Abstract
The establishment of transcriptional silencing in Saccharomyces cerevisiae requires progression through the cell cycle. We have previously found that transit through M-phase is necessary and sufficient to establish silencing at telomeres following induction of the Sir3 silencing factor. In this study we find that halting cell-cycle progression in either G1 or at the beginning of M-phase limits the ability of Sir3 to associate with a telomere-linked reporter gene and prevents the changes in histone modifications associated with gene repression. Deletion of genes coding for the histone variant H2A.Z (Htz1 in yeast) and histone acetyltransferase Sas2 abolish the cell-cycle progression requirement for the establishment of silencing. Cells blocked in telophase (but not at metaphase) are also able to establish silencing. We show that H2A.Z binds to the promoter of our telomere-linked reporter gene and that this binding diminishes in silenced cells. Finally, we observe a specific displacement of H2A.Z from chromatin in telophase-blocked cells, regardless of the silencing status of the reporter gene. These results suggest that the requirement for M-phase in the establishment of silencing may reflect a cell-cycle regulated relaxation of heterochromatin barriers.
YEAST uses the formation of heterochromatin to control the transcription of key determinants of cell development. Silent chromatin in yeast is associated with a histone modification pattern commonly linked to gene repression in all eukaryotes, including decreases in the acetylation of the histone H3 and H4 N-terminal tails (reviewed in Kurdistani and Grunstein 2003), and demethylation of lysine 4 and 79 of histone H3 (reviewed in Kouzarides 2007; Shahbazian and Grunstein 2007). A Sir protein complex that includes the Sir2, Sir3, and Sir4 proteins mediates the establishment of silent chromatin in yeast. Sir2 is a protein deacetylase that acts on histone H3 and H4 (Imai et al. 2000; Tanner et al. 2000; Tanny and Moazed 2001), while Sir3 preferentially binds the deacetylated tails of H3 and H4 (Carmen et al. 2002; Liou et al. 2005). Reiterative cycles of Sir3 binding to deacetylated histones, recruitment of Sir2, and deacetylation of adjacent histones provides a model for the spreading of both deacetylated chromatin and the Sir proteins. This spreading of silent chromatin may be limited by the presence of boundary or barrier factors (reviewed in Valenzuela and Kamakaka 2006). Sir protein action leads to an extremely efficient and stable repression of transcription; this stability is aided by an epigenetic mechanism (Pillus and Rine 1989; Mahoney et al. 1991).
While Sir-mediated gene repression is very stable, there is evidence that silent chromatin in yeast undergoes dynamic transitions that are coupled to cell-cycle progression. For instance, a trans-activator is better able to activate a gene repressed by telomeric heterochromatin when expressed at the start of M-phase than it is in G1 (Aparicio and Gottschling 1994). In addition, specific conditional alleles of the SIR2 silencing factor gene cause an M-phase-specific silencing defect (Matecic et al. 2006). Most significantly, the establishment of silencing in yeast requires progression through the cell cycle (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001; Lau et al. 2002; Kirchmaier and Rine 2006).
We have previously found that progression through M-phase is necessary and sufficient to establish silencing at telomeres following induction of the Sir3 silencing factor (Martins-Taylor et al. 2004). Understanding how the control of gene expression is regulated by and coordinated with cell division is crucial in understanding the assembly and propagation of transcription states. In this study we address three basic questions: First, how does cell-cycle progression regulate the onset of molecular events associated with silencing? Second, what discrete interval of mitosis is required for the establishment of silencing? Finally, what factors restrict the establishment of silencing to specific intervals of the cell cycle? Our results suggest that cell-cycle progression is required for Sir-mediated changes in histone modifications, that silencing is established in the anaphase–telophase transition, and that factors involved in demarcating transitions between euchromatin and heterochromatin, including H2A.Z (Htz1), Sas2, and the Mcd1 cohesin (also known as Scc1), have a key role in imposing the cell-cycle restriction on the establishment of silencing at telomeres in yeast.
MATERIALS AND METHODS
Media:
For telomeric silencing experiments, cultures were grown at 30° in YPraf media (1% Bacto-yeast extract, 2% Bacto-peptone extract, 2% raffinose). To induce expression of the GAL–SIR3 construct, galactose was added to YPraf media to 2%. For solid media, Bacto-agar was added to 2%.
Strains:
Yeast strains used in this study are listed in Table 1. Strain YSH505 was used to examine silencing at the telomere and has been previously described (Martins-Taylor et al. 2004). YSH645 is identical to YSH505 except that both the endogenous and galactose-inducible SIR3 genes have been fused at the C terminus to a nine-myc epitope tag. YSH730 was created from YSH645 by fusing a six-HA epitope tag to the C terminus of the HTZ1 gene. YSH814 is identical to YSH505 except that the MCD1 gene has been fused at the C terminus to a six-HA epitope tag. YSH1021 was created from YSH556 by fusing a nine-myc epitope tag to the C terminus of the SIR3 gene. The nine-myc and six-HA tags were derived from pYM6 and pYM3, respectively (Knop et al. 1999).
TABLE 1.
Strains used in this study
| Strain no. | Genotype | Reference |
|---|---|---|
| YSH505 | MATa ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 Δppr1∷HIS3 URA3-TELVR trp1Δ63∷GAL10p-SIR3-TRP1 | Martins-Taylor et al. (2004) |
| YSH530 | YDS634; Δgal4∷kanMX4 | |
| YSH549 | YSH505; Δscc1∷scc1-73-LEU2MX4 | |
| YSH556 | MATa ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 Δppr1∷HIS3 URA3-TEL-VR | |
| YSH573 | YSH505; Δsir2∷hphMX4 | |
| YSH645 | YSH505; trp1Δ63∷GAL10p-SIR3-9Xmyc-kanMX4/natMX4-TRP1 SIR3-9Xmyc-kanMX4/natMX4 | |
| YSH666 | YSH645; Δsir2∷hphMX4 | |
| YSH673 | YSH505; Δhml∷hphMX4 | |
| YSH674 | YSH673; Δsas2∷natMX4 | |
| YSH675 | YSH673; Δdot1∷natMX4 | |
| YSH676 | YSH673; Δhho1∷natMX4 | |
| YSH679 | YSH556; Δhml∷natMX4 trp1Δ63∷GAL10p-SIR3-TRP1 ycs4-1 | |
| YSH682 | YSH556; Δhml∷natMX4 trp1Δ63∷GAL10p-SIR3-TRP1 top2-4 | |
| YSH683 | YSH673; Δrsc2∷natMX4 | |
| YSH690 | YSH673; Δhtz1∷kanMX4 | |
| YSH709 | YSH673; Δset1∷natMX4 | |
| YSH727 | YSH505; Δhml∷LEU2MX4Δubp8∷kanMX4 | |
| YSH728 | YSH505; Δhml∷LEU2MX4Δrad6∷kanMX4 | |
| YSH729 | YSH645; Δset1∷hphMX4 | |
| YSH730 | YSH645; HTZ1-6HA-hphMX4 | |
| YSH734 | YSH645; Δdot1∷hphMX4 | |
| YSH738 | YSH673; Δubp3∷kanMX4 | |
| YSH739 | YSH673; Δubp10∷kanMX4 | |
| YSH772 | YSH673; Δdls1∷kanMX4 | |
| YSH773 | YSH673; Δyta7∷kanMX4 | |
| YSH774 | YSH673; Δitc1∷kanMX4 | |
| YSH775 | YSH673; Δchl1/ctf1∷kanMX4 | |
| YSH776 | YSH673; Δisw2∷kanMX4 | |
| YSH777 | YSH673; Δsas3∷kanMX4 | |
| YSH795 | YSH505; Δcdc14∷cdc14-1-natMX4 | |
| YSH813 | YSH505; Δcdc16∷cdc16-1-natMX4 | |
| YSH814 | YSH505; SCC1-6HA-hphMX4 | |
| YSH862 | YSH795; HTZ1-6FLAG-kanMX4 | |
| YSH869 | YSH505; HTZ1-6FLAG-kanMX4 | |
| YSH1020 | YSH556; HTZ1-6FLAG-kanMX4 | |
| YSH1021 | YSH556; SIR3-9Xmyc-kanMX4 | |
| YDS631 | MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 adh4∷URA3-telVIIL | Chien et al. (1993) |
| YDS634 | MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 adh4∷URA3-4xUASG-telVIIL | Chien et al. (1993) |
To create strain YSH549, hybrid PCR (Horton et al. 1989) was used to link the scc1-73 allele (Michaelis et al. 1997) (amplified from strain KN5832, provided by Kim Nasmyth) to the LEU2 gene from pKMT1, a LEU2 vector designed on the MX-series scaffold (Wach et al. 1994; Goldstein and McCusker 1999). The DNA fragment was transformed into YSH505. Leu+ transformants were then screened for those able to promote growth at 23°, but not at 37°; these strains were also found to contain the LEU2 directly downstream of the MCD1 locus.
