Abstract
Early detection is an effective means of reducing cancer mortality. Here, we describe a highly sensitive high-throughput screen that can identify panels of markers for the early detection of solid tumor cells disseminated in peripheral blood. The method is a two-step combination of differential display and high-sensitivity cDNA arrays. In a primary screen, differential display identified 170 candidate marker genes differentially expressed between breast tumor cells and normal breast epithelial cells. In a secondary screen, high-sensitivity arrays assessed expression levels of these genes in 48 blood samples, 22 from healthy volunteers and 26 from breast cancer patients. Cluster analysis identified a group of 12 genes that were elevated in the blood of cancer patients. Permutation analysis of individual genes defined five core genes (P ≤ 0.05, permax test). As a group, the 12 genes generally distinguished accurately between healthy volunteers and patients with breast cancer. Mean expression levels of the 12 genes were elevated in 77% (10 of 13) untreated invasive cancer patients, whereas cluster analysis correctly classified volunteers and patients (P = 0.0022, Fisher's exact test). Quantitative real-time PCR confirmed array results and indicated that the sensitivity of the assay (1:2 × 108 transcripts) was sufficient to detect disseminated solid tumor cells in blood. Expression-based blood assays developed with the screening approach described here have the potential to detect and classify solid tumor cells originating from virtually any primary site in the body.
Keywords: breast cancer, hematogeneous dissemination of tumor cells, cDNA , prognostic markers, differential display
Early detection is one of the most effective means of reducing cancer mortality. Randomized screening trials have shown that mammography reduces the relative risk of breast cancer death by 15–20% (1). Early detection greatly improves treatment options and the chances for successful treatment. Although highly effective, mammography has significant barriers as seen by the fact that only one-third of women age 40 and older had a mammogram in 1997 (2). A blood-based assay for the detection of disseminated breast tumor cells potentially represents a more accessible screening tool. Further, a blood-based assay would not be limited to breast cancer but would have the potential to detect and distinguish solid tumor cells originating from virtually any primary tumor site in the body.
Solid tumors as small as 2 mm in diameter typically display active angiogenesis (3) and hence are capable of releasing tumor cells into peripheral blood. Numerous PCR and immunocytochemistry studies performed with individual breast tumor cell markers have reported the detection of disseminated tumor cells in the blood of breast cancer patients, including patients with localized disease (4–6). In earlier stages of disease, disseminated cells are not yet capable of forming metastases. However, these cells represent an easily sampled source for cancer detection.
The development of a robust assay for detecting disseminated solid tumor cells requires a highly sensitive high-throughput screening technique that can identify panels of informative markers. Assays based on individual markers, including the studies cited above, generally report the detection of circulating tumor cells in fewer than half of cancer patients (6). These low detection rates are likely the result of genetic heterogeneity of most cancers. This heterogeneity is best accommodated by assays that incorporate numerous markers. Gene expression signatures composed of detailed information from panels of informative genes have the potential to lead to highly accurate diagnoses (7). Our study is significant in that it represents a highly sensitive high-throughput screen useful for the identification of panels of genes that can provide clinically useful information enabling the detection and characterization of disseminated solid tumor cells. Here, we demonstrate the utility of the two-step screening approach by identifying a panel of 12 markers that together identify 77% of patients with invasive breast cancer.
Materials and Methods
Cells and Tissues.
Normal breast myoepithelial and luminal epithelial cells were sorted from primary cultures of mammoplasty tissue by immunomagnetic methods (8). Cultured 76N breast epithelial cells were obtained from mammoplasty tissue (9). Metastatic breast tumor cell line MDA-MB-435 was obtained from the American Type Culture Collection. Cells were grown in DFCI-1 medium (9) and harvested at 70% confluence. RNA from cultured cells was prepared by the CsCl-cushion method (10, 11).
Differential Display (DD) Reverse Transcription–PCR.
