Abstract
Accurate chromosome segregation is dependent on the centromere-specific histone H3 isoform known generally as CenH3, or as Cse4 in budding yeast. Cytological experiments have shown that Cse4 appears at extracentromeric loci in yeast cells deficient for both the CAF-1 and HIR histone H3/H4 deposition complexes, consistent with increased nondisjunction in these double mutant cells. Here, we examined molecular aspects of this Cse4 mislocalization. Genome-scale chromatin immunoprecipitation analyses demonstrated broader distribution of Cse4 outside of centromeres in cac1Δ hir1Δ double mutant cells that lack both CAF-1 and HIR complexes than in either single mutant. However, cytological localization showed that the essential inner kinetochore component Mif2 (CENP-C) was not recruited to extracentromeric Cse4 in cac1Δ hir1Δ double mutant cells. We also observed that rpb1-1 mutants displayed a modestly increased Cse4 half-life at nonpermissive temperatures, suggesting that turnover of Cse4 is partially dependent on Pol II transcription. We used genome-scale assays to demonstrate that the CAF-1 and HIR complexes independently stimulate replication-independent histone H3 turnover rates. We discuss ways in which altered histone exchange kinetics may affect eviction of Cse4 from noncentromeric loci.
THE budding yeast centromere is minimally defined by 125 bp of DNA sequence (Fitzgerald-Hayes et al. 1982; Hieter et al. 1985), which serves as the site of kinetochore assembly and microtubule attachment. Three sequence elements, termed CDE I, CDE II, and CDE III have been characterized at the centromere (Sharp and Kaufman 2003). CDE I is a transcription factor binding site, which improves chromosome stability, but is not essential for function (Bram and Kornberg 1987). In contrast, CDE II, an AT-rich region (Gaudet and Fitzgerald-Hayes 1987) and CDE III, the specific recognition sequence for the centromere binding factor (CBF3) protein complex, are both essential for centromere function (McGrew et al. 1986). Therefore, budding yeast, unlike most other eukaryotes, has a centromere dependent on a short, specific DNA sequence.
In addition to these sequence elements, a fully functional budding yeast centromere is also characterized by the surrounding, specialized chromatin structure. Although budding yeast do not have many of the hallmarks of centromeric heterochromatin found in other eukaryotes (Morris and Moazed 2007), the CDE I, CDE II, and CDE III sequence elements are flanked by 2 kb of a highly phased array of nucleosomes, which provide the proper chromatin context for chromosome segregation (Bloom and Carbon 1982). Proper centromere function is also dependent on the core histones, as mutations in histone H2A (Pinto and Winston 2000), histone H2B (Maruyama et al. 2006), and histone H4 (Smith et al. 1996) decrease fidelity in chromosome segregation and disrupt centromere function. Most importantly, chromatin at centromeres is also characterized by the incorporation of a histone H3 variant, termed Cse4 in yeast and CENP-A in mammals, into nucleosomes (Meluh et al. 1998). This histone variant is essential for centromere function (Stoler et al. 1995; Meluh et al. 1998) and is specifically localized to centromeres in all eukaryotes (Bloom 2007). Together, these observations illustrate the central role of chromatin in centromere biology.
The conserved histone chaperone complexes chromatin assembly factor 1 (CAF-1) and histone regulator (HIR) mediate replication-dependent and replication-independent histone H3/H4 deposition, respectively (Smith and Stillman 1989; Kaufman et al. 1997; Ray-Gallet et al. 2002; Tagami et al. 2004; Green et al. 2005). In mammalian cells, these complexes deposit distinct histone H3 isoforms (Tagami et al. 2004), but in budding yeast, which express a single noncentromeric H3 isoform (besides Cse4), biological roles for these histone chaperones are significantly overlapping. For example, the growth and chromatin-mediated gene silencing defects in cells lacking both CAF-1 and HIR subunits are significantly more pronounced than those in either single mutant (Kaufman et al. 1998). CAF-1 and the HIR complex also make largely overlapping contributions to kinetochore function. CAF-1 and HIR both localize to kinetochores, and cells lacking subunits of both complexes (cac1Δ hir1Δ double mutants) display increased rates of chromosome loss and nondisjunction and have disordered nucleosomal ladders flanking the centromere (Sharp et al. 2002). Although Cse4 localizes to centromeres in cac1Δ hir1Δ cells, it is also found at extracentromeric loci (Sharp et al. 2002). Therefore, the normal localization of Cse4 is altered when both replication-dependent and -independent chromatin assembly factors are simultaneously disrupted.
Multiple mechanisms contribute to the specific localization of CenH3 to centromeric chromatin. In several eukaryotes, proteins have been identified that interact physically or genetically with CenH3s and are required for their centromeric targeting. These include histone binding protein RbAp48/Mis16 in Drosophila melanogaster (Furuyama et al. 2006) and Schizosaccharomyces pombe (Hayashi et al. 2004), the Myb domain-containing protein KNL2 in Caenorhabditis elegans (Maddox et al. 2007), the related Mis18 protein complex in humans (Fujita et al. 2007), and the NASP-(N1/N2)-related protein Sim3 in S. pombe (Dunleavy et al. 2007). Recently, HJURP in humans has been shown to be required for CENP-A deposition (Dunleavy et al. 2009; Foltz et al. 2009). Interestingly, HJURP appears to be a distant homolog of the budding and fission yeast Scm3 protein (Sanchez-Pulido et al. 2009), a CENP-A binding protein required for CenH3 localization and kinetochore function (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007; Pidoux et al. 2009; Williams et al. 2009).
In addition to centromere-targeting proteins, specific CenH3 localization is also regulated by proteolysis. For example, in budding yeast and Drosophila, ubiquitin-mediated proteolysis is responsible for removing any CenH3 molecules not localized to the centromere (Collins et al. 2004; Moreno-Moreno et al. 2006). To understand this centromeric-specific protection from degradation, we and others have studied mutants that misregulate the normal localization pattern of Cse4 (Sharp et al. 2002; Collins et al. 2004, 2007; Crotti and Basrai 2004; Au et al. 2008).
This study investigates the extracentromeric deposition of Cse4 observed in the absence of histone H3/H4 deposition proteins. We show that extracentromeric Cse4 was not sufficient to recruit the kinetochore protein Mif2. Moderate increases in Cse4 protein stability were observed in the absence of CAF-1 and/or HIR proteins. However, cytological and microarray-based detection of Cse4 mislocalization was only observed in the absence of both complexes, indicating that moderate Cse4 stabilization alone (as seen in the single mutants) is insufficient for Cse4 mislocalization. Additionally, thermal inactivation of RNA polymerase II modestly increased Cse4 half-life, suggesting that eviction of mislocalized Cse4 molecules partially occurs in a transcription-dependent manner. Finally, genomic analyses of Cse4 localization and H3/H4 turnover in chromatin assembly mutants suggest that altered histone eviction may contribute to aberrant localization of Cse4 when multiple pathways for histone H3/H4 deposition are compromised.