To replace the wild-type alleles of YCS4 and TOP2, the temperature-sensitive alleles ycs4-1 and top2-4 were amplified from strains SPY746 and SPY9, respectively (provided by Sue Biggins) and cloned into pRS404 (Sikorski and Hieter 1989) to produce plasmids pKMT2 and pKMT3. These plasmids include 300 bp of wild-type sequences both 5′ and 3′ to the open reading frame. To direct integration at the YCS4 or TOP2 loci, pKMT2 was cut with NruI, while pKMT3 was cut with MluI. Cut plasmids were then transformed into YSH556. Trp+ transformants were then grown on media containing 5-fluoroanthranilic acid (Toyn et al. 2000), to select for excision of TRP1 and either the wild-type or mutated allele. Candidates were then screened for those able to promote growth at 23°, but not at 37°. A galactose-inducible SIR3 allele was then integrated at the TRP1 locus using plasmid pAR83 (Holmes et al. 1997). To ensure that these strains would be sensitive to α-factor, HML was deleted via PCR-mediated gene deletion using pAG25 as a template (Goldstein and McCusker 1999), creating strains YSH679 and YSH682.
To create YSH813, a PCR fragment containing the cdc16-1 allele was linked to the selectable marker natMX (amplified from pAG25) by hybrid PCR and then introduced into YSH505, replacing the endogenous CDC16 gene by homologous recombination. To create YSH795, independent DNA fragments bearing the cdc14-1 allele and the natMX selectable marker were generated with short regions of homology to each other and to the CDC14 locus. These fragments were then simultaneously transformed into YSH505, where recombination with each other and with the chromosome yielded drug-resistant colonies bearing the natMX gene downstream of the CDC14 gene. Drug-resistant transformants were selected for at the permissive temperature (23°) and then screened for those unable to grow at nonpermissive temperature (37°). To confirm integration of the correct CDC allele, cells were arrested in the cell cycle by growth at the nonpermissive temperature and then examined for the expected cell morphology both by light microscopy and fluorescent microscopy with DAPI staining. YSH862, YSH869, and YSH1020 were created from YSH795, YSH505, and YSH556, respectively, by integrating a FLAG-kanMX construct amplified from pJR2659 (Babiarz et al. 2006) at the C terminus of the HTZ1 gene.
Cell cultures:
Cell-cycle blocks were performed as described (Martins-Taylor et al. 2004). α-Factor (10 μg/ml) or nocodazole (15 μg/ml) was used to block cells in G1 or G2/M, respectively. Cell-cycle blocks were routinely achieved in 2–3 hr with α-factor and 4–6 hr with nocodazole. Unless noted, cells exhibited at least a 90% arrest in the cell cycle in the presence of blocking agents. Cell-cycle arrest was determined by microscopic examination of cell morphology and confirmed by FACS analysis on representative cultures (see supporting information, Figure S1). FACS was performed at the Yale Cell Sorter Facility using a FACSCaliber instrument. Yeast cells were prepared for FACS as described (Hutter and Eipel 1978). Data were analyzed using the CellQuest and FlowJo software programs. FACS data presented in the manuscript were created by using the Dean–Jett–Fox model (Fox 1980).
cdc14-1 and cdc16-1 strains were blocked in telophase and metaphase, respectively, by shifting cells grown to log phase at 23° to 37° for approximately 6 hr. Unless noted, these cells exhibited at least a 95% arrest in the cell cycle. Cell-cycle arrest was determined by microscopic examination of cell morphology, FACS, and in initial experiments by fluorescence microscopy of DAPI-stained cells. Viability assays indicated that ∼77% of the cdc14-1 and 69% of the cdc16-1 cells could reenter the cell cycle after 6 hr at the nonpermissive temperature. For cell-cycle experiments, cultures were initially grown in media containing 2% raffinose. Sir3 induction was carried out by addition of galactose to a final concentration of 2%. Cells were collected for analysis after 6–8 hr of galactose addition, when complete repression of URA3 is achieved (Martins-Taylor et al. 2004).
RT–PCR:
RNA extraction, cDNA synthesis, and PCR were performed as previously described (Martins-Taylor et al. 2004). Results from 8% acrylamide gels were stained using Sybr gold dye (Invitrogen), and the gels were scanned using a Storm 840 PhosphorImager (GE Healthcare). Each band was quantified using ImageQuant TL (GE Healthcare) and converted to tiff files. Identical results were achieved in at least two independent experiments and in repeated determinations from RNA collected from individual experiments.
Chromatin immunoprecipitation assays:
Chromatin immunoprecipitations were performed essentially as described (Suka et al. 2001), with the following exceptions. Prior to the addition of antibody for precipitation, 50 μl of lysate was precleared with 7 μl of Protein A magnetic beads (New England Biolabs) by incubating at 4° for 30–60 min on a Labquake tube rotator. The samples were applied to a magnet to separate the beads from the supernatant; the supernatant was transferred to a new eppendorf tube and one of the following antibodies were added for an overnight incubation at 4°: 1 μl myc-epitope antibody (9B11; Cell Signaling Technology), 2.5 μl anti-acetyl-histone H4 antibody (Upstate Cell Signaling Solutions), 2.5 μl anti-acetyl-histone H3 antibody (Upstate), 2.5 μl anti-acetyl-histone H4 (K16) antibody (Upstate), 2.5 μl anti-trimethyl-histone H3 (K4) antibody (Upstate), 2.5 μl anti-dimethyl-histone H3 (K79) antibody (Upstate or Abcam), 2.5 μl anti-HA antibody (12CA5; Roche Applied Science), 0.5 μl anti-FLAG antibody (F1804; Sigma). Protein A magnetic beads (15 μl) were added to precipitate the chromatin.
DNA samples from chromatin immunoprecipitation experiments were quantified using either real-time PCR or quantitative PCR. For real-time PCR primers were designed using the Primer Express software package that accompanies the Applied Biosystems 7300 Real Time PCR System and are listed in Table 2. Regions of approximately 100 bp in size were amplified from the immunoprecipitated samples and the unenriched, whole cells control samples using Power Sybr green master mix (Applied Biosystems). To determine the enrichment values, relative standard curve method for quantification was used (Applied Biosystems). Standard curves were generated for each primer pair using data from a standard dilution series. Relative values for target abundance in each experimental sample were extrapolated from the standard curve generated from the reference standard. Relative yields in uninduced cells vs. galactose-induced cultures were determined as described (Pfaffl 2001). Each chromatin immunoprecipitation (ChIP) value was normalized to a reference locus to control for differential recovery of chromatin between strains and conditions. The ACT1 coding region (Rudner et al. 2005) or sequences adjacent to MAT (Kirchmaier and Rine 2006) were used as controls for the histone modifications, the PRP8 coding region was used as a control for Sir3 and H2A.Z (Kobor et al. 2004; Zhang et al. 2004; Babiarz et al. 2006; Shia et al. 2006), and the DCC1 coding region, a cohesin-free region, was used for Mcd1 (Laloraya et al. 2000).
TABLE 2.
Primers used for PCR
| Primer no. | Primer sequence | Region of hybridization | Distance from telomere VR in YSH505 (kb) |
|---|---|---|---|
| SP632 | TGTTGACATTGCGAAGAGCG | URA3 | 2.8–2.9 |
| SP633 | CACCGGGTGTCATAATCAACC | ||
| SP638 | ATCGTTATGTCCGGTGGTACC | ACT1 | |
| SP639 | TGGAAGATGGAGCCAAAGC | ||
| SP817 | GGAACGTGCTGCTACTCATCC | URA3 | 3.3–3.4 |
| SP818 | TGGTACGAACATCCAATGAAGC | ||
| SP819 | TCAGGGTAGTGCGCGTATTTG | Chr V 565042-063 | 3.7–3.8 |
| SP820 | CGTCCTTTTTCCGCCAATT | Chr V 565124-143 | |
| SP821 | GTGGGCAAATATGCGTATGATT | Chr V 564380-402 | 4.3–4.4 |
| SP822 | GGTTTCCTGTAAAAGCTGCTAAAC | Chr V 564456-480 | |
| SP823 | AAATCGTGGCAGCGGTACC | Chr V 566239-258 | 1.3–1.4 |
| SP824 | TGATATCCATGAGGTGGCGA | Chr V 566319-339 | |
| SP886 | TTGTAATGACGAGCATATCGGTG | Chr V 566603-626 | 1.0–1.1 |
| SP887 | CGTCAACAGTTCTTAATTTCGGGT | Chr V 566680-704 | |
| SP888 | CACCACATACGCGCTGATTG | Chr V 567001-021 | 0.6–0.7 |
| SP889 | GCATGTAACCAGATGAGTTATATTTGATAAT | Chr V 567072-103 | |
| SP890 | TATTTATTGCGATAGACGCACTACTTG | Chr V 567305-332 | 0.3–0.4 |
| SP891 | AAGTCTGATTGATTCTGTTGAGATTGTT | Chr V 567378-406 | |
| SP892 | GGTGGCTCTGGAGGCTCAT | Chr V 567537-556 | 0.0–0.1 |
| SP893 | CATCAACTGCACATAATTGGCG | Chr V 567617-639 | |
| SP976 | GCCCCTGGACTACGAAAACTT | MAT | |
| SP977 | ACAATTCATCATTTGCGTTCGTT | ||
| SP978 | TGGCAGAGAATCCAGATCCAA | PRP8 | |
| SP979 | ACTGCTCGCCCTAGGTTAACG | ||
| SP980 | CCCTAGGTCTTGGCAACTGG | DCC1 | |
| SP981 | ACCATCGTGGTGCTCTTTGTC |
Alternatively, immunoprecipitated and input DNAs were analyzed by quantitative PCR analysis. All amplifications were performed in the linear range; samples were then run on 8% acrylamide gels, were stained with Sybr gold dye, and were scanned as described above. Enrichments relative to an endogenous control were calculated as described (Noma et al. 2001). We determined the abundance ratios for each locus of interest relative to the endogenous control. These ratios were in turn normalized by the corresponding input ratio to determine relative enrichment values. Each reported value represents the average of at least three independent ChIP experiments, each analyzed in triplicate by real-time PCR or in duplicate by quantitative PCR.
