Abstract
Basal-like breast cancer (BBC) is a subtype of breast cancer with poor prognosis1–3. Inherited mutations of BRCA1, a cancer susceptibility gene involved in double-strand DNA break (DSB) repair, lead to breast cancers that are nearly always of the BBC subtype3–5; however, the precise molecular lesions and oncogenic consequences of BRCA1 dysfunction are poorly understood. Here we show that heterozygous inactivation of the tumor suppressor gene Pten leads to the formation of basal-like mammary tumors in mice, and that loss of PTEN expression is significantly associated with the BBC subtype in human sporadic and BRCA1-associated hereditary breast cancers. In addition, we identify frequent gross PTEN mutations, involving intragenic chromosome breaks, inversions, deletions and micro copy number aberrations, specifically in BRCA1-deficient tumors. These data provide an example of a specific and recurrent oncogenic consequence of BRCA1-dependent dysfunction in DNA repair and provide insight into the pathogenesis of BBC with therapeutic implications. These findings also argue that obtaining an accurate census of genes mutated in cancer will require a systematic examination for gross gene rearrangements, particularly in tumors with deficient DSB repair.
Advances in methodologies for high-throughput DNA sequencing and copy number analysis have hastened the global cataloguing of genetic abnormalities in cancer (for example, see refs. 6–8). Although such studies have identified classical coding mutations and large amplifications and deletions in known and putative oncogenes and tumor suppressors, some types of genetic change, such as chromosome breaks, gene translocations and microduplications or microdeletions, are difficult to detect. Indeed, recurrent fusions between TMPRSS2 and ETS family transcription factor genes have been identified in prostate carcinoma by a systems biology approach9, and they suggest that oncogenic gene rearrangements may be common in epithelial neoplasia. Global gene expression profiling studies of breast carcinoma have delineated several molecular subtypes that have distinct pathological and clinical characteristics1–5. Of these, basal-like breast cancer (BBC) comprises 10–20% of all breast cancer and is one of the subtypes with the worst prognosis2–5. The term BBC was coined because these tumors express cytokeratin markers typical of basally oriented epithelial cells of the normal mammary gland, such as CK5, CK14 and CK17 (refs. 1,3,5). In addition to having characteristic cytokeratin expression, BBCs are highly proliferative, poorly differentiated and genomically unstable, and they pose clinical challenges because they rarely express the three most common therapeutically targeted ‘Achilles’ heels’ of breast cancer: the estrogen receptor (ER), progesterone receptor and HER2 receptor (refs. 1,3,5,7).
Intriguingly, breast tumors initiated by an inherited mutation of BRCA1 are nearly always basal-like3,5. BRCA1 dysfunction is thought to be tumorigenic primarily owing to defective BRCA1-dependent DSB repair, which precipitates an accumulation of secondary mutations10; however, only general genomic patterns at relatively low resolution have been described (reviewed in ref. 5). Despite these advances in delineating BBC, the molecular lesions and oncogenic signaling pathways that drive BBC and the precise oncogenic consequences of BRCA1-mediated basal-like tumorigenesis are incompletely known. The phosphatidylinositol 3-kinase (PI3K) pathway is a potent oncogenic signaling cascade that promotes cell transformation, proliferation, migration, angiogenesis and genomic instability; inhibits apoptosis; maintains stem cell compartments; and is associated with poor prognosis in carcinoma11–14. PTEN is the crucial tumor suppressor in this pathway because it catalyzes the precise opposite reaction to PI3K, thereby inhibiting downstream signaling12. In breast cancer, documented coding mutations of PTEN are rare (<5%); however, ~25% of tumors have significantly diminished amounts of PTEN protein, resulting in unrestrained signaling by the PI3K pathway15.