MATERIALS AND METHODS
Yeast strains and plasmids:
All yeast strains used in this study (Table 1) are isogenic with W303 (Thomas and Rothstein 1989). All gene deletions used in this study were previously described (Sharp et al. 2002). Construction of the NDC10–GFP integrated allele and pCSE4–HA plasmids were described previously (Meluh et al. 1998; Zeng et al. 1999). The pCSE4–GFP plasmid was a gift from M. Fitzgerald-Hayes (Chen et al. 2000) and the pGAL1–CSE4–myc12 integrating construct (pSB245) was obtained from S. Biggins (Collins et al. 2004).
TABLE 1.
Yeast strains used
| Strain number | Genotype | Reference |
|---|---|---|
| PKY3054 | MATα; lys2Δ; hmlΔ∷LEU2; cse4Δ∷kanMX + pCSE4-GFP-TRP1 | This study |
| PKY3057 | MATa; cse4Δ∷kanMX; cac1Δ∷LEU2; hir1Δ∷HIS3 + pCSE4-GFP-TRP1 | This study |
| PKY2443 | MATα; cse4Δ∷kanMX; ndc10∷NDC10-GFP∷HIS3 + pCSE4-HA-TRP1 | Sharp et al. (2002) |
| PKY2453 | MATa; cse4Δ∷kanMX; ndc10∷NDC10-GFP∷HIS3; cac1Δ∷hisG-URA3-hisG; hir1Δ∷HIS3 + pCSE4-HA-TRP1 | Sharp et al. (2002) |
| PKY3412 | MATa; ura3∷GALp-CSE4-myc12∷URA3 | This study |
| PKY3413 | MATα; cac1Δ∷LEU2; ura3∷GALp-CSE4-myc12∷URA3 | This study |
| PKY3414 | MATa; hir1Δ∷HIS3; ura3∷GALp-CSE4-myc12∷URA3 | This study |
| PKY3415 | MATa; cac1Δ∷LEU2; hir1Δ∷HIS3; ura3∷GALp-CSE4-myc12∷URA3 | This study |
| PKY4233 | MATα; ura3∷GALp-CSE4-myc12∷URA3; rpb1-1 | This study |
| PKY4323 | MATa; bar1-1; hmlΔ∷TRP1 + pHHF1-GAL10/1p-FLAG-HHT1 | This study |
| PKY4324 | MATa; bar1-1; hmlΔ∷TRP1; cac1Δ∷hisG-URA3-hisG + pHHF1-GAL10/1p-FLAG-HHT1 | This study |
| PKY4325 | MATa; bar1-1; hmlΔ∷TRP1; hir1Δ∷HIS3 + pHHF1-GAL10/1p-FLAG-HHT1 | This study |
| PKY4326 | MATa; bar1-1; hmlΔ∷TRP1; cac1Δ∷hisG-URA3-hisG; hir1Δ∷HIS3 + pHHF1-GAL10/1p-FLAG-HHT1 | This study |
| PKY2299 | MATa; cse4Δ∷kanMX + pCSE4-HA-TRP1 | This study |
| PKY2295 | MATa; cac1Δ∷hisG-URA3-hisG; cse4Δ∷kanMX; hmlΔ∷LEU2 + pCSE4-HA-TRP1 | This study |
| PKY2297 | MATa; hir1Δ∷HIS3; cse4Δ∷kanMX; hmlΔ∷LEU2 + pCSE4-HA-TRP1 | This study |
| PKY2300 | MATa; cac1Δ∷hisG-URA3-hisG; hir1Δ∷HIS3; hmlΔ∷LEU2; cse4Δ∷kanMX + pCSE4-HA-TRP1 | This study |
All strains are isogenic with the W303 (Thomas and Rothstein 1989) background and are leu2-3, 112; ura3-1; his3-11,15; trp1-1; ade2-1; can1-100 in addition to the genotypes indicated.
Immunoblotting:
Detection of Cse4–Myc in alkaline lysates of cells (Kushnirov 2000) was performed with the 9E10 anti-Myc monoclonal antibody (Santa Cruz). PCNA was detected using a rabbit polyclonal antibody (Daganzo et al. 2003) at 1:10,000 dilution. Histone H3 was detected using a rabbit polyclonal antibody (Abcam) at 1:2000 dilution.
Chromosome spreads:
Chromosome spreads of yeast strains were processed as previously described (Sharp et al. 2002). Cultures were grown to an optical density (600 nm) of 1 in yeast extract-peptone 2% dextrose media (YPD) for cells expressing Cse4–HA or in YP + 2% galactose for cells expressing GAL1-driven Cse4–myc. Cse4 detection was performed using a polyclonal anti-HA antibody (Clontech) or the 9E11 anti-myc antibody (Delta Biolabs) at 0.02 μg/ml. The rabbit anti-Mif2 antibody was a generous gift from P. Meluh and used at a dilution of 1:750. Secondary antibodies were Cy3-coupled anti-rabbit IgG (Jackson Labs) or FITC anti-mouse IgG at 1 μg/ml. Counterstaining was performed with DAPI at 1 μg/ml.
For chromosome spreads of cells expressing GAL1–CSE4–myc12, yeast cells were grown in YP + 2% galactose until mid-log phase. The cells were then washed and resuspended with YP + 5% glucose for continued growth at 30°. At each time point, a sample of growing culture was removed and added to ice cold 7.5 mm Tris–Cl pH 7.5, 0.01% sodium azide. After mixing, cells were harvested and processed for chromosome spreads and immunoflourescence as previously described (Loidl et al. 1998; Sharp et al. 2002).
Chromatin association:
Yeast strains were grown to mid-log phase in rich yeast extract-peptone (YP) + 2% raffinose. Cultures were centrifuged and resuspended into YP + 2% galactose media to induce GAL promoter-driven CSE4–myc12 expression. After 3 hr, the cells were centrifuged and transferred into 200 ml YP + 2% dextrose media containing 400 μm cycloheximide to simultaneously repress CSE4–myc12 transcription and inhibit protein translation. At 0, 30, 60, and 120 min after shutoff, 50-ml samples were collected into 1% sodium azide and centrifuged at 4° for 5 min at 1500 relative centrifugal force (rcf). Pellets were frozen in liquid nitrogen and stored at −80° until processing.
Cultures were grown at 30° except those comparing the rpb1-1 temperature-sensitive strain to wild type (wt). These were grown at 23° in YP + raffinose, then transferred to prewarmed, 37° YPD + 400 μm cycloheximide for the dextrose/cycloheximide shutoff.