Construction of Gal4 binding domain fusion constructs:
To create the Gal4 binding domain (GBD)–Mcd1 fusion construct, the MCD1 open reading frame was amplified from a wild-type yeast strain using primers with overhangs containing restriction enzyme recognition sites allowing subcloning into vector pOBD2 (Uetz et al. 2000), creating pMH2. pMH2 complements the lethality observed in strains bearing the temperature-sensitive scc1-73 allele, grown at the nonpermissive temperature (not shown). pMH2 was transformed into yeast strains YDS631 (from David Shore) (Chien et al. 1993) and YSH530. To create YSH530, GAL4 was deleted from YDS634 (Chien et al. 1993) via PCR-mediated gene deletion using pFA6a–kanMX4 (Wach et al. 1994) as a template. The GBD–Yku70 fusion construct was created via gap repair. YKU70 was amplified from a wild-type strain using primers with tails homologous to the multiple cloning site of pOBD2 (Uetz et al. 2000). pOBD2 was cut with PvuII and NcoI, and then cotransformed with the YKU70 fragment into YDS631 and YSH530. PCR on DNA extracted from the transformants was used to verify successful gap repair.
RESULTS
Limiting event in the assembly of silent chromatin:
We have previously shown that the establishment of silencing of a telomere-linked gene requires progression through mitosis (Martins-Taylor et al. 2004). In this system, the URA3 gene is located approximately 3.5 kb from the telomere (Figure 1A), where it is not subject to silencing (Martins-Taylor et al. 2004). This strain contains the endogenous SIR3 gene, as well as an integrated, galactose-inducible copy of SIR3. Upon addition of galactose, increased Sir3 levels cause spreading of telomeric heterochromatin and silencing of URA3 (Martins-Taylor et al. 2004). Silencing at specific loci in yeast correlates with an increase in Sir protein association (Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002; Strahl-Bolsinger et al. 1997), a decrease in the acetylation of histones H3 and H4 (Braunstein et al. 1993; Braunstein et al. 1996), and a decrease in the methylation of histone H3 on lysine 4 (H3K4) (Briggs et al. 2001; Bernstein et al. 2002; Bryk et al. 2002) and on lysine 79 (H3K79) (Singer et al. 1998; San-Segundo and Roeder 2000; Lacoste et al. 2002; Ng et al. 2002; van Leeuwen et al. 2002). A variety of genetic and biochemical experiments indicate that these events correlate with the establishment and maintenance of silencing, while deletion of the genes responsible for these modifications cause silencing defects. To examine the possibility that Sir protein association or silencing-specific histone modifications are regulated as a function of the cell cycle, we monitored the changes in chromatin associated with silencing using ChIP assays.
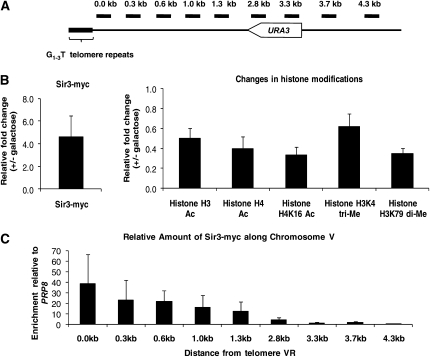
Figure 1.—
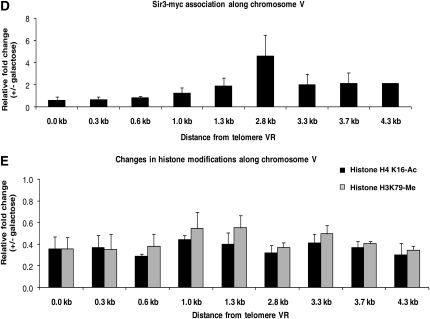
Marks of inducible silencing at the telomere. (A) Probes along chromosome VR. Primer pairs hybridize roughly every 300 bp along chromosome VR from the telomere repeat sequence, amplifying a region of approximately 100 bp. The distance depicted above each probe represents the position of the primer closest to the telomere within each pair. Primer sequences can be found in Table 2. (B) Defining the marks of silencing at telomere VR. YSH645 was grown to steady state (>16 hr) in raffinose media with or without galactose. ChIP assays were used to compare the association of Sir3–myc and the modification state of histones in galactose-induced vs. uninduced cultures. All determinations were made using the 2.8-kb primer pair, within the URA3 open reading frame, and are expressed as a ratio of the galactose-induced vs. uninduced signal. The relative enrichment of Sir3 at this region was determined relative to PRP8, a Sir3-free region (Lieb et al. 2001), while the change in histone modifications was determined relative to ACT1 (Rudner et al. 2005). (C) Sir3 association along chromosome VR. ChIP assays were performed on strain YSH645 grown to steady state in raffinose (noninducing) media. The data are expressed relative to the input (whole-cell lysate) signal, normalized to the signal from the PRP8 locus. (D) Changes in Sir3 association. YSH645 was grown to steady state in raffinose media with or without galactose. The primers in Figure 1A were used to examine the change in the association of Sir3 in galactose-induced vs. uninduced cultures. (E) Changes in histone modifications. Cultures of YSH645 were grown in raffinose media with or without galactose. To examine the change in the acetylation of histone H4K16 and the methylation of histone H3K79 along the chromosome upon induction with galactose, the primers in Figure 1A were used.
To determine the association of Sir3 with the telomere, both the endogenous and galactose-inducible SIR3 genes were tagged at their C terminus with the myc epitope. Modification-specific antibodies were used to examine the acetylation of histones H3 and H4 and the methylation of histone H3K4 and H3K79. For our initial experiments, we used a probe to the URA3 coding region (2.8-kb probe, Figure 1A). We compared the change in the association of Sir3 or changes in histone modifications in cells grown in noninducing raffinose media vs. those grown in raffinose media containing galactose. Under these conditions we observe that URA3 mRNA is reduced to 20–25% of uninduced levels (Martins-Taylor et al. 2004); (Figure 2A; see also Figure S3). As shown in Figure 1B, we observe an increase in the association of Sir3 with URA3 in silenced cells relative to unsilenced cells. This increase in the association of Sir3 is accompanied by the deacetylation of lysines in histones H3 and H4, including H4K16. The methylation of histone H3K79 also significantly decreases in silenced vs. unsilenced cells, while only a modest decrease in the methylation of histone H3K4 is observed. The qualitative and quantitative characteristics of chromatin we observed in our induced system for Sir3 association, histone acetylation, and H3 lysine 79 methylation are similar to those reported for genes subject to noninduced telomeric silencing (Braunstein et al. 1996; Ng et al. 2003; Katan-Khaykovich and Struhl 2005), while the decrease in H3 K4 methylation is less than previously seen for a reporter gene placed adjacent to telomere repeats (Santos-Rosa et al. 2004). Control experiments demonstrated that the change in Sir3 association and histone acetylation depended on the presence of the GAL–SIR3 gene and were not due simply to the addition of galactose (Figure S2).
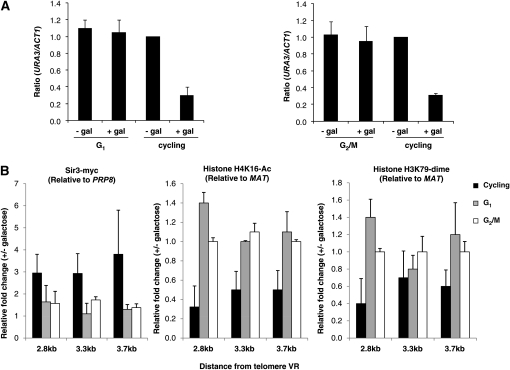
Figure 2.—
Changes in chromatin cannot occur in the absence of silencing. (A) Silencing does not occur in cells blocked in G1-phase or at the G2/M boundary. A culture of strain YSH645 was grown to early log phase in raffinose media. This culture was divided and half was blocked either in G1 with α-factor or in G2/M with nocodazole. Following cell-cycle arrest, the blocked culture was further divided into induced (addition of galactose) and uninduced (no galactose addition) cultures. Galactose was also added to the unblocked cycling cells at this time. Following induction, cells were collected to quantify URA3 expression (see materials and methods). The URA3/ACT1 ratio is shown relative to the uninduced (no galactose) cycling control. Cells from these experiments were also collected for the ChIP assays shown in B. (B) Changes in chromatin cannot occur in the absence of cell-cycle progression. The change in the acetylation of histone H4K16, methylation of histone H3K79, and the association of Sir3–myc was determined using the 2.8-, 3.3-, and 3.7-kb primers depicted in Figure 1A, as described in the legend of Figure 1B.