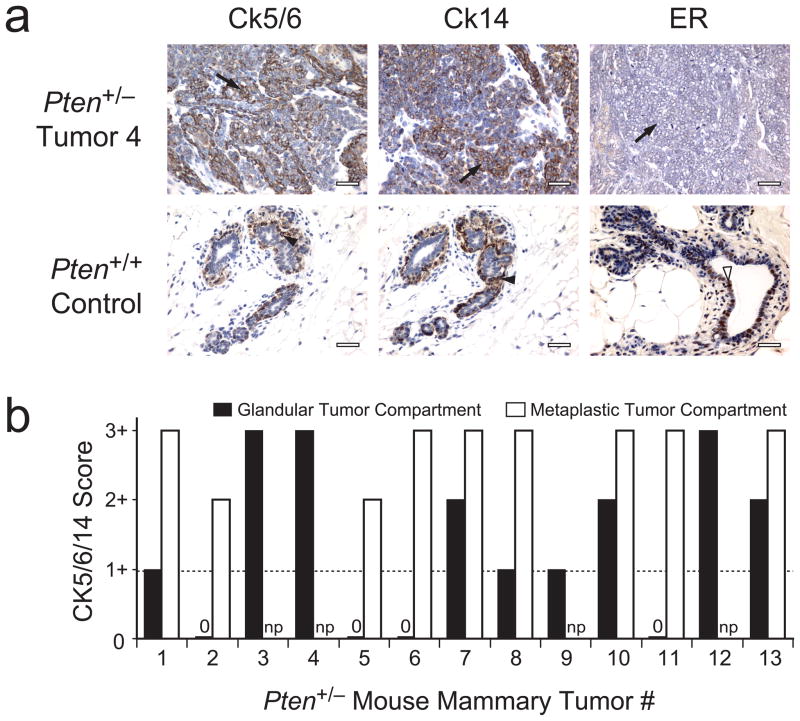
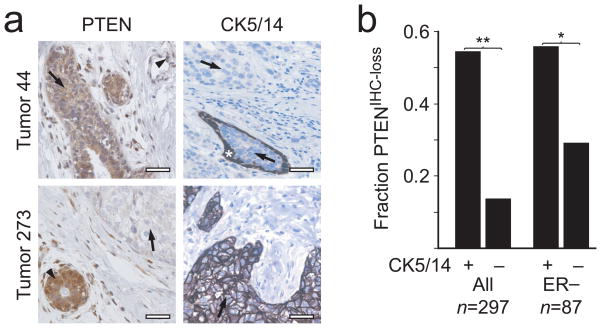
While reviewing breast cancer subtypes in mouse models, we noted that all 13 mammary tumors derived from 12 mice with heterozygous inactivation of one Pten allele16 showed basal-like characteristics (positive staining for CK5, CK6 or CK14 (hereafter CK5/6/14) by immunohistochemistry (IHC); Fig. 1 and Supplementary Table 1 online). To investigate the role of PTEN in human BBC, we therefore characterized PTEN and CK5/14 protein expression by IHC in non-hereditary breast tumors from 297 individuals (Fig. 2a). A reduction in PTEN protein immunostaining (PTENIHC-loss) of tumor cells as compared with adjacent non-neoplastic cells was observed significantly more frequently in CK5/14-positive BBC than in CK5/14-negative non-BBC (54.3 versus 13.4%, respectively; P = 4 × 10−9; Fig. 2b and Supplementary Table 2 online). Because both the BBC phenotype1,3,5 and reduced PTEN15 have been associated with lack of ER expression, we tested the relationship between PTEN and basal-like status in the ER-negative subset and found that it remained significant (56.3 versus 29.1%; P = 0.0122; Fig. 2b and Supplementary Table 2 online). Loss of PTEN thus defines a significant subset of non-hereditary BBC (NHBBC).
Figure 1.
IHC analysis of basal-like phenotype markers in Pten+/− mouse mammary tumors. (a) Representative section showing IHC staining for CK5/6, CK14 and ER in a Pten+/− mammary tumor (top) and normal breast tissue from a wild-type (Pten+/+) control mouse (bottom). Arrows indicate tumor cells; filled arrowheads indicate non-neoplastic basal-layer epithelial cells; open arrowhead indicates non-neoplastic luminal-layer epithelial cells. Scale bars, 25 μm. (b) CK5/6/14 IHC staining scores for 13 Pten+/− mammary tumors. The maximal score for CK5/6 or CK14 staining (Methods) is plotted for each tumor compartment; a score of 1+ (horizontal broken line) is defined as basal-like. 0, tumor compartment present but no staining; np, respective component not present in tumor. See also Supplementary Table 1 online.
Figure 2.
Immunohistochemical analysis of PTEN and CK5/14 basal-like phenotype markers in 297 non-hereditary human breast carcinomas. (a) Representative PTEN and CK5/14 IHC in two non-hereditary breast tumors. Top, a PTENIHC-positive and CK5/14-negative example; bottom, a PTENIHC-loss and CK5/14-positive example. Arrows indicate tumor cells; arrowheads indicate non-neoplastic cells. Asterix denotes the non-neoplastic CK5/14-positive basal epithelial layer enclosing a portion of in situ breast carcinoma, shown here as an example of an internal positive control. Scale bars, 25 μm. (b) Correlation analysis between PTEN loss and the CK5/14-positive basal-like subtype among all tumors (left) and in the ER-negative group (right). *P = 0.0122; **P = 4 × 10−9.
