steroid hormones across biomembranes: a simplex search with implicit solvent model calculations. Biophys J 87: 768–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Phillips AL, Hood SR, Gibson GG, Plant NJ (2003) CYP3A and nuclear receptor messenger RNA expression in liver and HuH7 cells. Drug Metabol Rev 35: 83 [Google Scholar]
- Poon IKH, Jans DA (2005) Regulation of nuclear transport: central role in development and transformation? Traffic 6: 173–186 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Sanchez ER, Bresnick EH, Meshinchi S, Scherrer LC, Dalman FC, Welsh MJ (1989) Interaction of the glucocorticoid receptor with the Mr 90,000 heat shock protein: an evolving model of ligand-mediated receptor transformation and translocation. Cancer Res 49: 2222s–2229s [PubMed] [Google Scholar]
- Prüfer K, Boudreaux J (2007) Nuclear localization of liver X receptor alpha and beta is differentially regulated. J Cell Biochem 100: 69–85 [DOI] [PubMed] [Google Scholar]
- Racz A, Barsony J (1999) Hormone-dependent translocation of vitamin D receptors is linked to transactivation. J Biol Chem 274: 19352–19360 [DOI] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM (2009) Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res 19: 2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketson D, Hostick U, Fang L, Yamamoto KR, Darimont BD (2007) A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticold receptor on Hsp90. J Molecul Biol 368: 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara IG (2005) A systems analysis of importin-alpha-beta mediated nuclear protein import. J Cell Biol 168: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara IG (2007) The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol Sys Biol 3: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova EI (2002) Protein transport in plant cells. Russ J Plant Physiol 49: 255–268 [Google Scholar]
- Snoep JL, Olivier BG (2002) Java Web Simulation (JWS); a web based database of kinetic models. Mol Biol Rep 29: 259–263 [DOI] [PubMed] [Google Scholar]
- Spona J, Leibl H, Bieglmayer C (1980) Nuclear translocation of estrogen-receptor complex and stimulation of Rna-synthesis by estrogens of different biological potencies in the female rat pituitary. Biochim Biophys Acta 607: 189–200 [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Wiench M, John S, Conway-Campbell BL, McKenna MA, Pooley JR, Johnson TA, Voss TC, Lightman SL, Hager GL (2009) Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat Cell Biol 11: 1093–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Nishi M, Morimoto M, Sugimoto T, Kawata M (2005) Imaging analysis of mineralocorticoid receptor and importins in single living cells by using GFP color variants. Cell Tiss Res 320: 447–453 [DOI] [PubMed] [Google Scholar]
- Tran EJ, Wente SR (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Van Steensel B, Brink M, Van der Meulen K, Van Binnendijk EP, Wansink DG, De Jong L, De Kloet ER, Van Driel R (1995) Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci 108 (Pt 9): 3003–3011 [DOI] [PubMed] [Google Scholar]
- Von Knethen A, Tzieply N, Jennewein C, Brune B (2010) Casein-kinase-II-dependent phosphorylation of PPAR gamma provokes CRM1-mediated shuttling of PPAR gamma from the nucleus to the cytosol. J Cell Sci 123: 192–201 [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Johnson TA, John S, Hager GL (2009) Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity. J Cell Sci 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisiger RA (2002) Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands. Mol Cell Biochem 239: 35–43 [PubMed] [Google Scholar]
- Westerhoff HV (1985) Thermodynamics and control of proton motive free-energy transduction. Biomedica Biochimica Acta 44: 929–941 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.