We next used additional probes that hybridize roughly every 300 bp to examine the distribution of Sir3 across the subtelomeric region encompassing URA3. In cells grown in noninducing raffinose media we find that the greatest association of Sir3 occurs at the regions adjacent to the telomere repeat sequence (Figure 1C), consistent with previous studies (Strahl-Bolsinger et al. 1997; Kimura et al. 2002; Suka et al. 2002). The association of Sir3 decreases as the distance from the telomere repeats sequence increases such that Sir3 association is weakly detected at distances greater than 1.3 kb from the telomere repeat sequence (Figure 1C). These results suggest that telomeric heterochromatin is limited to approximately 1 kb under noninducing conditions.
Focusing on the chromatin alterations that were most significantly associated with the onset of silencing at this locus, we used the probes depicted in Figure 1A in additional ChIP experiments to compare induced vs. uninduced cultures. Upon galactose induction of Sir3 we observe an increase in Sir3 association at all positions tested that are greater than 1 kb from the telomere repeat sequence (Figure 1D). We also find that the acetylation of histone H4K16 and the methylation of histone H3K79 decreases at all positions along the chromosome (Figure 1E). We note that we observe alterations in histone modifications adjacent to the telomere repeat sequences, at positions where we fail to observe increased Sir3 association. This may indicate that downstream Sir3 association increases or stabilizes Sir3-dependent modifications. Alternatively, the ChIP technique may have an insufficient dynamic range to detect further increases in Sir3 association at these sequences. For instance, if immunoprecipitation of chromatin is highly efficient then it may not be possible to distinguish between chromatin in which a single Sir3 molecule binds per nucleosome vs. chromatin in which two or more Sir3 molecules bind each nucleosome.
Next we asked whether the onset of one or more of these alterations in chromatin required cell-cycle progression. We examined the association of Sir3, the deacetylation of histone H4K16, and the demethylation of histone H3K79 in cells blocked in G1 phase with α-factor or blocked at the G2/M boundary with nocodazole. Cells were grown to log phase in noninducing raffinose media and cell-cycle progression was blocked. Galactose was then added to induce SIR3 overexpression. Control cultures were blocked but were not induced or were allowed to continue progression through the cell cycle in the presence of galactose. Following a period of induction that was more than sufficient to silence URA3 in cycling cells, cells were collected for ChIP analysis. Cells were also collected from each of the cultures to determine the levels of URA3 mRNA. As reported previously, silencing is not observed in cells blocked in G1 or G2/M, but is observed in parallel unblocked cultures (Martins-Taylor et al. 2004) (Figure 2A).
Over this time, course-cycling cells induced to overexpress Sir3 exhibit significant decreases in H4 K16 acetylation and H3 K79 methylation (Figure 2B). However, we find that cells overexpressing Sir3 while blocked in G1 or G2/M fail to exhibit decreases in H4 K16 acetylation or H3 K79 methylation (Figure 2B). Our experiments suggest that Sir3 association is also regulated by cell-cycle progression; however, the variability in our data prevents us from drawing firm conclusions about the relative association of Sir3 in cycling vs. blocked cells. Overall, we find that cell-cycle progression is required for the Sir-protein-mediated changes in histone modifications that are associated with gene repression.
Timing of silencing:
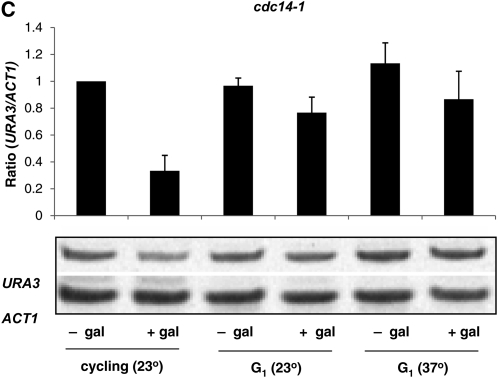
Our prior experiments indicated that silencing is established in this system between the G2/M boundary (as defined by a nocodazole block) and the subsequent G1 phase (as defined by an α-factor block) (Martins-Taylor et al. 2004). To identify the discrete interval in mitosis required for silencing we introduced temperature-sensitive alleles of specific CDC genes into our strain background. CDC genes code for proteins required to transit specific points of the cell cycle. We used the temperature-sensitive cdc16-1 allele, coding for a component of the anaphase-promoting complex, to block cells in metaphase, and the temperature-sensitive cdc14-1 allele, coding for a phosphatase required to exit mitosis, to block cells in telophase. Cells were grown to log phase at the permissive temperature and then shifted to the nonpermissive temperature. Control cultures were maintained at the permissive temperature. Sir3 production was then induced, and the expression status of URA3 was monitored. Consistent with our prior experiments examining nocodazole-blocked cells (Martins-Taylor et al. 2004), we find that cells blocked at metaphase in the cdc16-1 strain are unable to establish silencing, while URA3 transcription is significantly repressed in an unblocked parallel culture grown at the permissive temperature (Figure 3A). In contrast, we find that the cdc14-1 cells blocked in telophase are able to establish silencing at levels equivalent to the cycling control (Figure 3B). This indicates that cell-cycle progression is not an absolute requirement for establishing silencing and suggests that an event coinciding with the anaphase–telophase transition is crucial in allowing repression of URA3.
Figure 3.—
Silencing can be established in telophase-blocked cells. (A) Strain YSH813 bearing the cdc16-1 allele was grown to early log phase in raffinose media at permissive temperature (23°). The culture was divided, with one-half shifted to the nonpermissive temperature (37°). After cell-cycle arrest at the nonpermissive temperature each culture was subsequently divided, when half was induced with galactose. URA3 mRNA levels were measured as described in the Figure 2A legend. The figure shows a histogram representing the cumulative data for at least three independent experiments; results from a representative experiment are shown below the histogram. Values are expressed relative to the uninduced control. (B) Strain YSH795, bearing the cdc14-1 allele, was analyzed as described in A. (C) Inactivation of Cdc14 at a G1 block does not permit the establishment of silencing. Strain YSH795 was grown to early log phase in raffinose media at 23°. The culture was divided, and half was arrested in G1-phase with α-factor. This blocked culture was further divided, with half remaining at 23° and the other half shifted to 37°. Finally, each of these three cultures was divided into galactose-induced and uninduced samples. URA3 mRNA levels were determined for each of these conditions. The cumulative results of at least three independent experiments are shown in the histogram, along with a gel of a representative experiment.
An alternative explanation for these results is that Cdc14 has a specific role in inhibiting the establishment of silencing that is independent of the cell-cycle block that its inactivation causes. If this is true, then inactivation of Cdc14 at other points in the cell cycle should also allow silencing. To test this possibility we grew the cdc14-1 cells at the permissive temperature, blocked them in G1 phase with α-factor, and raised the culture to the nonpermissive temperature. As seen in Figure 3C, inactivation of Cdc14 in G1 does not relieve the inhibition of silencing seen in wild-type cells. This experiment does not rule out the possibility that Cdc14 directly influences the establishment of silencing, but indicates that such an influence is likely restricted to telophase.
Inhibitors of silencing:
The establishment of silencing may require progression through M-phase due to a mitosis-specific factor or event that positively promotes the formation of yeast heterochromatin. Alternatively, the action of an inhibiting factor may be specifically relieved in mitosis. We conducted a search for such inhibitors by assessing the timing of silencing in a collection of strains lacking candidate proteins (Table 3). These candidates included histone-modifying enzymes, Sir3- or Sir4-interacting factors, proteins proposed to function as barriers between heterochromatin and euchromatin, and proteins whose association with chromatin changes dramatically as a function of the cell cycle. To examine the influence of these factors on silencing in our inducible system we constructed strains in which each of the genes encoding nonessential proteins was deleted. We also created strains bearing temperature-sensitive alleles of the essential genes coding for topoisomerase II (top2-4), a condensin subunit (ycs4-1), and the Mcd1 cohesin subunit (scc1-73).
TABLE 3.