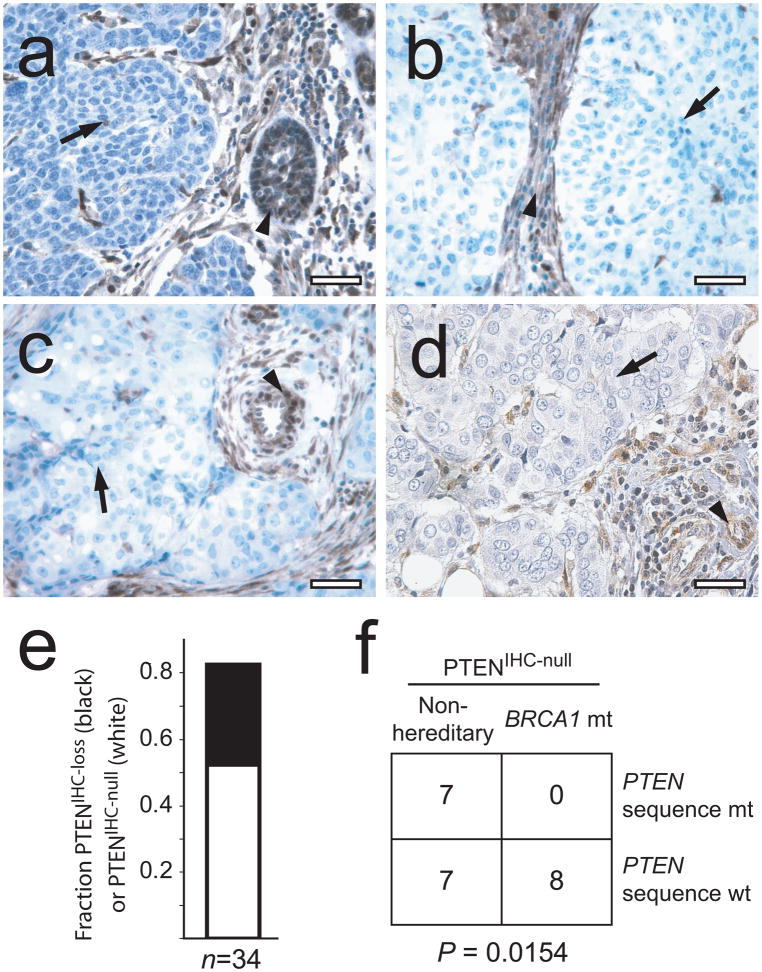
Given that nearly all BRCA1-associated hereditary breast cancers are basal-like3,5,17, we next investigated PTEN expression in 34 breast tumor biopsies from individuals with inherited BRCA1 mutations (hereditary BBC (HBBC); Supplementary Table 3 online). Notably, 82.4% (28/34) of tumors showed loss of PTEN by IHC (P = 4 × 10−12; Fig. 3a). In addition, suggestive of a genetic mechanism for PTEN loss, 52.9% (18/34) of tumors showed a marked staining pattern; namely, PTEN protein was completely undetectable in tumor cells (PTENIHC-null) but strongly expressed in adjacent normal cells (Fig. 3a–c,e). To test for an association between the PTENIHC-null phenotype and mutation of PTEN, we screened 14 PTENIHC-null non-hereditary breast tumors (Fig. 3d) and 8 PTENIHC-null HBBC tumors (Fig. 3a–c) for mutations in PTEN by DNA sequencing. Whereas half of the PTENIHC-null non-hereditary tumors had coding mutations in PTEN, unexpectedly, no sequence alterations were found in the HBBC samples (P = 0.0154; Fig. 3f). These results, together with the fact that the HCC-1937 BRCA1-mutant cell line is known to have homozygous deletion of PTEN18, suggested that BRCA1-mutant tumors may have a distinct type of PTEN genetic lesion.
Figure 3.
PTEN status in human BRCA1-mutant hereditary breast cancers. (a–d) PTEN IHC results for three representative PTENIHC-null BRCA1-associated hereditary breast tumor specimens (a–c) and one non-hereditary PTENIHC-null breast tumor with a PTEN fs108X mutation (d). Scale bars, 25 μm. (e) Fraction of PTENIHC-loss (filled) and PTENIHC-null (open) tumors among 34 BRCA1-associated hereditary breast tumors. (f) Analysis of PTEN nucleotide sequence mutations in PTENIHC-null non-hereditary breast cancers versus PTENIHC-null BRCA1-mutant hereditary breast cancers.
To investigate this possibility, we examined two model breast cancer cell lines, MDA-MB-436 and SUM-149, that are known to have mutations in BRCA1 (ref. 19) and to be basal-like20. These cell lines are ideal models because both carry the wild-type PTEN gene, as determined by DNA sequencing, but neither expresses PTEN protein, as assessed by protein blotting. In addition, PTEN expression cannot be induced by treatment with demethylation agents, indicating that PTEN silencing is not due to promoter methylation (data not shown). Conventional tiling BAC array comparative genomic hybridization (aCGH) experiments (data not shown) indicated that both cell lines show DNA copy number loss at the PTEN locus on chromosome 10q23.31, consistent with loss of one allele (loss of heterozygosity; LOH).