Cell pellets were processed for chromatin association as previously described (Donovan et al. 1997; Liang and Stillman 1997). Briefly, cells were washed with water and resuspended into 1.5 ml 10 mm DTT, 100 mm PIPES, pH 9.6 and incubated at 30° for 30 min. Cells were centrifuged at 1500 rcf and resuspended with 2 ml spheroplast buffer (0.6 m sorbitol, 50 mm KPO4, pH 7.5) with 0.5 mm PMSF (phenylmethylsulfonyl fluoride). Cell wall digestion was performed with 80 μg/ml T20 zymolyase (Seikagaku) at room temperature. Digestion was monitored by reading A600 of cells diluted in 1% SDS and allowed to proceed until A600 reached 10% of the starting material. Spheroplasts were placed on ice and centrifuged for 10 min at 4°, 1500 rcf. The pellets were gently resuspended into 1 ml lysis buffer (0.4 m sorbitol, 150 mm KOAc, 3 mm MgAc) with protease inhibitors (0.5 mm PMSF, 1 μg ml−1 aprotinin, 0.16 mg ml−1 benzamidine, 0.5 μg ml−1 leupeptin and 0.7 μg ml−1 pepstatin) and centrifuged at 1500 rcf in a microcentrifuge for 5 min at 4°. The nuclear pellets were resuspended with 600 μl lysis buffer, 1% Triton X-100, mixed and placed on ice for 5 min. A total of 200 μl was stored as “total extract.” The rest was centrifuged at 20,000 rcf for 10 min at 4°. The supernatant was collected and stored as the soluble supernatant fraction. The pellet was washed again in lysis buffer at 20,000 rcf for 10 min and resuspended in 200 μl and stored as the chromatin-associated pellet fraction. Samples were analyzed on 15% SDS PAGE gels (18% when H3 analyzed). Chemiluminescent signals were measured on a Fuji LAS4000 densitometer, and the ratios of the MYC–Cse4 signal were normalized to the PCNA or histone H3 loading controls using MultiGauge software.
Live cell imaging:
Yeast cells expressing Cse4–GFP were grown to log phase in YPD. Cells were treated with Hoechst to stain the DNA and spotted onto an agar pad for imaging with the ×100 objective of Nikon Eclipse E600 microscope equipped with a Hamamatsu CCD camera controlled by ImageQuant software. For quantification, >100 cells were analyzed for each strain in two independent experiments.
Histone turnover measurement:
Strains were transformed with plasmid pHHF1–GAL10/1–FLAG–HHT1. Cell growth, galactose induction, micrococcal nuclease digestion and microarray analyses were all performed as described (Kaplan et al. 2008).
Localization analysis of Cse4-HA:
ChIP/Chip was carried out as previously described in Liu et al. (2005) with the following exceptions: Cse4–HA strains (PKY2299–wt, 2295–cac1Δ, 2297–hir1Δ, 2300–cac1Δ hir1Δ) were grown to mid-log phase (OD600 of between 0.38 and 0.54) in YPD and then fixed with formaldehyde, spheroplasted and lysed, and digested to ∼80% mononucleosomal DNA with MNase. Samples were immunoprecipitated with 7.5 μl of anti-HA antibody (Abcam ab-9110). Amplification, labeling, hybridization, and analysis were carried out as described in Liu et al. (2005). Microarray data sets are available in supporting information, File S1.
RESULTS
Extracentromeric deposition of Cse4 in live cells:
When subunits of the chromatin assembly factors CAF-1 and the HIR complex are both deleted, Cse4 localizes not only to the centromere but also to noncentromeric chromatin (Sharp et al. 2002). This had previously been detected via “chromosome spreads,” during which cells are lysed onto glass slides, allowing for analysis of the spread chromatin by indirect immunofluorescence (Loidl et al. 1998). In budding yeast, centromeres cluster and appear as one or two foci per nucleus (Guacci et al. 1997). In wild-type cells, Cse4 therefore appears in one or two centromeric foci in each nucleus (Meluh et al. 1998). In contrast, in cac1Δ hir1Δ double mutants, Cse4 is dispersed throughout chromatin (Sharp et al. 2002). Cells containing single cac1Δ or hir1Δ mutations do not display this phenotype (Sharp et al. 2002).
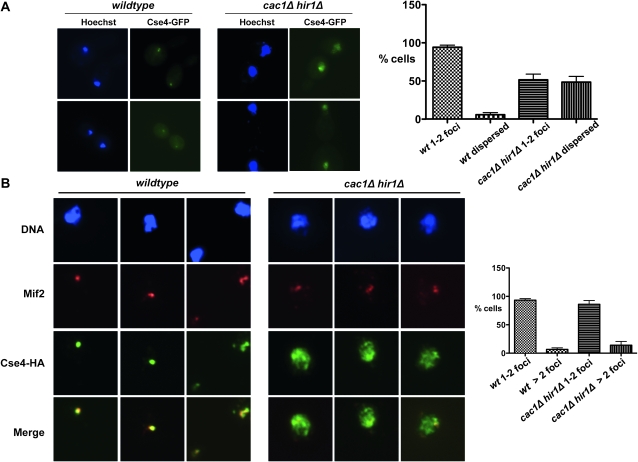
The previously reported chromatin spread experiments could not distinguish whether Cse4 first becomes mislocalized in vivo or only after the harsh cell lysis and fixation steps inherent to the chromosome spread protocol. Therefore, we examined the localization of a Cse4–GFP fusion protein in live cells to avoid artifacts due to lysis and fixation. We observed that Cse4–GFP localized to one or two small, discrete foci in >90% of wild-type cells, indicative of proper centromere localization (Figure 1A). This was also true for cac1Δ and hir1Δ single mutant cells (data not shown), consistent with results with lysed, fixed cells (Sharp et al. 2002). However, ∼50% of nuclei in cac1Δ hir1Δ double mutants displayed a larger, more dispersed area of Cse4–GFP, overlapping much of the DAPI-stained nuclei. These data indicate that Cse4 mislocalization indeed occurs in live cac1Δ hir1Δ cells. Additionally, the chromatin immunoprecipitation studies discussed below support this conclusion (Figure 6).