Inducible silencing in mutant yeast backgrounds
| Strain | Relative URA3 mRNA levels (cycling induced/cycling uninduced) | G1 mRNA (blocked induced/cycling uninduced) | G2/M mRNA (blocked induced/cycling uninduced) |
|---|---|---|---|
| Wild type | 0.2 ± 0.08 | 1.0 ± 0.09 | 1.1 ± 0.13 |
| Δsir2 | 1.0 ± 0.11 | ND | ND |
| Δhtz1 | 0.2 ± 0.01 | 0.4 ± 0.00 | 0.5 ± 0.14 |
| Δsas2 | 0.1 ± 0.01 | 0.3 ± 0.14 | 0.1 ± 0.00 |
| Δset1 | 0.4 ± 0.21 | 1.1 ± 0.14 | 1.0 ± 0.28 |
| Δdot1 | 1.1 ± 0.19 | ND | ND |
| Δubp3 | 0.5 ± 0.10 | 0.9 ± 0.00 | 1.0 ± 0.00 |
| Δubp 8 | 0.3 ± 0.17 | 1.1 ± 0.00 | 1.1 ± 0.07 |
| Δubp 10 | 0.5 ± 0.14 | 1.1 ± 0.00 | 1.0 ± 0.00 |
| Δrad6 | 0.7 ± 0.07 | 1.0 ± 0.00 | 0.9 ± 0.14 |
| Δhho1 | 0.2 ± 0.08 | 1.1 ± 0.07 | 1.0 ± 0.21 |
| Δdls1 | 0.2 ± 0.07 | 1.0 ± 0.07 | 1.1 ± 0.00 |
| Δyta7 | 0.4 ± 0.07 | 1.1 ± 0.07 | 0.9 ± 0.07 |
| Δitc1 | 0.1 ± 0.00 | 1.1 ± 0.07 | 1.1 ± 0.07 |
| Δisw2 | 0.1 ± 0.00 | 1.1 ± 0.00 | 0.9 ± 0.00 |
| Δsas3 | 0.3 ± 0.07 | 1.1 ± 0.07 | 1.0 ± 0.14 |
| Δrsc2 | 0.4 ± 0.14 | 0.9 ± 0.14 | 0.9 ± 0.07 |
| Δdcc1 | 0.9 ± 0.20 | ND | ND |
| Δchl1/ctf1 | 0.5 ± 0.00 | 1.1 ± 0.00 | 1.1 ± 0.07 |
| scc1-73 (PT) | 0.5 ± 0.10 | 0.9 ± 0.05 | 1.0 ± 0.00 |
| ycs4-1 (PT) | 0.9 ± 0.19 | 1.2 ± 0.07 | 1.2 ± 0.00 |
| top2-4 (PT) | 0.9 ± 0.15 | ND | ND |
Strains bearing the indicated gene deletions were grown to steady state in raffinose media or raffinose media plus galactose and assayed for expression of the telomere-linked URA3 gene. The URA3/ACT1 ratio is shown for the galactose-induced cells relative to its uninduced control. YSH556 (lacking the galactose-inducible SIR3 gene) and YSH505 served as negative and positive controls, respectively. For G1 or G2/M experiments cells were grown in raffinose media, arrested in the cell cycle with α-factor or nocodazole, and induced with galactose (see materials and methods). Strains bearing temperature-sensitive alleles were assayed at 23° (PT) for steady-state determinations. ND, not determined.
Assessing these factors' influence on the timing of silencing required that strains lacking these factors permit inducible silencing. Therefore we first examined the establishment of silencing in cells allowed to progress through the cell cycle. Cultures were grown to log phase in noninducing media, Sir3 was induced, and URA3 mRNA levels were measured. In most of the knockout strains tested, the degree of repression of URA3 was comparable to the wild-type strain YSH505 (Table 3, first data column). The absence of some of these proteins, including Set1 (Nislow et al. 1997), and Dot1 (Ng et al. 2002, 2003; van Leeuwen et al. 2002) was previously reported to cause silencing defects; in most of these cases we also observed a defect in inducible silencing. We also found that deletion of genes involved in cohesin deposition or function, including Dcc1 (Mayer et al. 2001), Chl1 (Mayer et al. 2004; Petronczki et al. 2004; Skibbens 2004), and Rsc2 (Baetz et al. 2004) consistently led to silencing defects in this assay. Finally, introduction of the temperature-sensitive alleles ycs4-1 and top2-4 also resulted in a compromised ability to induce repression of URA3. Strains that permitted inducible silencing in cycling cells were next examined for their ability to repress URA3 transcription in the absence of cell-cycle progression. We examined silencing in cells blocked in G1 phase with α-factor or blocked at the G2/M boundary with nocodazole. We found that in most of these strains the timing of silencing was unaltered compared to the wild-type control; when Sir3 production was induced, silencing was established in cycling cells but not in cells blocked from progressing through the cell cycle (Table 3). However, we found that when either the SAS2 or HTZ1 gene was deleted the requirement for cell-cycle progression was eliminated, and URA3 was repressed in cells blocked in G1 or at the G2/M boundary (Table 3).
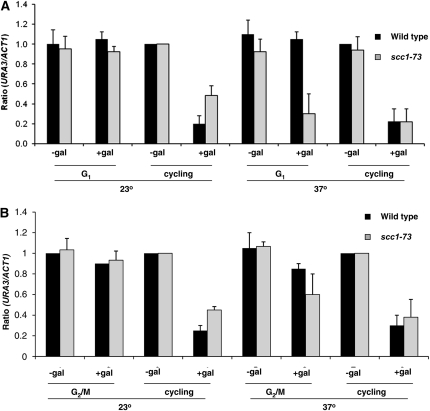
A previous study using a conditional SIR3 allele to study the establishment of silencing found that the cohesin subunit Mcd1 affected the timing of silencing at the HMR locus. In this study silencing required passage from G1 phase through mitosis; however, the controlled repression of MCD1 transcription allowed silencing to occur following progression from G1 to the end of S-phase, prior to mitosis (Lau et al. 2002). To determine if Mcd1 affected the timing of silencing in our system we incorporated the temperature-sensitive scc1-73 allele into our strain background. Cells were grown at the permissive temperature and blocked in G1, when a portion of the culture was shifted to the nonpermissive temperature for 2 hr to inactivate Scc1–73p. SIR3 expression then was induced by the addition of galactose. In parallel control experiments silencing was not observed in wild-type G1-blocked cells at either low or high temperatures (Figure 3). However, when Mcd1p is inactivated by shifting cultures to the nonpermissive temperature in G1-blocked cells, URA3 is silenced (Figure 4A). We were unable to observe a significant effect of inactivating Mcd1p in nocodazole-blocked cells (Figure 4B).
Figure 4.—
Mcd1 affects the timing of silencing. Cultures were grown to early log phase in raffinose media and then divided; half the culture was blocked in G1 with α-factor (A) or at the G2/M boundary with nocodazole (B). Following cell-cycle block, a portion of the culture was shifted to the nonpermissive temperature (37°) for 2 hr to inactivate Mcd1p. SIR3 overexpression was then induced by the addition of galactose and URA3 mRNA levels were examined. The URA3/ACT1 ratio is shown relative to the uninduced (no galactose) cycling control. Shifting the cells to a higher temperature does not affect the silencing of the wild-type strain; silencing is not observed in cells blocked in G1 or G2/M at either temperature. When Mcd1p is inactivated at nonpermissive temperature in G1 blocked cells, URA3 is silenced.
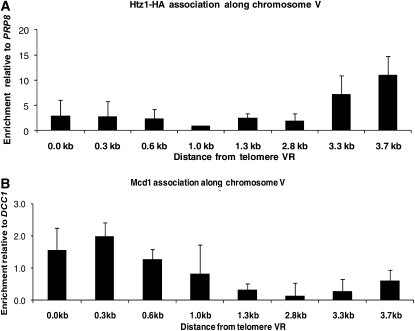
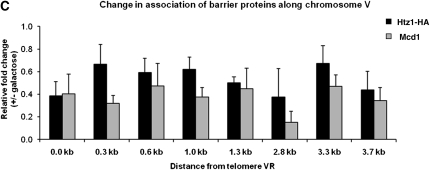
H2A.Z and Mcd1 are widely dispersed chromatin proteins that have broad effects on transcription, DNA repair, and chromosome segregation (Krogan et al. 2004; Keogh et al. 2006; Zlatanova and Thakar 2008). If H2A.Z and Mcd1 directly affect the establishment of silencing at URA3, these factors may exhibit a silencing-dependent dissociation with the telomere. To examine the association of H2A.Z and Mcd1 along the chromosome region encompassing URA3, we grew cells in noninducing raffinose media and examined their association by ChIP assays, using the probes depicted in Figure 1A. We find a significant enrichment of H2A.Z over this region, with a peak at the URA3 promoter (Figure 5A). H2A.Z association with this promoter is also seen when URA3 is at its endogenous location (Guillemette et al. 2005; Zhang et al. 2005; Millar et al. 2006). We observe that Mcd1 is also associated with this region, binding at levels above background adjacent to the telomere repeat sequences (Figure 5B). To determine if H2A.Z and Mcd1 binding varied in silenced vs. unsilenced cells we assessed their association in cells grown to steady state either in raffinose media or in raffinose media containing galactose. We observe a decrease in the association of both H2A.Z and Mcd1 at all positions along telomere VR in silenced cells relative to unsilenced cells (Figure 5C).