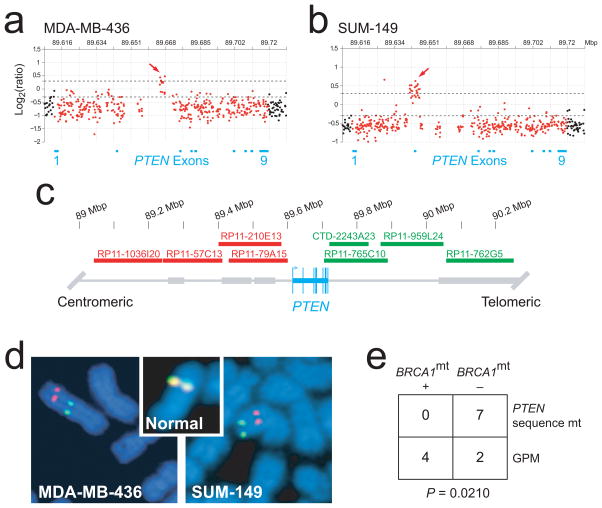
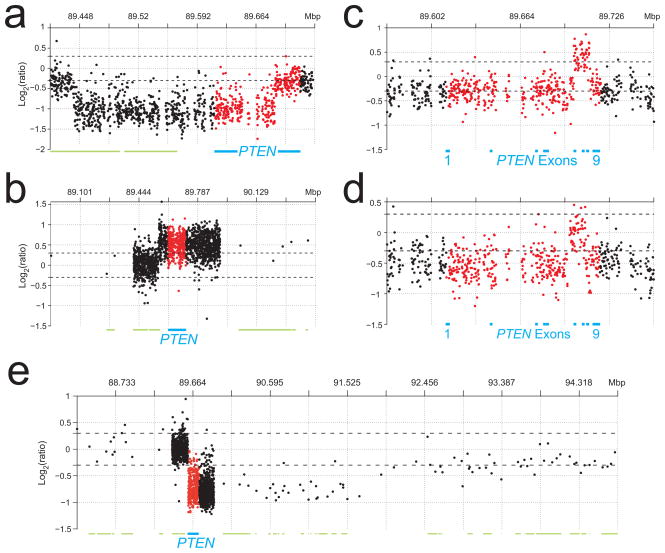
To look for smaller genetic lesions, we used a custom-designed high-density oligonucleotide aCGH (HD-aCGH) platform containing 1,747 probes spanning a region of ~500 kb centered on PTEN with an average interprobe spacing of 288 bp. HD-aCGH analysis showed that these regions of apparent PTEN LOH in fact contained highly focal, intragenic copy number increases (CNIs) in PTEN, affecting a portion of intron 2 (MDA-MB-436) and spanning exon 2 (SUM-149; Fig. 4a,b). Given the role of BRCA1 in DSB repair, we considered that the intragenic PTEN CNIs may be associated with gross chromosomal rearrangements. We therefore designed a two-color FISH assay (Fig. 4c) and identified distinct rearrangements of chromosome 10 that physically split the single remaining PTEN allele in each cell line (Fig. 4d). These results were corroborated for both cell lines by 5′-to-3′ exon-walking using RT-PCR, by which we could not amplify specific PTEN transcripts beyond exon 2 (data not shown). These data are most consistent with a mechanism involving multiple DSBs and inappropriate repair and reduplication of DNA segments at or near the breakpoints.
Figure 4.
Gross PTEN mutations in BRCA1-mutant human cell lines and xenografts. (a,b) HD-aCGH analyses of two model BRCA1-mutant breast cancer cell lines. The location of PTEN exons 1–9 on chromosome 10q23.31 are indicated by blue bars. The log2 ratios for probes localized in the PTEN gene are plotted in red; those for all other probes are plotted in black (gaps with no probes are present in several regions that contain highly repetitive sequence elements such as intron 2). Arrows indicate regions of focal CNI. Prominent broken horizontal lines are placed at log2 ratios of 0.3 and −0.3. (c) Two-color FISH assay. The PTEN gene and exons are indicated in blue. Centromeric BAC probes were labeled with spectrum orange–dUTP (red); telomeric BAC probes were labeled with spectrum green–dUTP (green). (d) FISH results for the BRCA1-mutant breast cancer cell lines in a,b. Inset, representative FISH results for one chromosome of a normal lymphocyte metaphase spread. (e) Analysis of BRCA1 mutation (mt) and GPM among 13 breast cancer xenografts and cell lines with any type of PTEN mutation. The basal-like status of these cell lines and xenografts is given in Table 1.