Figure 1.—
Extracentromeric Cse4 in the absence of CAF-1 and HIR complexes. (A) Live cell imaging of wild-type (PKY3054) and cac1Δ hir1Δ (PKY3057) cells that produce Cse4–GFP. Cells were treated with Hoechst to stain the DNA. Percentages of cells with either dispersed Cse4–GFP localization or 1–2 small foci are indicated on the right, with mean and SEM bars shown for two experiments, each with n > 100 cells. 1–2 foci: wild-type 94.25 ± 2.75%, cac1Δ hir1Δ 51.5 ± 7.5%; dispersed: wild-type 5.75 ± 2.75%, cac1Δ hir1Δ 48.5 ± 7.5%. Unpaired, two-tailed t-tests showed that the wild-type and cac1Δ hir1Δ samples were significantly different (P < 0.035 for both 1–2 foci and dispersed classes). (B) Kinetochore protein Mif2 is not recruited to extracentromeric Cse4 in cac1Δ hir1Δ cells. Chromosome spreads were prepared from wild-type (PKY2443) and cac1Δ hir1Δ (PKY2453) cells expressing Cse4–HA under the control of its endogenous promoter. Mouse anti-Mif2 and rabbit anti-HA were used to detect endogenous Mif2 and Cse4–HA. DNA is stained with DAPI. Percentages of cells with 1–2 or > 2 Mif2 foci is indicated on the right, with average and SEM bars shown for three experiments, each with n > 100 cells. 1–2 foci: wild-type 93.33 ± 2.73%, cac1Δ hir1Δ 86 ± 6.66%; >2 foci: wild-type 6.67 ± 2.73%, cac1Δ hir1Δ 14 ± 6.66%. Mann–Whitney nonparametric, two-tailed t-tests showed that the wild-type and cac1Δ hir1Δ samples were not significantly different (P > 0.5 for both 1–2 foci and > 2 foci classes).
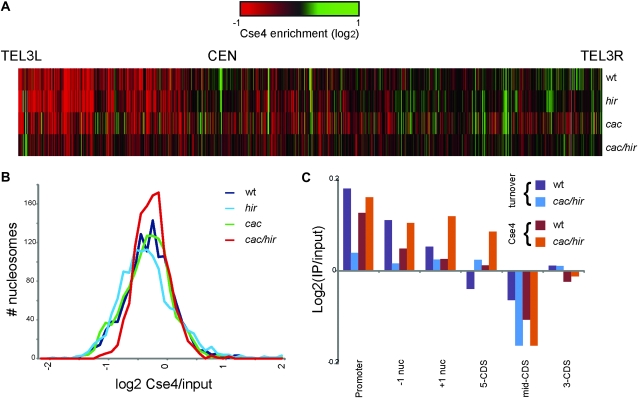
Figure 6.—
Genome-scale localization of Cse4–HA in wt (PKY2299), cac1Δ (PKY2295), hir1Δ (PKY2297), and cac1Δ hir1Δ (PKY2300) cells detected via chromatin IP/microarray analysis. (A) Heat map of Cse4 enrichment along yeast chromosome III. (B) Distribution of Cse4 signal per nucleosome in each strain. cac1Δ hir1Δ cells display a narrower distribution than the other three strains. (C) Correlation of Cse4 localization with rates of H3 turnover (Figure 5) at various types of genomic locus.
Extracentromeric Cse4 does not recruit kinetochore protein Mif2:
In human and Drosophila cells, extracentromeric deposition of CenH3 can be achieved by overexpression of the protein (Van Hooser et al. 2001; Tomonaga et al. 2003; Heun et al. 2006). When CenH3 is localized to noncentromeric chromatin under these conditions, it is able to recruit inner kinetochore proteins (Van Hooser et al. 2001; Heun et al. 2006) and form ectopic kinetochores in some cases, even in the absence of centromere-specific DNA (Heun et al. 2006). In particular, in these metazoan cells, CENP-A is able to recruit the essential kinetochore protein CENP-C to DNA. Because ectopic kinetochores could contribute to the chromosome loss and nondisjunction observed in cac1Δ hir1Δ cells (Sharp et al. 2002, 2003; Sharp and Kaufman 2003), we examined the localization of Mif2, the CENP-C homolog in budding yeast (Meluh and Koshland 1995; Figure 1B). In chromatin from wild-type cells, Mif2 and Cse4–HA colocalized in >90% of cells at centromere clusters observed as one or two foci per nuclei. However, in cac1Δ hir1Δ mutants, Cse4 was dispersed throughout the chromatin, but Mif2 remained confined to one or two foci per nuclei. Quantitative analysis of three experiments showed that the percentage of cells with Mif2 in one to two foci was not significantly different in wild-type or cac1Δ hir1Δ cells (Figure 1B). Therefore, we favor the idea that the increased chromosome loss and nondisjunction observed in cac1Δ hir1Δ cells (Sharp et al. 2002, 2003; Sharp and Kaufman 2003) results from centromeric chromatin defects rather than titration of kinetochore components, although mislocalization of kinetochore proteins other than Mif2 has not been ruled out in these studies.
Protein stability of Cse4 in chromatin assembly mutants:
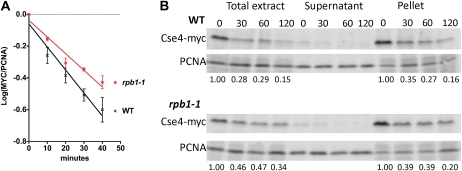
Studies in budding yeast and fruit flies have shown that CenH3 is subject to ubiquitin-mediated proteolysis, and that preferential degradation of CenH3 at noncentromeric loci contributes to its specific localization at centromeres (Collins et al. 2004; Moreno-Moreno et al. 2006). In budding yeast, when proteolysis of Cse4 is inhibited by either loss of proteasome function or by mutation of Cse4 lysine residues, Cse4 remains localized to the centromere, but is also observed at noncentromeric sites (Collins et al. 2004, 2007). In addition, overexpression of certain cse4 alleles results in spindle checkpoint activation (Collins et al. 2007) reminiscent of that observed in cac1Δ hir1Δ cells (Sharp et al. 2002). We therefore tested whether Cse4 protein is stabilized in cac1Δ hir1Δ mutants, analyzing the stability of Cse4 tagged with a 12×-myc epitope and expressed under the control of the GAL1 promoter. Yeast were grown in galactose and then treated simultaneously with glucose to repress CSE4–myc12 gene transcription and cycloheximide to repress translation. Cells were harvested at the indicated times, and nuclei were prepared and separated into soluble and chromosome-bound fractions and analyzed via immunoblotting (Figure 2). As observed for canonical histone H3 (Gunjan and Verreault 2003), very little Cse4 was found in the soluble fractions, precluding accurate quantitation. Focusing on the chromatin-associated proteins in the pellet fractions, we observed that modestly elevated levels of Cse4–myc persisted in cac1Δ hir1Δ and cac1Δ hir1Δ mutant cells over the 2-hr period analyzed. Because similar differences were observed in both the single mutants that do not display Cse4 mislocalization and the cac1Δ hir1Δ cells that do, these data suggest that a moderate increase in Cse4 protein stability is insufficient to explain mislocalization in cac1Δ hir1Δ cells.