DD was performed (12, 13) to compare normal breast epithelial cells (either 76N cells or sorted myoepithelial and luminal epithelial cells) with MDA-MB-435 tumor cells. Both up-regulated and down-regulated cDNA bands were selected for analysis. Approximately 70 primer pairs were used to identify and clone 170 genes (13). A listing of all genes is available (http://mbcf.dfci.harvard.edu/labs/pardee/expression_patterns.html).
Blood Collection and RNA Isolation.
Blood was drawn at the time of chemotherapy treatments or immediately before surgery. All donors were female. Age distributions of volunteers and patients were similar. To reduce contamination of samples with skin epithelial cells from the needle stick, 3 ml of blood was drawn into a first tube, which was discarded. Then, 5–10 ml of whole blood was drawn into a second EDTA tube. White cells were isolated within 4 h by using a red cell lysis procedure (Ambion, Austin, TX), and cell pellets were stored at −80°C. Total RNA was purified by using Trizol reagent (Life Technologies, Rockville, MD). Agarose gel electrophoresis and densitometry determined RNA quality and verified its concentration.
Follow-up samples were drawn from three healthy volunteers. Samples N0b and N0c were drawn from the same individual, as was initial sample N0a, after intervals of 5 months and 5 months plus 1 week. N2b and N2c were from the same individual as N2a following intervals of 2 weeks and 5 months. N2c(1) and N2c(2) were run on different days with different membranes by using the same N2c RNA preparation. N4b was from the same individual as N4a following an interval of 2 weeks. N4a(1) and N4a(2) are repeated experiments performed with the same RNA preparation.
High-Sensitivity cDNA Arrays.
cDNA arrays were used to test a set of 196 candidate tumor marker genes for their ability to distinguish the blood of breast cancer patients from that of healthy volunteers. One-hundred seventy of these candidate marker genes were isolated by DD (13), and 26 genes were obtained from the literature. Arrays were prepared (13) by spotting cDNA onto positively charged nylon membranes (μm Separations, Westboro, MA). Forty replicate membranes were prepared, and radiolabeled cDNA probes were prepared (11, 13). Membranes were prehybridized 3 h in formamide-based buffer (11) at 41°C. Probe was then added to buffer, and membranes were hybridized 18 h at 41°C. Membranes were washed (11), exposed to phosphorimaging screens for 2 days, and analyzed by using the Storm system and ImageQuant software (Molecular Dynamics). Membranes were stripped and reused three times. Profiles represent individual experiments.
Control experiments were performed to test array reproducibility. Repeated analyses of a single preparation of MDA-MB-435 RNA on different days performed with different membranes showed that 95% of data points fell within 2.5-fold limits. Similar results were obtained with other RNA preparations.
Data Analysis.
Signal intensities were quantified and normalized (13). Cluster analysis was performed by using software written by M. Eisen (14). Data sets were logarithmically transformed. Average linkage hierarchical clustering was performed by using an uncentered correlation for both array and gene-clustering dimensions. Hierarchical cluster analysis performed by using alternate similarity metrics gave similar conclusions, as did K means clustering. Full cluster diagram with gene identities and other materials are available (see Figs. 4 and 5, which are published as supplemental data on the PNAS web site, www.pnas.org).
Quantitative Real-Time PCR.
Two micrograms of total RNA from white cells were denatured at 70°C for 10 min and then reverse-transcribed in a 30-μl reaction mixture containing 250 μM of each dNTP, 50 units of reverse transcriptase enzyme (Superscript II, Life Technologies), 80 ng/μl oligo(dT), 12–18 primers (Life Technologies), 1 × PCR buffer, 2.0 mM MgCl2 (Applied Biosystems) at 42°C for 50 min. cDNAs were purified on Sepharose G-50 columns (Boehringer Mannheim), dried, and resuspended in 50 μl of dH2O. Reactions omitting enzyme or RNA were used as negative controls.