To investigate further the association of these types of PTEN locus structural mutation (hereafter termed ‘gross’ PTEN mutations; GPMs) with BRCA1 status, we sequenced BRCA1 in three breast cancer xenografts with identified homozygous deletions of PTEN21 (Table 1, Supplementary Table 3 and Supplementary Fig. 1 online). One xenograft, MX-1 (also known as Bx11), was found to have the 2795del4 BRCA1 mutation. (Although matched normal DNA was not available, this mutation is likely to be germ line because 2795del4 is a known BRCA1 founder mutation and the xenograft was established from a 29-year-old individual22.) Next, to test the relationship between BRCA1 and GPM rigorously, we selected all breast cancer cell lines and xenografts with any type of PTEN mutation from 31 samples that were comprehensively annotated for BRCA1 and PTEN mutational status (all cell lines positive for PTEN protein by protein blotting were assumed to not have GPMs; Table 1). Among the 13 samples with any type of PTEN mutation, GPMs were significantly associated with mutations in BRCA1; by contrast, PTEN coding mutations were specifically associated with wild-type BRCA1 (P = 0.0210; Fig. 4e). We therefore conclude that BRCA1 dysfunction is associated with GPM.
Table 1.
BRCA1 and PTEN status in 31 breast cancer cell lines and xenografts
| Sample ID | BRCA1 mutation statusa | PTEN mutation status | PTEN protein blot | Subtypeb |
|---|---|---|---|---|
| HCC-1937 | 5382insC (fs>1829X) | GPM (HD) | Null | Basal-like |
| MDA-MB-436 | 5396+1G>A (splice donor) | GPM (micro-CNI) | Null | Basal-like |
| MX-1c | 2795delT (fs>999X) | GPM (HD) | n.d. | n.a. |
| SUM-149 | 2288delT (fs>735X) | GPM (micro-CNI) | Null | Basal-like |
| BT-549 | WT | 822delG (L295X) | Null | Basal-like |
| Bx15c | WT | GPM (HD) | n.d. | n.a. |
| Bx33c | WT | GPM (HD) | n.d. | n.a. |
| CAMA-1 | WT | 274G>C (D92H) | Positive | Luminal |
| EVSA-T | WT | 951del4 (T319X) | n.d. | n.a. |
| MDA-MB-415 | WT | 407G>A (C136Y) | Positive | Luminal |
| MDA-MB-453 | WT | 919G>A (E307K) | Positive | Luminal |
| MDA-MB-468 | WT | IVS4+1G>T (A72fsX5) | Null | Basal-like |
| ZR75-1 | WT | 323T>G (L108R) | Positive | Luminal |
| BT-20 | WT | WT | Positive | Basal-like |
| BT-474 | WT | WT | Positive | Luminal |
| BT-483 | WT | WT | Positive | Luminal |
| DU-4475 | WT | WT | Positive | n.a. |
| Hs578T | WT | WT | Positive | Basal-like |
| MCF-7 | WT | WT | Positive | Luminal |
| MDA-MB-134VI | WT | WT | Positive | Luminal |
| MDA-MB-157 | WT | WT | Positive | Basal-like |
| MDA-MB-175VII | WT | WT | Positive | Luminal |
| MDA-MB-231 | WT | WT | Positive | Basal-like |
| MDA-MB-361 | WT | WT | Positive | Luminal |
| MDA-MB-435S | WT | WT | Positive | Basal-like |
| SK-BR-3 | WT | WT | Positive | Luminal |
| SUM-159PT | WT | WT | Positive | Basal-like |
| T-47D | WT | WT | Positive | Luminal |
| UACC-812 | WT | WT | Positive | Luminal |
| UACC-893 | WT | WT | Positive | n.a. |
| ZR75-30 | WT | WT | Positive | Luminal |
All mutations are relative to the +1 adenine nucleotide of the ATG start codon, with the amino acid change given in parentheses. GPM, gross PTEN mutation; HD, homozygous deletion; CNI, DNA copy number increase; n.d., not done; n.a., not available.
For cell line samples, BRCA1 mutational status is taken from ref. 19.
Breast cancer subtype is taken from ref. 20.
Xenograft.
These results were corroborated in breast tumor biopsies from women with hereditary BRCA1 mutations. Of 17 HBBC tumors analyzed for conventional PTEN mutations by DNA sequencing, all had wild-type PTEN (Supplementary Table 3 online). When analyzed on the HD-aCGH platform, however, five (71.4%) of seven PTENIHC-null HBBC tumors had specific GPMs associated with micro-CNIs (n = 2), macro-CNI (n = 1) and homozygous deletions (n = 2; Fig. 5a–e, Supplementary Table 3 and Supplementary Fig. 1 online). Seven of these HBBC samples, in addition to one PTENIHC-loss and three PTENIHC-positive HBBC tumors, were also analyzed on the conventional BAC aCGH platform (data not shown). Indicating the increased sensitivity of the HD-aCGH platform for detecting GPMs, one homozygous deletion was difficult to discern and both micro-CNIs were undetectable by BAC aCGH (data not shown). In addition, beyond copy number loss consistent with LOH, no obvious GPMs were detected in the PTENIHC-loss or PTENIHC-positive HBBCs.