Figure 2.—
Protein stability and chromosome association of Cse4. The stability of Cse4–myc was monitored in wild-type (PKY3412), cac1Δ (PKY3413), hir1Δ (PKY3414), and cac1Δ hir1Δ (PKY3415) strains. Strains were grown in raffinose, and CSE4–myc12 expression was induced by galactose for 3 hr. At time 0, cells were shifted to media containing dextrose and cycloheximide to shutoff CSE4–myc12 expression. At the indicated time points (in minutes), cells were harvested, and nuclei were prepared, separated into soluble and chromosome-bound pellet fractions, and analyzed by immunoblotting. PCNA serves as the loading control for the soluble extracts, and histone H3 is the loading control for the chromosome-bound pellet material. For the pellets, the ratios of background-subtracted Cse4–myc and PCNA signals were calculated and normalized to 1.0 at time 0; values at each time point are shown beneath each lane.
In wild-type cells, Cse4 localized to euchromatin is degraded, but Cse4 associated with centromeres appears to be protected from proteolysis (Collins et al. 2004). To address whether the centromeric and euchromatic pools of Cse4 are degraded differently in chromatin assembly mutants, we performed chromosome spreads to analyze the localization of Cse4–myc during a time course after transcriptional repression, comparing wild-type and cac1Δ hir1Δ cells (Figure 3). In wild-type cells, the large Cse4 pool resulting from GAL1-driven overexpression was initially associated with noncentromeric chromatin, as evidenced by the enlarged Cse4 foci relative to the Mif2 foci in many nuclei. However, this pool of Cse4 appeared to be rapidly degraded because within 1–2 hr of transcriptional repression, Cse4–myc remained only at the centromeres, overlapping the Mif2 foci. In cac1Δ hir1Δ mutants, Cse4–myc localized throughout the chromatin in almost all nuclei. However, the noncentromeric Cse4 was degraded relatively rapidly in almost all nuclei, whereas the centromeric Cse4 was largely protected from proteolysis, similar to the wild-type cells. These results show that euchromatin-associated Cse4 is degraded more readily than centromeric Cse4 even in cac1Δ hir1Δ cells. Together with the protein stability measurements (Figure 2), these results lead us to conclude that Cse4 proteolysis in cac1Δ hir1Δ cells is largely normal at noncentromeric loci.
Figure 3.—
Chromosome spreads were performed on wild-type (PKY3412) and cac1Δ hir1Δ (PKY3415) cells that express CSE4–myc12 driven by the GAL1 promoter. Cells were grown in galactose-containing media and switched to glucose-containing media. Samples were taken at 0, 1, and 2 hr after repression with glucose. Immunofluorescence was performed with a rabbit anti-Mif2 antibody to visualize centromeres and a mouse anti-myc antibody to detect Cse4–myc. DNA was stained with DAPI. To image Cse4–myc in cac1Δ hir1Δ cells, the camera exposure time was half as long as that for Cse4–myc images for other cell types.
Effect of RNA polymerase II inactivation on Cse4 half-life:
Exchange of histones H3 and H4 into and out of chromatin is mediated by histone chaperones (Schwabish and Struhl 2006; Rufiange et al. 2007; Kaplan et al. 2008; Park and Luger 2008). Histone H3/H4 exchange also often occurs with the passage of RNA polymerase II at high transcription rates, although histone exchange events apparently unrelated to RNA polymerase movement are also common at many loci (Dion et al. 2007; Rufiange et al. 2007; Jamai et al. 2009). We hypothesized that ongoing transcription might also affect the half-life of the centromeric histone, Cse4. To test this, we monitored the rate of degradation of Cse4–myc in both wild-type yeast and yeast harboring a temperature-sensitive allele of RNA polymerase II, rbp1-1. Cells were first grown at the permissive temperature in galactose to induce overexpression of CSE4–myc, and then transferred to glucose media containing cycloheximide at the nonpermissive temperature to repress simultaneously GAL-driven CSE4 expression, global RNA pol II transcription, and protein translation. We observed that the stability of Cse4–MYC was modestly increased in rbp1-1 cells, both at early (Figure 4A) and later time points (Figure 4B). Cell fractionation detected Cse4 stabilization in the rbp1-1 cells in whole cell extracts and in chromatin pellet fractions, although the differences from wild-type levels were greater in the former. These data suggest that nucleosome disruption related to RNA pol II transcription contributes to the eviction of Cse4 molecules misdeposited at noncentromeric loci, at least in wild-type cells. The relationship between histone turnover and Cse4 localization is explored in more detail below.
Figure 4.—
Thermal inactivation of RNA polymerase II mildly increases Cse4 stability. The stability of Cse4–myc was monitored in wild-type (PKY3412) and rpb1-1ts (PKY4233) cells as described for Figure 2A, except that cells were grown at the permissive temperature (23°) during galactose treatment and then transferred to prewarmed YP + 2% dextrose + 400 μm cycloheximide at the restrictive temperature (37°) to shut off Cse4–myc synthesis. (A) Loss of Cse4–myc in total extracts was quantitatively analyzed in two experiments and shown to differ significantly (P-value = 0.028). Cells were grown in galactose for 2 hr and samples were collected at 0, 10, 20, 30, and 40 min after shutoff. Ratios of Cse4–myc to PCNA were quantified and normalized as in Figure 2. The log10 ratios and standard error of the mean (SEM) were graphed using GraphPad Prism software. (B) Cells were grown at the permissive temperature (23°) during galactose treatment for 3 hr prior to transfer to YPD + cycloheximide at 37°. Samples were collected at 0, 30, 60, and 120 min after shutoff, and whole cell and fractionated extracts were analyzed by immunoblotting. For the whole cell and pellet fractions, the ratios of background-subtracted Cse4–myc and PCNA signals were calculated using MultiGauge software and normalized to 1.0 at time 0; values at each time point are shown beneath each lane.
Genome-scale analysis of histone turnover in chromatin assembly mutants:
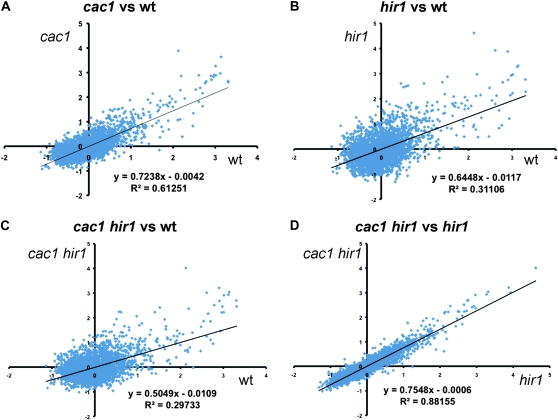
Because the CAF-1 and HIR complexes are both involved in chromatin assembly, we hypothesized that altered histone dynamics would be observed in cac1Δ hir1Δ double mutant cells. To test this, we compared the extent of replication-independent FLAG–H3 incorporation (Schermer et al. 2005; Rufiange et al. 2007; Dion et al. 2007; Jamai et al. 2009) in wild-type, cac1Δ, hir1Δ and cac1Δ hir1Δ cells arrested in G1, after 90 min of pGAL-driven transcriptional induction of the tagged histone (Figure 5). Several aspects of the data were striking. First, cac1Δ, hir1Δ and cac1Δ hir1Δ cells all displayed reduced FLAG–H3 incorporation relative to wild-type cells, demonstrating among other things a replication-independent role for CAF-1 in histone dynamics (Figures 5, A–C). Second, H3 turnover slows to a greater extent in the hir1Δ cells than in the cac1Δ cells (Figure 5B). Similar results were observed at t = 45 min (not shown). We therefore conclude that both CAF-1 and HIR complexes accelerate the rate of histone H3 incorporation in vivo, although it is important to note that in this assay we cannot distinguish between effects on new histone deposition and effects on old histone eviction.