Specific primers for human maspin, mdm-2, and gro-alpha genes (see Table 1, which is published as supplemental data on the PNAS web site) were designed to work in the same cycling conditions (95°C for 10 min followed by 45 cycles at 95°C for 15 s and 60°C for 1 min) generating products with sizes 100 to 150 bp. Glyceraldehyde-3-phosphate dehydrogenase, β-actin, ribosomal protein PO, and cyclophilin (see Table 1, which is published as supplemental data on the PNAS web site) were tested as reference genes by using a group of four normal blood samples and four patients' blood. Glyceraldehyde-3-phosphate dehydrogenase and ribosomal protein PO levels were similar in tested samples (and consistent with cDNA amounts as determined by absorbance (A) at 260 nm and fluorescence of the single-strand DNA-specific dye OliGreen, Molecular Probes), whereas β-actin and cyclophilin showed high variations (data not shown). PCR was performed by using an Applied Biosystems PRISM 7700 Sequence Detector and SYBR Green reagents. For each reaction, standard curves for both target and reference gene were made by using six 4-fold serial dilutions of N27 cDNA. All samples were run in triplicate. Relative amounts of maspin, mdm-2, and gro-alpha transcripts were calculated by comparison with standard curves. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase or the amount of cDNA in the reaction. All samples were resolved in a 1.8% DNA agarose gel to confirm the PCR specificity.
Results
High-Sensitivity Array Screening of Candidate Markers in Blood.
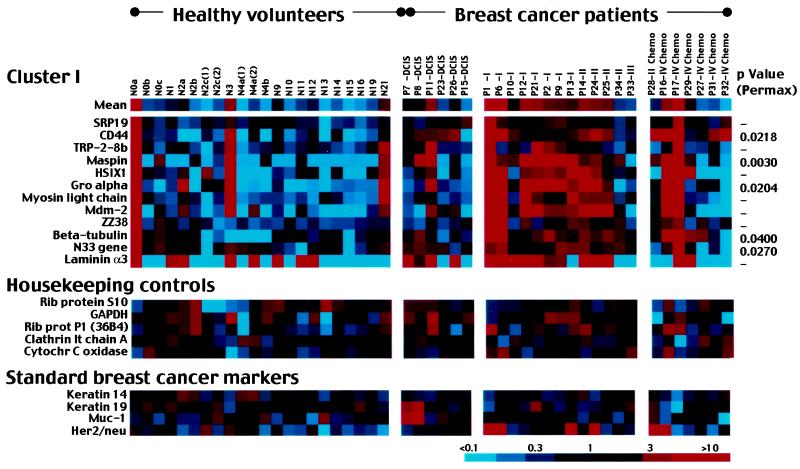
Blood samples were collected from a total of 41 different women (15 normal volunteers, 26 breast cancer patients). Five follow-up blood samples were collected from three of the normal volunteers, and two normal blood samples were analyzed in duplicate, resulting in a total of 48 different array analyses performed. Cluster analysis allowed identification of a single group of 12 breast cancer marker genes that were expressed at higher levels in the blood of breast cancer patients than that of healthy volunteers. The 12 cancer marker genes as a group were elevated in 77% (10 of 13) of untreated invasive cancer patients (Fig. 1). These marker genes were generally not elevated in patients treated with chemotherapy (29%, 2 of 7). The genes were also not generally elevated in patients with local disease (ductal carcinoma in situ; 17%, 1 of 6). Three false positives initially resulted among 15 healthy volunteers (19%). Follow-up tests of three (N0, N2, and N4) confirmed two originally negative results (N2 and N4) and corrected one originally positive result (N0), bringing the false positive rate to 13% (2 of 15).
Figure 1.
View of gene cluster I and control gene expression levels for 48 blood samples. Healthy volunteers included 15 women with no history of breast disease; patients included 26 women with a variety of stages of breast cancer. Gene expression levels are color-coded according to key. Blood samples N0a, N0b, and N0c were drawn from the same individual at successive time points, as were samples N2a, N2b, N2c, N4a, and N4b. N2c(1) and N2c(2) represent two different experiments performed with the same sample of N2c RNA, as do N4a(1) and N4a(2). Cancer patients are ordered by stage of disease at the time of blood collection. Patients who had received any chemotherapy treatments before blood collection are indicated. (Middle) Five “housekeeping” genes as nondifferential controls. (Lower) Four genes commonly used as breast tumor markers. Full cluster diagram with all gene identities is available (see Fig. 5, which is published as supplemental data on the PNAS web site).