Figure 5.
Gross PTEN mutations in five BRCA1-mutant breast tumor biopsies in vivo. (a–e) Analysis of PTENIHC-null BRCA1-associated breast tumors by HD-aCGH, revealing PTEN homozygous deletions (a,e), macro-CNI (b) and micro-CNIs (c,d). Probes and the PTEN locus are plotted as in Figure 4a,b. Green bars indicate neighboring genes.
Combining our patient tumor data with our data from cell lines and xenografts, we analyzed in total 28 BRCA1-mutant samples by conventional BAC aCGH, custom HD-aCGH and/or conventional DNA sequencing, and found nine cancers (32.1%) with GPMs. We conclude that PTEN loss is highly associated with both NHBBC and HBBC and can result from gene rearrangements involving DNA DSBs, intragenic inversions or insertions, homozygous deletions and focalized CNIs. Because chromosomal breaks, CNIs and small microdeletions were not detected by conventional aCGH or DNA sequencing, we are probably underestimating the true rate of GPMs in HBCC. Indeed, our PTENIHC-null data would suggest a rate of more than 50%. In addition, we identified two NHBBC samples with homozygous deletion of PTEN and, notably, both had hypermethylation of the BRCA1 promoter (data not shown), suggesting that BRCA1-dependent PTEN disruption may also occur in some sporadic breast tumors.
Our results have important implications for the pathogenesis and treatment of BBC and raise significant considerations for future mutation-cataloguing efforts in disease. The basal-like phenotype of Pten+/− mammary tumors and the high rate of PTEN loss in NHBBC and HBBC imply that the PTEN pathway is directly involved in transformation of basal-like progenitor cells. Notably, conventional sequence mutations of the TP53 tumor suppressor gene occur in >80% of NHBBC and HBBC (reviewed in ref. 5). Together, our results suggest that all three tumor suppressors are inactivated in a considerable proportion of BBC. Although the spectrum of TP53 mutations seems to differ between non-hereditary and hereditary breast cancer23, the observed mutational profiles of TP53 are not as specific or as markedly divergent as we have found here for PTEN, indicating that the selective pressure and/or mutational mechanism on TP53 and PTEN are likely to be distinct. Our data are most consistent with a model of HBBC oncogenesis in which selected stochastic coding mutation of TP53 in a basal-like breast cancer progenitor cell precedes loss of the second BRCA1 allele, which is known to otherwise be lethal to cells (reviewed in ref. 24), and that the subsequent BRCA1-dependent DSB repair defect precipitates genetic disruption of PTEN, which is then clonally selected. This model implies that most BBCs may be addicted25 to aberrant PTEN-PI3K pathway signaling. As a result, therapy targeted to this pathway may be an effective way to treat and possibly to prevent some sporadic and hereditary BBC.
Our study demonstrates an example of a specific and recurrent oncogenic consequence of defective DSB repair in breast cancer, and it is possible that other grossly rearranged genes also contribute to tumor progression. Our observations in HBBC are analogous to those in hereditary nonpolyposis colon cancer, where microsatellite instability due to lack of mismatch repair leads to mutation of TGFBR2 and other genes that drive tumor progression26. Our results highlight the need for high-throughput genomic methods to screen for gross structural gene mutations at a sub-kilobase resolution. There may be a wealth of gross mutations in breast cancer and other types of carcinoma in which such changes were once thought to be rare.
METHODS
Human tumor samples and immunohistochemistry
All tumor specimens were studied after approval by the Lund University Hospital ethics committee and Columbia University Institutional Review Board. Tissue microarrays containing formalin-fixed paraffin-embedded tissues in replicates from 425 individuals diagnosed with stage II invasive breast cancer but without a significant family history of breast or ovarian cancer were subjected to IHC staining for CK5/14. CK5/14 IHC was performed and scored into CK5/14-positive (defining basal-like status) and CK5/14-negative groups as described17. Some of these individuals were evaluated for PTEN protein expression by IHC on whole-mount, formalin-fixed, paraffin-embedded tissue sections, and the tumors were scored as PTEN-positive (PTENIHC-positive) or PTEN-loss (PTENIHC-loss) relative to normal epithelial, endothelial and/or stromal cells in each tissue section as described15. Tumors with undetectable PTEN immunostaining in contrast to adjacent normal cells were scored as PTEN-null (PTENIHC-null). We analyzed 297 tumors for both PTEN and CK5/14. Thirty-four breast tumors from carriers of germline BRCA1 mutations were obtained for PTEN IHC and genetic analyses. PTEN IHC was performed and scored for these tumors as above, with the exception that the primary antibody to PTEN was monoclonal (138G6, Cell Signaling Technology; dilution, 1:200; incubation, 2 h), and the Dako EnVision Plus system was used for signal detection.