Figure 5.—
H2A.Z and Mcd1 association along chromosome VR. (A) H2A.Z association along chromosome VR. ChIP assays were performed on strain YSH730, grown to steady state in raffinose. The relative enrichment of H2A.Z was determined using the primers depicted in Figure 1A and was normalized to the PRP8 coding region (Babiarz et al. 2006; Shia et al. 2006). (B) Mcd1 association along chromosome VR. ChIP assays were performed on strain YSH730, which was grown to steady state in raffinose. The relative enrichment of Mcd1 was determined along chromosome VR using the primers depicted in Figure 1A and was normalized to the DCC1 coding region (Laloraya et al. 2000). In control experiments conducted at the same time Mcd1 showed robust association with CEN16 and CEN3 sequences (27-fold and 32-fold enrichment, respectively), which were previously shown to bind Mcd1 (Megee et al. 1999). (C) Changes in association of H2A.Z–HA and Mcd1. YSH730 was grown to steady state in raffinose media with or without galactose. The primers in Figure 1A were used to examine the relative fold change in the association of H2A.Z upon induction with galactose.
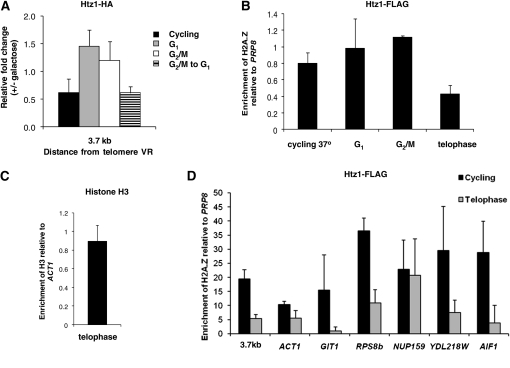
To further examine the correlation between the dissociation of H2A.Z and the onset of silencing we monitored its binding to the URA3 promoter at specific points in the cell cycle in the presence or absence of Sir3 induction, focusing on its site of maximum association. Control experiments conducted with the same cultures used for the ChIP experiments showed that URA3 is not repressed in G1- or G2/M-blocked cultures, but is repressed in cells progressing from G2/M to G1 (Figure S3), as previously reported (Martins-Taylor et al. 2004). When examined under these conditions, we find that the dissociation of H2A.Z with the URA3 promoter correlates closely with the onset of silencing, and not with the induction of Sir3. In cells blocked at G1 or G2/M, addition of galactose does not reduce H2A.Z binding; however, in cycling cells H2A.Z binding decreases (Figure 6A). We also observed this decrease in cells traversing only the G2/M to G1 interval (Figure 6A).
Figure 6.—
M-phase-specific removal of H2A.Z allows establishment of silencing. (A) Decreased association H2A.Z correlates with silencing. The relative association of H2A.Z and with telomere VR was determined in induced vs. uninduced cultures in G1 blocked cells, G2/M blocked cells, or in cells allowed to progress through the G2/M to G1 interval using the primers depicted in Figure 1A. Control experiments indicated that dissociation of H2A.Z required the presence of the GAL-SIR3 gene, and was not simply due to the addition of galactose (Figure S2). (B) Decrease in H2A.Z association at the telomeres in telophase. ChIP assays were performed to assess the association of H2A.Z–FLAG with the URA3 promoter (3.7-kb probe; see Figure 1A) in cycling cells and in cultures blocked at the indicated points in the cell cycle. Cultures were grown to log phase, then divided in two, with one-half blocked in a specific stage of the cell-cycle while the other half was maintained as a cycling control. Once the cultures were arrested in the desired cell-cycle stage, ChIP assays were performed. For each cell-cycle block the value of H2A.Z enrichment at the URA3 promoter is normalized to the PRP8 coding region and is expressed as the ratio of H2A.Z association in blocked cultures to that in a cycling cells control conducted on the same strain at the same time. For the 37° control experiment the ratio is of a culture shifted to 37° to a parallel culture maintained at 23°. Strain YSH869 was used to conduct cycling, G1, G2/M, and cycling 37° experiments; strain YSH862 was used for the telophase arrest experiment. The difference between telophase-blocked cells and each of the other conditions tested is statistically significant (Student's t-test, P < 0.05). (C) Histone H3 association remains unchanged in telophase. ChIP assays were performed on strain YSH862 to determine the association of histone H3 at the URA3 promoter. Cells were grown to log phase, when half the culture was shifted to nonpermissive temperature at 37°. Once the cells were blocked in telophase, ChIP assays were performed. H3 enrichment at the URA3 promoter is normalized to the ACT1 coding region and is expressed as a ratio of H3 enrichment in telophase-blocked cells to that in the cycling cells. (D) Dissociation of H2A.Z from nontelomeric genes in telophase. ChIP DNA samples from the experiment described in B were used to analyze the association of H2A.Z at the indicated loci in cells blocked in telophase and in cycling cells. Enrichment of H2A.Z is expressed relative to the PRP8 coding region.
Sir3-promoted silencing correlated with decreased H2A.Z association with the telomere, but these experiments couldn't determine a cause and effect relationship. H2A.Z loss could be a prerequisite for Sir3 association and transcriptional silencing; alternatively, the spreading of silencing that is specifically allowed in mitosis could actively displace H2A.Z. To distinguish these possibilities we used the cdc14-1 strain described above. If H2A.Z removal is a cause rather than an effect of silencing then its loss should be independent of the silencing status of the locus. We blocked cells in telophase and monitored the association of H2A.Z with the URA3 promoter. As shown in Figure 6B, we observe a decrease in H2A.Z association with the telomere in cells blocked in telophase, in the absence of Sir3 induction (i.e., when the locus is unsilenced). This decrease is not a function of shifting to a higher temperature and doesn't occur when these cells are blocked at other points in the cell cycle (Figure 6B). These results suggest there is an M-phase-specific loss of H2A.Z that is not dependent on the induction of Sir3 or on the establishment of silencing.
There are two basic possibilities for the fate of this H2A.Z-containing nucleosome in telophase. First, the nucleosome could remain in place, with H2A.Z–H2B dimers replaced with H2A–H2B dimers. Alternatively, displacement of H2A.Z could reflect a temporary loss of the entire nucleosome from chromatin. To distinguish between these possibilities we conducted chromatin immunoprecipitation experiments with an antibody to histone H3 using the same probes and conditions we used to track H2A.Z association (Figure 6C). We find that H3 association is unchanged in telophase-blocked cells. This suggests that the loss of H2A.Z we detect is not accompanied by a persistent loss of the nucleosome at this position.
We observed a cell-cycle-specific displacement of H2A.Z from a telomere-proximal URA3 gene. Is H2A.Z loss from chromatin a general feature of telophase-blocked cells? To examine this possibility we compared the association of H2A.Z in cycling and telophase-blocked cells at genes previously reported to bind H2A.Z at their promoters (Meneghini et al. 2003; Guillemette et al. 2005; Zhang et al. 2005). For these experiments we selected a diverse group of genes varying in chromosomal position and expression levels. We find that most, but not all, of these loci also exhibit a decrease in H2A.Z binding in telophase (Figure 6D). Thus, telophase-specific loss of H2A.Z does not occur exclusively on our reporter gene and may be widespread.
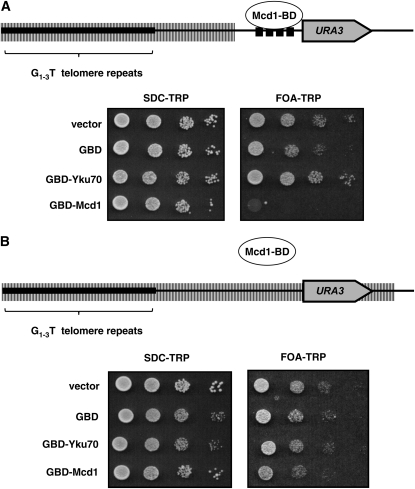
Our experiments suggest that H2A.Z, Sas2, and Mcd1 act as inhibitors of silencing, and either their removal or inactivation bypasses the cell-cycle-progression requirement for the establishment of silencing. Independent studies have suggested that Sas2 and H2A.Z act in a direct manner to define the transition between heterochromatin and euchromatin (Kimura et al. 2002; Suka et al. 2002; Meneghini et al. 2003). The localization of Mcd1 relative to silent chromatin has led to speculation that it may also help define these transitions (Blat and Kleckner 1999; Donze et al. 1999; Laloraya et al. 2000; Glynn et al. 2004; Chang et al. 2005). To examine the possibility that Mcd1 is able to limit the spread of heterochromatin, we expressed a fusion protein linking Mcd1 to the Gal4 DNA binding domain in a strain bearing binding sites for Gal4 adjacent to a telomere-linked URA3 gene (Figure 7). We find that this fusion protein can inhibit the silencing of URA3 in this strain, but not in a strain that lacks the Gal4 binding sites. The Mcd1–BD fusion does not activate transcription when tethered adjacent to a reporter gene located at an internal chromosome location (not shown), while a temperature-sensitive Mcd1 protein exhibits a temperature-dependent ability to inhibit heterochromatin spreading (Figure S4). Thus, tethered Mcd1 appears to specifically inhibit the spread of heterochromatin.