Figure 5.—
Altered rates of histone turnover in chromatin assembly mutants. Yeast strains (PKY4323-6) all carried a FLAG-tagged histone H3 gene driven by the inducible GAL1 promoter. Epitope tag incorporation into nucleosomes was measured 90 min after induction of FLAG–H3 expression in duplicate experiments via chromatin IP/microarray analysis. Normalized rates for two strains are graphed relative to each other. Linear regression fits to the data are shown. (A) cac1 vs. wt. (B) hir1 vs. wt. (C) cac1 hir1 vs. wt. (D) cac1 hir1 vs. hir1.
Since cac1Δ hir1Δ double mutants exhibit pronounced Cse4 mislocalization that is not seen in either single mutant (Sharp et al. 2002), we compared defects in H3 turnover in the cac1Δ hir1Δ double mutant to the hir1Δ single mutant to determine whether any loci were specifically affected only in cac1Δ hir1Δ cells (Figure 5D). In the absence of Hir1, further loss of Cac1 caused relatively uniform effects on turnover rates across the genome, with an average ∼25% reduction in FLAG–H3 incorporation (Figure 5D). Importantly, the good correlation between cac1Δ hir1Δ and hir1Δ data indicates that there are no genomic loci (over the 4% of the genome interrogated) for which Cac1 and Hir1 play fully redundant roles in turnover, as this would manifest as points off the diagonal in Figure 5D.
Genome-scale analysis of Cse4 localization in wild-type and mutant cells:
To determine the genomic location of the mislocalized Cse4, we performed mononucleosome-resolution chromatin immunoprecipitation experiments (Figure 6). These were analyzed on arrays designed to interrogate nucleosomes across all of chromosome III as well as selected promoter regions throughout the genome (Yuan et al. 2005). In wild-type, cac1Δ, and hir1Δ yeast, Cse4 was most strongly localized at CEN3, as expected, although as previously described (Camahort et al. 2009; Lefrancois et al. 2009) some minor sites of Cse4 localization could be observed even in wild-type yeast, such as tRNA genes (Figure 6A and analysis not shown). In stark contrast, Cse4 was extensively mislocalized at loci other than CEN in cac1Δ hir1Δ double mutants, extending our low-resolution chromosome spread results to single-nucleosome resolution. Because Cse4 is highly enriched at a small number of genomic loci in wild type, extensive mislocalization should manifest as a decrease in the dynamic range of microarray signals. Indeed, we observed a specific decrease in dynamic range in Cse4 localization in cac1Δ hir1Δ cells, but not in either single mutant (Figure 6B). Since Cse4 is maintained at CEN sequences in cac1Δ hir1Δ cells (Sharp et al. 2002), we therefore conclude that Cse4 appears at a larger number of extracentromeric loci in cac1Δ hir1Δ cells.
Where does Cse4 mislocalize in cac1Δ hir1Δ double mutants? Because of the role of histone exchange factors in Cse4 mislocalization, we compared the location of Cse4 incorporation relative to our histone turnover data. We specifically examined classes of nucleosomes at different positions relative to genes (Figure 6C). For example, previous analyses had shown that promoter nucleosomes and the +1 and −1 nucleosomes flanking transcriptional start sites tend to be rapidly exchanging in wild-type cells (Dion et al. 2007), and these exchange more slowly in cac1Δ hir1Δ cells (Figure 5C). Notably, the +1 and −1 nucleosomes also acquire higher levels of Cse4 in cac1Δ hir1Δ cells than in wt cells. However, Cse4 does not uniformly relocalize to genomic loci where turnover slows in the cac1Δ hir1Δ mutant. Specifically, nucleosomes associated with coding regions did not become associated with Cse4 despite very low H3/H4 exchange rates (Figure 6C). Therefore, different mechanisms may regulate Cse4 eviction at different locations, with chaperone-mediated turnover predominating as a Cse4 eviction mechanism at promoters.
DISCUSSION
The role of histone chaperones in CenH3 regulation:
CAF-1 is believed to be largely responsible for replication-coupled histone H3/H4 deposition, while the HIR complex is responsible for replication-independent H3/H4 deposition (Tagami et al. 2004; Green et al. 2005). Previously, defective localization of the centromeric histone variant, Cse4, was observed in cac1Δ hir1Δ double mutants, but not in single cac1Δ or hir1Δ mutants (Sharp et al. 2002). Therefore, Cse4 mislocalization occurs in cells in which both replication-coupled and replication-independent H3/H4 deposition have been compromised. Additionally, proper stoichiometry between Cse4 and histone H3 is important for the localization and function of Cse4 (Au et al. 2008). Together, these studies suggested that the disruption of normal histone deposition may promote extracentromeric deposition of Cse4 or prevents its removal from chromatin, or both.
A landmark discovery was that extracentromeric deposition of Cse4 is observed when degradation of the protein is compromised, suggesting that there may not be a specific mechanism for targeted deposition of Cse4 at centromeres (Collins et al. 2004). Instead, the observed enrichment of Cse4 at centromeres appears to result from protection of CEN-deposited Cse4 from proteolysis. Therefore, although deposition per se seemed not be a regulated event, we sought to understand why histone chaperones are required for the normal localization of Cse4.
Cse4 protein stability:
In previous work (Collins et al. 2004; Crotti and Basrai 2004), overexpression of CSE4 in wild-type budding yeast did not cause mislocalization of the protein. In contrast, in higher eukaryotes, overproduction of CenH3 (termed CENP-A in mammals and CID in flies) is sufficient to cause mislocalization of the protein (Van Hooser et al. 2001; Tomonaga et al. 2003; Heun et al. 2006). Therefore, in budding yeast a major contributor to the normal CenH3 localization is a proteolytic surveillance mechanism that degrades Cse4 molecules not shielded by centromeric chromatin. Although this type of mechanism has also been observed in flies (Moreno-Moreno et al. 2006), it appears to be especially important in budding yeast.