Cell lines and xenografts
Breast cancer cell lines were obtained from the American Type Culture Collection and S. Ethier and were cultured according to standard recommendations. The MX-1 (also known as Bx11), Bx15 and Bx33 breast cancer xenografts have been described21. PTEN protein blotting was performed as described14.
BRCA1 promoter hypermethylation
BRCA1 promoter region methylation was analyzed by using a PSQ HS 96 pyrosequencing system according to manufacturer protocols (Biotage). DNA was isolated with a Wizard Genomic DNA purification kit (Promega) and bisulfate-modified with a EZ DNA Methylation kit (Zymo Research). We designed an assay containing two CpG island regions with internal controls for bisulfite treatment (nucleotide positions are relative to the +1 adenine of the ATG translational start codon on the annotated Ensembl v36 BRCA1 genomic sequence): BRCA1 region 1, −1291 to −1267 (containing five CpGs); and region 2, −1332 to −1310 (four CpGs). We used the following primers: BRCA1_Region1/ 2_PCR_F, 5′-NNTATTTTGAGAGGTTGTTGTT TAG-3′; BRCA1_Region1/2_PCR_R, 5′-biotin-TAA AAAACCCCACAACCTATCC-3′; BRCA1_Region1 _Seq, 5′-GAATTATAGATAAATTAAAA-3′; and BRCA1_Region2_Seq, 5′-GGTAGTTTTTTGGTTT T-3′. In all runs, CpGenome Universal Methylated DNA (Chemicon) was analyzed as a positive control, and CpGenome Universal Unmethylated DNA and reactions with no DNA were used as negative controls.
DNA sequencing
PTEN exons including intronic boundaries were sequenced as described15. BRCA1 mutations were identified by standard techniques27.
aCGH
Experiments using the 32K BAC aCGH platform were performed by using described methods28. A custom-designed HD-aCGH microarray (Agilent) was also used as described29. In brief, this HD-aCGH microarray contains 1,747 probes in replicates covering a region of ~500 kb centered on the PTEN locus at a mean spacing of 288 bp, and an additional ~20,000 probes with wide genome coverage that are used for data normalization with the popLowess algorithm29. Labeling and hybridization were performed according to the manufacturer’s recommendations, and data were loaded into BASE30 for analysis.
Two-color split-probe FISH
The following BACs were obtained from the BACPAC Resource Center (Children’s Hospital Oakland Research Institute) or Invitrogen, and DNA was prepared by standard techniques: RP11-1036I20, RP11-57C13, RP11-210E13, RP11-79A15, RP11-765C10, CTD-2243A23, RP11-959L24 and RP11-762G5. The RP11-1036I20, RP11-57C13, RP11-210E13 and RP11-79A15 probes 5′ to PTEN were generated by nick translation and labeled with spectrum orange dUTP; the RP11-765C10, CTD-2243A23, RP11-959L24, RP11-762G5 probes 3′ to PTEN were labeled with spectrum green dUTP. Metaphase chromosome spreads were prepared, and FISH was carried out by standard methods; hybridization signals were scored on at least 20 metaphase spreads.
Pten+/− mouse mammary tumors
The Pten+/− mouse model has been described16. Mouse research was approved by the Columbia Animal Care and Use Committee. Female mice developed mammary tumors at an average age of 11 months. Thirteen tumors from 12 mice were collected, fixed in formalin and embedded in paraffin. For IHC, primary antibodies D5/16 B4 (monoclonal antibody to CK5 or CK6 (anti-CK5/6); Covance) and AF64 (polyclonal anti-CK14; Covance) were used at 1:500 dilution and SC542 (polyclonal anti-ER; Santa Cruz) was used at 1:400 dilution after microwave antigen retrieval in 0.01 M citrate buffer. D5/16 B4 was applied overnight at 4 °C, whereas AF64 and SC542 were applied for 1 h at 22 °C. A Vectastain kit (Vector Labs) was used to detect binding of the primary antibody to its target antigen in accordance with the manufacturer’s recommendation using 3,3′-diaminobenzidine for visualization. Cytokeratin immunostaining was scored as follows: 0, no staining; 1+, moderate staining; 2+, focally strong (<15% of cells); 3+, strong in >15% of cells. CK5/6/14-positive was defined as 1+ (one case), 2+ (two cases) or 3+ (ten cases) for CK5/6 or CK14 staining in either the gland-forming tumor epithelial cell or metaplastic tumor epithelial cell compartments. IHC staining of ER was scored as follows: 0, <1% positive nuclei; 1+, 1–4% positive nuclei; 2+, ≥5% positive nuclei.