Figure 7.—
Mcd1 can limit heterochromatin spreading. (A) Cells bearing plasmids expressing the indicated Gal4 DNA binding domain (GBD) fusion proteins were applied at 10-fold dilutions on the indicated media and allowed to grow for approximately 3 days at 30°. The SDC–TRP control plate selects for the plasmid and ensures equal platings between cultures. FOA–TRP media selects against URA3 expression; thus only silenced cells grow. Yku70 is a positive control promoting telomeric silencing. Strains bearing the Mcd1–BD plasmid exhibit decreased silencing compared to the vector control or to a control expressing only the DNA binding domain (GBD). The plasmid expressing the Gal4BD–Mcd1 fusion complements the lethality observed in strains bearing a temperature-sensitive allele of MCD1, grown at the nonpermissive temperature (not shown). (B) The experiment described in A was performed in a strain lacking Gal4 binding sites adjacent to URA3.
DISCUSSION
Independent studies have demonstrated that the establishment of Sir protein-dependent silencing in yeast requires progression through the cell cycle (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001; Lau et al. 2002; Martins-Taylor et al. 2004). Using strains expressing a temperature-sensitive Sir3 protein, Miller and Nasmyth reported that silencing is principally established in S-phase (Miller and Nasmyth 1984), a finding consistent with later studies that used an inducible tethered Sir1 protein to also examine the establishment of silencing at HMR (Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001). A subsequent study using the conditional sir3-8 strain concluded that progression through both S- and M-phases was needed to establish silencing at HMR, but that silencing was largely accomplished in M-phase (Lau et al. 2002). Finally, we have previously reported that progression through mitosis is both necessary and sufficient for the silencing of a telomere-linked URA3 gene (Martins-Taylor et al. 2004). In this study we conducted experiments to identify which event in the establishment of silencing at telomeres requires cell-cycle progression. Consistent with prior studies, we find that silent chromatin at our telomere-linked reporter gene is characterized by an increase in the association of Sir3, the deacetylation of histone H3 and H4, specifically H4K16, and a decrease in the methylation of histone H3K79. In cells blocked in the cell cycle, induced expression of Sir3 may lead to a modest increase in the association of Sir3 across the URA3 reporter gene, but does not lead to changes in histone modification or to changes in URA3 expression. Pior studies suggested that limited association of the Sir proteins occurred prior to the establishment of silencing at the HMR locus (Lau et al. 2002; Kirchmaier and Rine 2006). In one of these studies decreases in histone acetylation were observed at the HMR silencer sequence, and to a lesser extent at the a1 gene, prior to the establishment of silencing (Kirchmaier and Rine 2006), but the magnitude of these changes did not reach the level seen in silenced cells. Overall, these results indicate that cell-cycle progression regulates an early step in the establishment of silencing.
As shown in this study, at telomeres this regulation is manifested at a point in mitosis coincident with the anaphase–telophase transition; cells held at telophase (but not at metaphase) are able to establish silencing, in the absence of cell-cycle progression. This phase of the cell cycle features a significant reorganization of chromatin, including the loss of chromosome cohesion and decondensation of chromosomes. It is possible that these or concomitant events create a window of opportunity allowing the establishment and spreading of Sir proteins. To identify factors that restrict the establishment of silencing to mitosis, we screened a series of strains lacking candidate factors. We found that elimination of either H2A.Z or Sas2, or the inactivation of Mcd1, bypasses the cell-cycle-progression requirement for the establishment of silencing. Our Mcd1 results extend a prior observation showing that elimination of MCD1 transcription alters the timing of silencing at the HMR locus, allowing silencing to occur following only S-phase progression (Lau et al. 2002). The Mcd1 protein undergoes regulated removal from chromosomes at a time coincident with when we observe the establishment of silencing. Our observation that inactivation of Mcd1 in G1-blocked cells allows gene repression suggests that it may be the loss of Mcd1 binding or activity, and not loss of chromosome cohesion per se, that is the relevant event in allowing the establishment of silencing. While our results do not address whether the effect of Mcd1 on our reporter gene is direct or indirect, we show that tethering Mcd1 can block the spread of silent chromatin, a result that is consistent with prior studies suggesting a role for cohesins in the establishment of chromatin boundaries on the basis of its pattern of association with the HMR locus (Blat and Kleckner 1999; Donze et al. 1999; Laloraya et al. 2000; Glynn et al. 2004). ChIP experiments indicated that Mcd1 protein was only weakly bound to individual sites near the URA3 reporter gene; it is possible that the collective binding of Mcd1 at this telomere is sufficient to exert the effects we observed.
Previous studies have shown that both Sas2 and H2A.Z help demarcate transitions between heterochromatin and euchromatin (Kimura et al. 2002; Suka et al. 2002; Meneghini et al. 2003). Loss of H2A.Z leads to decreased expression of telomere-proximal genes, suggesting that it helps limit the spreading of silent chromatin from the telomere (Meneghini et al. 2003). Sas2 is a histone acetyltransferase that opposes the action of the Sir2 deacetylase; relative levels of the Sir2 and Sas2 proteins help determine the transition point between heterochromatin and euchromatin at telomeres (Kimura et al. 2002; Suka et al. 2002). In addition, Sas2-mediated acetylation of lysine 16 on histone H4 has been reported to be important for the deposition of H2A.Z at yeast telomeres (Babiarz et al. 2006; Shia et al. 2006). Therefore, H2A.Z and Sas2 appear to function in a direct manner to limit the spread of silent chromatin from the telomere. In our inducible silencing system, H2A.Z has a causal role in regulating the timing of silencing: deletion of the HTZ1 gene abolishes the requirement for cell-cycle progression in establishing silent chromatin. Is H2A.Z acting in a direct or indirect manner? Our ChIP experiments are consistent with H2A.Z playing a direct role. First, H2A.Z is present at the promoter of the URA3 reporter gene. Second, H2A.Z's association at the URA3 promoter is anticorrelated with the silencing status of the locus; H2A.Z association decreases under conditions in which URA3 is silenced. Finally, H2A.Z is displaced from the URA3 promoter in a silencing-independent manner at telophase, the single point in the cell cycle at which we know silencing can be established in our system.
There are at least two general models for how H2A.Z removal could directly affect the spreading of heterochromatin. H2A.Z-containing nucleosomes generally exhibit increased turnover; this frequent turnover has been proposed to limit heterochromatin spreading by disrupting the cycle of Sir2-mediated histone modification/Sir3/Sir4 histone binding. Rapid erasure of Sir2-mediated histone deacetylation via nucleosome turnover could prevent Sir3/Sir4 binding and inhibit further incursions of the Sir complex into subtelomeric sequences (Dion et al. 2007). Alternatively, the presence of H2A.Z may directly affect post-translational modifications of the nucleosome or Sir protein association with histones. In vitro the presence of H2A.Z in reconstituted nucleosomes has been shown to affect susceptibility of the nucleosome to histone modifying enzymes (Li et al. 2005). In each of these models the presence of an H2A-containing nucleosome in the URA3 promoter is a preferred substrate for Sir-mediated heterochromatin spreading.
Our limited survey suggests that dissociation of H2A.Z may occur at many genes in telophase. The mechanism for this removal is unclear. H2A.Z deposition in yeast is accomplished by the Swr complex (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004); this complex colocalizes with H2A.Z in yeast (Zhang et al. 2005) and could potentially also function in removing H2A.Z. H2A.Z is also removed from promoter sequences of many yeast genes upon induction of gene transcription; it is not clear if this occurs via a passive or active mechanism (Santisteban et al. 2000; Adam et al. 2001; Larochelle and Gaudreau 2003; Zhang et al. 2005). Since H2A.Z nucleosomes may be intrinsically unstable compared to H2A-containing nucleosomes (Suto et al. 2000; Abbott 2001; Zhang et al. 2005; Dion et al. 2007) the dynamic changes in chromosome structure occurring at telophase may act to preferentially displace H2A.Z-containing nucleosomes. Our H3 ChIP results suggest that these nucleosomes are rapidly replaced by ones containing conventional H2A.
We found that three discrete genetic alterations relieved the cell-cycle requirement for establishing silencing. Do these factors function in independent pathways to impose the cell-cycle-progression requirement on the establishment of silencing, or are their functions interrelated? As described above, histone acetylation appears to affect the deposition of H2A.Z. In addition, prior studies have identified a role for H2A.Z in regulating chromosome stability (Krogan et al. 2004; Daniel et al. 2006; Keogh et al. 2006). However, thus far no role for H2A.Z or Mcd1 in regulating each other's activity or chromosome association has been reported. We define the anaphase–telophase transition as the key point in the cell cycle allowing the establishment of silencing at yeast telomeres. Using a conditional SIR3 allele Lau et al. (2002) also reported that the establishment of silencing at HMR occurred primarily in M-phase. However, independent studies using an inducible, tethered Sir1 protein to establish silencing at HMR reported observing the onset of repression in S-phase (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001; Lau et al. 2002; Martins-Taylor et al. 2004). This suggests that the manner in which silencing is induced can influence the cell-cycle interval required for the establishment of silencing. In independent work we have also observed that dependence on cell-cycle progression for the establishment of silencing is locus dependent; in particular, we have found that that HML is far less subject to the cell-cycle progression requirement than is HMR (A. G. Lazarus and S. G. Holmes, unpublished results). We suggest that a common mechanism involving functionally interrelated factors acts to impose the cell-cycle progression requirement, but that sensitivity to both the system used to induce silencing and the specific locus examined can alter when in the cell cycle inhibition is lifted. In particular, our results from this study suggest that a cell-cycle-dependent relaxation of chromatin barriers can dictate the timing of the establishment of silencing; the causes and possible broader consequences of the cell-cycle-dependent displacement of H2A.Z remain to be determined.