Given that mutations in chromatin assembly factors and mutations that inhibit Cse4 proteolysis show similar extracentromeric localization of the protein, we speculated that Cse4 might be stabilized in the absence of functional CAF-1 and HIR complex. However, the rates of proteolysis of Cse4–MYC were similar in both single and double mutant backgrounds, indicating that the extracentromeric deposition of Cse4 in these mutants is not solely due to increased protein stabililty. Our immunofluorescence data also suggest that the euchromatin-associated Cse4 is not protected from proteolysis even in the mutant cells, suggesting that the main difference between strains is the steady-state levels of Cse4 in chromatin rather than Cse4 half-life. These data suggested that altered levels of histone turnover, rather than altered Cse4 protein stability, may contribute to the observed phenotypes.
Kinetochore protein recruitment:
We tested whether the ectopic sites of Cse4 in chromatin assembly mutants are able to recruit other kinetochore proteins by analyzing the localization of Mif2, the CENP-C homolog (Meluh and Koshland 1995). In cac1Δ hir1Δ cells, Mif2 is not recruited to the mislocalized Cse4 (Figure 1B). This result differs from what is observed in flies and mammals, where other kinetochore proteins are recruited to the sites of extracentromeric CenH3 deposition (Van Hooser et al. 2001; Tomonaga et al. 2003; Heun et al. 2006), suggesting that there are additional requirements for kinetochore protein recruitment in budding yeast. Consistent with this idea. de novo kinetochore assembly requires kinetochore protein Chl4 for the recruitment and function of several other kinetochore proteins, including Cse4 (Mythreye and Bloom 2003). We hypothesize that the different observations in different species may reflect the special nature of the budding yeast point centromere, which is specified not only by the association of CenH3 with the CDE II region, but also by the essential site-specific CBF III complex, which is required for formation of pericentric chromatin loops (Anderson et al. 2009).
The effect of histone chaperones on histone turnover:
We show here that the CAF-1 and HIR histone chaperones both affect the rate of replication-independent histone H3 incorporation in vivo. In particular, hir1Δ mutants display a nearly twofold reduction in turnover rates of slowly exchanging nucleosomes (Figure 5), which are particularly enriched over genic coding regions (Dion et al. 2007). These data are consistent with previous studies that implicate the yeast HIR complex in histone exchange during transcriptional elongation (Formosa et al. 2002), and for suppression of cryptic transcription from within ORFs (Cheung et al. 2008). The effects of a cac1Δ deletion are less pronounced and more uniform than a hir1Δ deletion, additionally reducing turnover in cells lacking the HIR complex by ∼25% across the genome (Figure 5D). This is consistent with CAF-1 having roles not only in genome-wide deposition during S phase (Smith and Stillman 1989; Tagami et al. 2004), but also in locus-specific histone replacement outside of S phase (Enomoto and Berman 1998). Together, these data further illustrate the different roles for the CAF-1 and HIR complexes in chromosome biology.
What regions of the genome are particularly susceptible to excessive accumulation of extracentromeric Cse4? In wild-type cells, prior studies have described extracentromeric Cse4 at ribosomal protein genes (Lefrancois et al. 2009), tRNA genes (Camahort et al. 2009), and other loci known to exhibit high rates of histone turnover (Dion et al. 2007). Consistent with this observation, we observe relatively high levels of extracentromic Cse4 at “hot” loci such as promoter nucleosomes (Figure 6C). Interestingly, at these genomic loci, reduced histone exchange in cac1Δ hir1Δ cells correlates with increased Cse4 association (Figure 6C), suggesting that Cse4 is normally incorporated at some frequency at promoter nucleosomes, and it is decreased eviction at promoters in the cac1Δ hir1Δ mutant that results in increased Cse4 association.
However, this simple mechanism cannot explain changes at all loci. For example, coding region nucleosomes are slowly exchanged both in wt and cac1Δ hir1Δ cells, yet they are not particularly enriched for Cse4 in either strain. Decreased histone replacement at coding regions in the double mutant also does not result in increased Cse4 binding, consistent with the possibility that Cse4 eviction mechanisms differ at promoters and coding regions. Since the rate of degradation of Cse4 increases somewhat in the absence of on-going transcription by RNA polymerase II (Figure 4), we therefore propose that RNA polymerase plays a role in Cse4 eviction over coding regions, whereas histone chaperones play the major role in Cse4 eviction at promoters. In this manner, non-CEN deposited Cse4 molecules may serve as useful probes of the complex events that exchange histones throughout the genome.
Acknowledgments
We thank Sue Biggins for strains and plasmids and Pam Meluh for the anti-Mif2 antisera. This work was supported by National Institutes of Health (NIH) R01 GM55712 (to P.D.K.), NIH F31 AI 078726 (to J.L.S.), and NIH R01 GM079205 and the Burroughs Wellcome Fund (O.J.R.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123117/DC1.
References
- Anderson, M., J. Haase, E. Yeh and K. Bloom, 2009. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol. Biol. Cell 20 4131–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au, W. C., M. J. Crisp, S. Z. DeLuca, O. J. Rando and M. A. Basrai, 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, K., 2007. Centromere dynamics. Curr. Opin. Genet. Dev. 17 151–156. [DOI] [PubMed] [Google Scholar]
- Bloom, K. S., and J. Carbon, 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29 305–317. [DOI] [PubMed] [Google Scholar]
- Bram, R. J., and R. D. Kornberg, 1987. Isolation of a Saccharomyces cerevisiae centromere DNA-binding proteins, its human homolog, and its possible role as a transcription factor. Mol. Cell. Biol. 7 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort, R., B. Li, L. Florens, S. K. Swanson, M. Washburn et al., 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26 853–865. [DOI] [PubMed] [Google Scholar]
- Camahort, R., M. Shivaraju, M. Mattingly, B. Li, S. Nakanishi et al., 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., R. E. Baker, K. C. Keith, K. Harris, S. Stoler et al., 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, V., G. Chua, N. Batada, C. Landry, S. Michnick et al., 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6 e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, K. A., S. Furuyama and S. Biggins, 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14 1968–1972. [DOI] [PubMed] [Google Scholar]
- Collins, K. A., R. Camahort, C. Seidel, J. L. Gerton and S. Biggins, 2007. The overexpression of a Saccharomyces cerevisiae centromeric histone H3 mutant protein leads to a defect in kinetochore biorientation. Genetics 175 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti, L. B., and M. A. Basrai, 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Sacchromyces cerevisiae. EMBO J. 23 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daganzo, S. M., J. P. Erzberger, W. M. Lam, E. Skordalakes, R. Zhang et al., 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13 2148–2158. [DOI] [PubMed] [Google Scholar]
- Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman et al., 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315 1405–1408. [DOI] [PubMed] [Google Scholar]
- Donovan, S., J. Harwood, L. S. Drury and J. F. Diffley, 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 94 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy, E. M., A. Pidoux, M. Monet, C. Bonilla, W. Richardson et al., 2007. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell 28 1029–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy, E. M., D. Roche, H. Tagami, N. Lacoste, D. Ray-Gallet et al., 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137 485–497. [DOI] [PubMed] [Google Scholar]