Gene expression levels of a set of five “housekeeping” control genes often used as nondifferential controls are shown (Fig. 1 Middle). These genes were generally expressed at nondifferential levels in the blood samples.
cDNA arrays also included genes reported in the literature to be useful expression markers for breast cancer, including keratins 14 and 19, muc-1 and her2/neu. Expression results for these are shown (Fig. 1 Bottom). Keratins are specific for epithelial cells. Keratin 14 and 19 distinguish the two epithelial cell types present in breast tissue, myoepithelial and luminal epithelial cells, respectively (15). Keratin 19 has been reported to be a useful marker for disseminated tumor cells in the blood in numerous studies (e.g., ref. 16), although others have reported elevated keratin 19 in similar percentages of healthy individuals and cancer patients (17). Both keratins in our study were found at elevated levels in the blood of most healthy individuals and cancer patients and hence were not useful markers.
muc-1 and her2/neu have also been reported as useful markers for breast cancer in some blood studies (4, 18) but not others (19, 20). Our results found muc-1 high in normal blood and hence not a useful marker (Fig. 1). In contrast, her2/neu was generally low in healthy individuals but was elevated in 4 of 13 (31%) untreated invasive breast cancer patients (Fig. 1). her2/neu would likely be a useful addition to an expression signature panel. Apparently conflicting results of different studies may reflect heterogeneity in relatively small cancer patient and control populations. Conclusive studies to determine marker utility will require large numbers (hundreds) of samples. Variations in blood collection, RNA preparation, and marker detection procedures may also generate differing results.
Statistical Analysis of Expression Results.
The statistical significance of individual gene expression levels was tested by permutation analysis (21). The software used, permax, performs permutation 2-sample t tests on large arrays. The significance level for each gene is determined by comparing its statistic to the permutation distribution of the max statistic over all genes. The main use is to determine genes that are most different between two groups. We applied this test to a group of blood samples that included the first sample collected from each of the 15 healthy volunteers and all of 26 breast cancer patients. permax identified five “most significant” genes that drive distinction among the 12 genes of cluster I (Fig. 1). These genes include CD44, maspin, gro-alpha, tubulin, and N33. P values are indicated (Fig. 1). These five genes were found to be most distinguished between cancer patient and healthy volunteer groups. A cross-validation procedure done by dropping one tissue at each test confirmed these five most significant genes. Other significant genes (P < 0.05) identified by permax but not represented in cluster I included β-actin (P = 0.0014), doc-1 (P = 0.0124), mac25 (P = 0.0126), unknown 28/13 (P = 0.0180), unknown TG90D (P = 0.0188), desmoglein 2 (P = 0.0240), c-fos (P = 0.0262), interferon γ (P = 0.0278), and chondroitin sulfate proteoglycan (P = 0.0372).
Classification of Blood Samples by Hierarchical Cluster Analysis.
Hierarchical cluster analysis was used to classify blood samples based on array results. This analysis was performed by using expression information from only the 12 cluster I genes. Clinical information for the blood samples and the tree generated by this analysis are available (see Fig. 6, which is published as supplemental data on the PNAS web site). Cluster analysis sorted blood samples into three classes: A, B and C. Class A included samples with a high percentage of overexpressed (red) cluster I genes, class B included samples with a mix of overexpressed and underexpressed (red and blue) genes, and class C included samples with underexpressed (blue) genes. Classification of blood samples was in close agreement with the disease status of the blood donors (P = 0.0022; Fisher's exact test). Class A was predominantly composed of samples from patients with untreated invasive ductal breast cancer. It included 69% (9 of 13) patients with untreated invasive ductal carcinoma and only 14% (3 of 22) healthy volunteers. Classes B and C together included predominantly healthy volunteers (86%, 19 of 22). Patients with localized disease (ductal carcinoma in situ) generally fell into class B (75%, 3 of 4), indicating that the markers did not effectively detect noninvasive cancer. Patients who had been treated with chemotherapy before blood sampling were split between class A (33%, 2 of 6) and class C (67%, 4 of 6). This division between cancer-predominant and normal-predominant classes may reflect the extent of treatment efficacy, although data are not currently available to confirm this.