Acknowledgments
We thank Sue Biggins, Kim Nasmyth, David Shore, Joshua Babiarz, Jasper Rine, and Dan Burke for yeast strains and plasmids and Vincent Guacci for antibody to Mcd1. We also thank Marc Meneghini and the members of the Holmes lab for valuable discussions, Merrit Hickman for assistance in plasmid construction, and Lewis Lukens for comments on the manuscript. Finally, we are grateful to Ewa Menet at the Yale Cell Sorter Facility for her help with FACS analysis. T.R. gratefully acknowledges support from a Pfizer SURF award and the Howard Hughes Medical Foundation. This work was supported by a grant from the National Science Foundation (MCB-0617986) to S.G.H.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123844/DC1.
References
- Abbott, D., 2001. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 276 41945–41949. [DOI] [PubMed] [Google Scholar]
- Adam, M., F. Robert, M. Larochelle and L. Gaudreau, 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21 6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, O. M., and D. E. Gottschling, 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8 1133–1146. [DOI] [PubMed] [Google Scholar]
- Babiarz, J. E., J. E. Halley and J. Rine, 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz, K. K., N. J. Krogan, A. Emili, J. Greenblatt and P. Hieter, 2004. The ctf13–30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman et al., 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat, Y., and N. Kleckner, 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98 249–259. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis and J. R. Broach, 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7 592–604. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., C. D. Allis, B. M. Turner and J. R. Broach, 1996. Transcriptional silencing in yeast requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie et al., 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis et al., 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12 165–170. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., L. Milne and M. Grunstein, 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277 4778–4781. [DOI] [PubMed] [Google Scholar]
- Chang, C. R., C. S. Wu, Y. Hom and M. R. Gartenberg, 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19 3031–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, C. T., S. Buck, R. Sternglanz and D. Shore, 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75 531–541. [DOI] [PubMed] [Google Scholar]
- Daniel, J. A., B. E. Keyes, Y. P. Ng, C. O. Freeman and D. J. Burke, 2006. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics 172 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman et al., 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315 1405–1408. [DOI] [PubMed] [Google Scholar]
- Donze, D., C. R. Adams, J. Rine and R. T. Kamakaka, 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C., A. Ehrenhofer-Murray, S. Loo and J. Rine, 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276 1547–1551. [DOI] [PubMed] [Google Scholar]
- Fox, M. H., 1980. A model for the computer analysis of synchronous DNA distributions obtained by flow cytometry. Cytometry 1 71–77. [DOI] [PubMed] [Google Scholar]
- Glynn, E. F., P. C. Megee, H. G. Yu, C. Mistrot, E. Unal et al., 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2 E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette et al., 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3 e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh et al., 1997. Hyperactivation of the silencer proteins Sir2p and Sir3p causes chromosome loss. Genetics 145 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie et al., 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen and L. R. Pease, 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77 61–68. [DOI] [PubMed] [Google Scholar]
- Hutter, K. J., and H. E. Eipel, 1978. Flow cytometric determinations of cellular substances in algae, bacteria, moulds and yeasts. Antonie Van Leeuwenhoek 44 269–282. [DOI] [PubMed] [Google Scholar]
- Imai, S., C. M. Armstrong, M. Kaeberlein and L. Guarente, 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403 795–800. [DOI] [PubMed] [Google Scholar]
- Katan-Khaykovich, Y., and K. Struhl, 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan et al., 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A., T. Umehara and M. Horikoshi, 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32 370–377. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2001. DNA replication-independent silencing in S. cerevisiae. Science 291 646–650. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2006. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor et al., 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 963–972. [DOI] [PubMed] [Google Scholar]
- Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2 E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T., 2007. Chromatin modifications and their function. Cell 128 693–705. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan et al., 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12 1565–1576. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., K. Baetz, M. C. Keogh, N. Datta, C. Sawa et al., 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani, S. K., and M. Grunstein, 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4 276–284. [DOI] [PubMed] [Google Scholar]
- Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier and J. Cote, 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277 30421–30424. [DOI] [PubMed] [Google Scholar]
- Laloraya, S., V. Guacci and D. Koshland, 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle, M., and L. Gaudreau, 2003. H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 22 4512–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., S. G. Pattenden, D. Lee, J. Gutiérrez, J. Chen et al., 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102 18385–18390. [DOI] [PMC free article] [PubMed]
- Li, Y. C., T. H. Cheng and M. R. Gartenberg, 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291 650–653. [DOI] [PubMed] [Google Scholar]
- Lieb, J. D., X. Liu, D. Botstein and P. O. Brown, 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28 327–334. [DOI] [PubMed] [Google Scholar]
- Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz and D. Moazed, 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121 515–527. [DOI] [PubMed] [Google Scholar]
- Luo, K., M. A. Vega-Palas and M. Grunstein, 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, D. J., R. Marquardt, G. J. Shei, A. B. Rose and J. R. Broach, 1991. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 5 605–615. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor, K., M. L. Dula and S. G. Holmes, 2004. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics 168 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic, M., K. Martins-Taylor, M. Hickman, J. Tanny, D. Moazed et al., 2006. New alleles of SIR2 define cell-cycle-specific silencing functions. Genetics 173 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. L., S. P. Gygi, R. Aebersold and P. Hieter, 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7 959–970. [DOI] [PubMed] [Google Scholar]
- Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas et al., 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P. C., C. Mistrot, V. Guacci and D. Koshland, 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4 445–450. [DOI] [PubMed] [Google Scholar]
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk and K. Nasmyth, 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 35–45. [DOI] [PubMed] [Google Scholar]
- Millar, C. B., F. Xu, K. Zhang and M. Grunstein, 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A., and K. Nasmyth, 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312 247–251. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen et al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303 343–348. [DOI] [PubMed]
- Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst et al., 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger and K. Struhl, 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., E. Ray and L. Pillus, 1997. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K., C. D. Allis and S. I. Grewal, 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293 1150–1155. [DOI] [PubMed] [Google Scholar]
- Petronczki, M., B. Chwalla, M. F. Siomos, S. Yokobayashi, W. Helmhart et al., 2004. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 117 3547–3559. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W., 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcription states in S. cerevisiae. Cell 59 637–647. [DOI] [PubMed] [Google Scholar]
- Rudner, A. D., B. E. Hall, T. Ellenberger and D. Moazed, 2005. A nonhistone protein–protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25 4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo, P. A., and G. S. Roeder, 2000. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11 3601–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban, M. S., T. Kalashnikova and M. M. Smith, 2000. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103 411–422. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., A. J. Bannister, P. M. Dehe, V. Geli and T. Kouzarides, 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 279 47506–47512. [DOI] [PubMed] [Google Scholar]
- Shahbazian, M. D., and M. Grunstein, 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76 75–100. [DOI] [PubMed] [Google Scholar]
- Shia, W. J., B. Li and J. L. Workman, 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20 2507–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson et al., 1998. Identification of high-copy disruptors of telomeric silenicng in Saccharomyces cerevisiae. Genetics 150 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R. V., 2004. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., A. Hecht, K. Luo and M. Grunstein, 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11 83–93. [DOI] [PubMed] [Google Scholar]
- Suka, N., Y. Suka, A. A. Carmen, J. Wu and M. Grunstein, 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8 473–479. [DOI] [PubMed] [Google Scholar]
- Suka, N., K. Luo and M. Grunstein, 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32 378–383. [DOI] [PubMed] [Google Scholar]
- Suto, R. K., M. J. Clarkson, D. J. Tremethick and K. Luger, 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7 1121–1124. [DOI] [PubMed] [Google Scholar]
- Tanner, K. G., J. Landry, R. Sternglanz and J. M. Denu, 2000. Silent information regulator 2 family of NAD-dependent histone/protein deaceylases generates a unique product 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny, J. C., and D. Moazed, 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn, J. H., P. L. Gunyuzlu, W. H. White, L. A. Thompson and G. F. Hollis, 2000. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast 16 553–560. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403 623–627. [DOI] [PubMed] [Google Scholar]
- Valenzuela, L., and R. T. Kamakaka, 2006. Chromatin insulators. Annu. Rev. Genet. 40 107–138. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., P. R. Gafken and D. E. Gottschling, 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109 745–756. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Zhang, H., D. O. Richardson, D. N. Roberts, R. Utley, H. Erdjument-Bromage et al., 2004. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 24 9424–9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., D. N. Roberts and B. R. Cairns, 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova, J., and A. Thakar, 2008. H2A.Z: view from the top. Structure 16 166–179. [DOI] [PubMed] [Google Scholar]