- Enomoto, S., and J. Berman, 1998. Chromatin assembly factor I contributes to the maintenance, but not the reestablishment, of silencing at the yeast silent mating loci. Genes Dev. 12 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke and J. Carbon, 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29 235–244. [DOI] [PubMed] [Google Scholar]
- Foltz, D. R., L. E. T. Jansen, A. O. Bailey, J. R. Yates, 3rd, E. A. Basset et al., 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson et al., 2002. Defects in SPT16 or POB3 (yFACT) cause dependence on the Hir/Hpc pathway: accessing DNA may degrade chromatin structure. Genetics 162 1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y., T. Hayashi, T. Kiyomitsu, Y. Toyoda, A. Kokubu et al., 2007. Priming of centromere for CENP-A recruitment by human Mis18alpha, hMis18beta and M18BP1. Dev. Cell 12 17–30. [DOI] [PubMed] [Google Scholar]
- Furuyama, T., Y. Dalal and S. Henikoff, 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. USA 103 6172–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet, A., and M. Fitzgerald-Hayes, 1987. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol. Cell. Biol. 7 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu et al., 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., E. Hogan and D. Koshland, 1997. Centromere position in budding yeast: evidence for anaphase A. Mol. Biol. Cell 8 957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan, A., and A. Verreault, 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115 537–549. [DOI] [PubMed] [Google Scholar]
- Hayashi, T., Y. Fujita, O. Iwasaki, Y. Adachi, K. Takahashi et al., 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118 715–729. [DOI] [PubMed] [Google Scholar]
- Heun, P., S. Erhardt, M. D. Blower, S. Weiss, A. D. Skora et al., 2006. Mislocalization of the Drosophila centromere-specific histone cid promotes formation of functional ectopic kinetochores. Dev. Cell 10 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter, P., D. Pridmore, J. H. Hegemann, M. Thomas, R. Davis et al., 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42 913–921. [DOI] [PubMed] [Google Scholar]
- Jamai, A., A. Puglisi and M. Strubin, 2009. Histone chaperone Spt16 promotes redeposition of the original H3–H4 histones evicted by elongating RNA polymerase. Mol. Cell 35 377–383. [DOI] [PubMed] [Google Scholar]
- Kaplan, T., C. L. Liu, J. A. Erkmann, J. Holik, M. Grunstein et al., 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4 e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, P. D., R. Kobayashi and B. Stillman, 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cell lacking chromatin assembly factor-I. Genes Dev. 11 345–357. [DOI] [PubMed] [Google Scholar]
- Kaufman, P. D., J. L. Cohen and M. A. Osley, 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of Chromatin Assembly Factor I. Mol. Cell. Biol. 18 4793–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov, V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16 857–860. [DOI] [PubMed] [Google Scholar]
- Lefrancois, P., G. M. Euskirchen, R. K. Auerbach, J. Rozowsky, T. Gibson et al., 2009. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, C., and B. Stillman, 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber et al., 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J., F. Klein and J. A. Engebrecht, 1998. Genetic and morphological approaches for the analysis of meitotic chromosomes in yeast. Methods Cell Biol. 53 257–285. [DOI] [PubMed] [Google Scholar]
- Maddox, P. S., F. Hyndman, J. Monen, K. Oegema and A. Desai, 2007. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, T., T. Nakamura, T. Hayashi and M. Yanagida, 2006. Histone H2B mutations in inner region affect ubiquitination, centromere function, silencing and chromosome segregation. EMBO J. 25 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew, J., B. Diehl and M. Fitzgerald-Hayes, 1986. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 6 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., and D. Koshland, 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland and M. M. Smith, 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94 607–613. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., H. Xiao, J. Wisniewski, M. M. Smith and C. Wu, 2007. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 2007 1153–1164. [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno, O., M. Torras-Llort and F. Azorin, 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34 6247–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, C. A., and D. Moazed, 2007. Centromere assembly and propagation. Cell 128 647–650. [DOI] [PubMed] [Google Scholar]
- Mythreye, K., and K. S. Bloom, 2003. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y. J., and K. Luger, 2008. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 18 282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux, A., E. Choi, J. Abbott, X. Liu, A. Kagansky et al., 2009. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, I., and F. Winston, 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19 1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski et al., 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9 1091–1100. [DOI] [PubMed] [Google Scholar]
- Rufiange, A., P.-É. Jacques, W. Bhat, F. Robert and A. Nourani, 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27 393–405. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido, L., A. Pidoux, C. Ponting and R. Allshire, 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137 1173–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer, U. J., P. Korber and W. Horz, 2005. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19 279–285. [DOI] [PubMed] [Google Scholar]
- Schwabish, M. A., and K. Struhl, 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22 415–422. [DOI] [PubMed] [Google Scholar]
- Sharp, J. A., and P. D. Kaufman, 2003. Chromatin proteins are determinants of centromere function. Curr. Top. Microbiol. Immunol. 274 24–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., A. A. Franco, M. A. Osley and P. D. Kaufman, 2002. Chromatin assembly Factor-I and Hir proteins contribute to building functional kinetochores in Saccharomyces cerevisiae. Genes Dev. 16 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. A., D. C. Krawitz, K. A. Gardner, C. A. Fox and P. D. Kaufman, 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 17 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein et al., 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S., and B. Stillman, 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication. Cell 58 15–25. [DOI] [PubMed] [Google Scholar]
- Stoler, S., K. C. Keith, K. E. Curnick and M. Fitzgerald-Hayes, 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9 573–586. [DOI] [PubMed] [Google Scholar]
- Stoler, S., K. Rogers, S. Weitze, L. Morey, M. Fitzgerald-Hayes et al., 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA 104 10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami, H., D. Ray-Gallet, G. Almouzni and Y. Nakatani, 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116 51–61. [DOI] [PubMed] [Google Scholar]
- Thomas, B., and R. Rothstein, 1989. Elevated recombination rates in transcriptionally active DNA. Cell 58 619–630. [DOI] [PubMed] [Google Scholar]
- Tomonaga, T., K. Matsushita, S. Yamaguchi, T. Oohashi, H. Shimada et al., 2003. Overexpression and mistargeting of centromere Protein-A in human primary colorectal cancer. Cancer Res. 63 3511–3516. [PubMed] [Google Scholar]
- Van Hooser, A. A., I. I. Ouspenski, H. C. Gregson, D. A. Starr, T. J. Yen et al., 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114 3529–3542. [DOI] [PubMed] [Google Scholar]
- Williams, J., T. Hayashi, M. Yanagida and P. Russell, 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 13 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu et al., 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309 626–630. [DOI] [PubMed] [Google Scholar]
- Zeng, X., J. A. Kahana, P. A. Silver, M. K. Morphew, J. R. McIntosh et al., 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]