Statistical analyses
Correlations between variables in 2 × 2 tables were assessed with Pearson’s χ2 test, and tables with cells with fewer than five observations were assessed with Fisher’s exact test (sum of small P values). Tests were two-sided, and a P value of <0.05 was considered significant. Assuming a 25% prior probability rate (the approximate rate of PTEN loss in breast cancer), a one-sided P value for PTEN protein loss in BRCA1-associated hereditary breast cancer was calculated as the probability of having 28 or more PTENIHC-loss samples out of 34 cases with a binomial distribution.
Supplementary Material
Acknowledgments
We thank the patients whose contributions made this work possible. We thank B. Giovanella (Stehlin Foundation for Cancer Research) for breast cancer xenografts; J. Valcich, L. Tellhed and C. Strand for technical support; and R. Szalasny for administrative assistance. We regret our inability to cite all references germane to this work owing to space limitations. Funding was provided in part by US National Institutes of Health Scientist training grant 5T32 GM07367-29 (L.H.S.); grants CA082783 and CA097403 (R.P.); Department of Defense Breast Cancer Research Program Era of Hope Award BC061955 (S.K.G.-S.); the Avon Foundation (H.H., R.P.); the OctoberWoman Foundation (R.P.); the Swedish Cancer Society, Mrs. Berta Kamprad Foundation, Gunnar Nilsson Cancer Foundation, and Ingabritt and Arne Lundberg Foundation (Å.B.); and the Knut and Alice Wallenberg Foundation via the SWEGENE program (M.K., Å.B.).
Footnotes
AUTHOR CONTRIBUTIONS
L.H.S., S.K.G.-S., R.P. and Å.B conceived and designed the study; L.H.S., S.K.G.-S., J.S., G.J., K.H., S.K., J.V.-C., H.O., T.S., L.M., S.P.E., H.H., R.P. and Å.B. collected the samples; L.H.S., K.L., M.J., L.M., T.L., M.S., J.I. and H.H. performed and analyzed immunohistochemistry experiments; L.H.S. and C.P. designed and performed methylation analyses; L.H.S., C.P., M.M. and K.H. performed nucleotide sequencing experiments; L.H.S., J.S., G.J. and K.H. performed and analyzed aCGH experiments; L.H.S., M.M.P., S.S. and V.V.V.S.M. designed and performed FISH experiments; L.H.S., S.K.G.-S. and M.K. performed statistical analyses; R.P. and Å.B. supervised the study; and L.H.S. wrote the paper with assistance from R.P. and Å.B. and input from all coauthors.
Note: Supplementary information is available on the Nature Genetics website.
Published online at http://www.nature.com/naturegenetics
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva LD, Clarke C, Lakhani SR. Basal-like breast cancer. J Clin Pathol. doi: 10.1136/jcp.2006.041731. (published online 11 May 2007) [DOI] [Google Scholar]
- 4.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 6.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 10.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 11.Puc J, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 13.Janzen V, Scadden DT. Stem cells: good, bad and reformable. Nature. 2006;441:418–419. doi: 10.1038/441418b. [DOI] [PubMed] [Google Scholar]
- 14.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saal LH, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 16.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laakso M, Loman N, Borg A, Isola J. Cytokeratin 5/14-positive breast cancer: true basal phenotype confined to BRCA1 tumors. Mod Pathol. 2005;18:1321–1328. doi: 10.1038/modpathol.3800456. [DOI] [PubMed] [Google Scholar]
- 18.Tomlinson GE, et al. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 19.Elstrodt F, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 20.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 22.Giovanella BC, Stehlin JS, Williams LJ., Jr Heterotransplantation of human malignant tumors in ‘nude’ thymusless mice II Malignant tumors induced by injection of cell cultures derived from human solid tumors. J Natl Cancer Inst. 1974;52:921–930. doi: 10.1093/jnci/52.3.921. [DOI] [PubMed] [Google Scholar]
- 23.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 2001;61:4092–4097. [PubMed] [Google Scholar]
- 24.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 25.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz S, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 27.Loman N, Johannsson O, Kristoffersson U, Olsson H, Borg A. Family history of breast and ovarian cancers and BRCA1 and BRCA2 mutations in a population-based series of early-onset breast cancer. J Natl Cancer Inst. 2001;93:1215–1223. doi: 10.1093/jnci/93.16.1215. [DOI] [PubMed] [Google Scholar]
- 28.Jonsson G, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosom Cancer. 2007;46:543–558. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 29.Staaf J, et al. Detection and precise mapping of germline rearrangements in BRCA1, BRCA2, MSH2 and MLH1 using zoom-in array CGH. Hum Mutat. doi: 10.1002/humu.20678. (in the press) [DOI] [PubMed] [Google Scholar]
- 30.Saal LH, et al. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 2002;3:SOFTWARE0003. doi: 10.1186/gb-2002-3-8-software0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.