We have considered possible sources of artifacts that could impact our system. First, cancer patients will be subjected to a level of stress that may result in alterations in gene expression by white cells. In this regard, it is significant that patients with localized disease, which would likely generate similar stress responses, did not produce elevated markers. Also, recent breast biopsy or surgery could increase circulating breast cells potentially generating marker responses. The timing of recent procedures (see Fig. 6, which is published as supplemental data on the PNAS web site) indicated that the length of time postoperation did not relate to the level of expression of cluster I markers.
Confirmation of Array Results by Real-Time PCR.
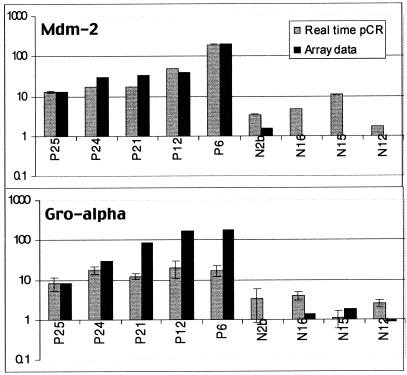
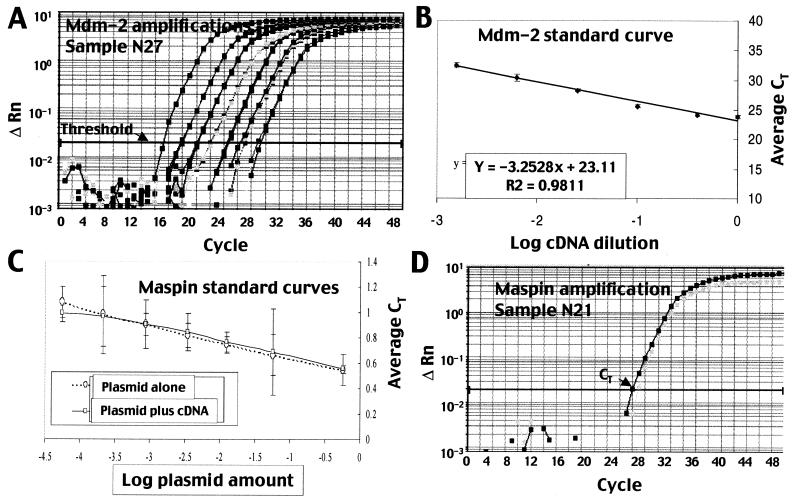
Quantitative real-time PCR was used to check the accuracy of representative cDNA array measurements (Fig. 2). Real-time PCR of three markers (mdm-2, gro-alpha, and maspin) was performed in a subset of both cancerous and healthy blood samples by using the standard curve method. Cycle times exceeding a threshold value (CT) for a set of dilutions (Fig. 3A) were used to generate a standard curve (Fig. 3B) from which expression levels of experimental samples were interpolated. Representative data for mdm-2 are shown (Fig. 3 A and B); similar procedures were followed for gro-alpha and maspin. For all three genes, real-time PCR confirmed array results (Fig. 2 and data not shown). In all cases, markers were expressed at higher levels in the blood of cancer patients than that of healthy individuals.
Figure 2.
Real-time PCR confirmed cDNA array expression levels. mdm-2 and gro-alpha expression levels were measured in the blood of breast cancer patients and healthy volunteers. PCR results were normalized to total cDNA determined by OD260 and expressed relative to sample N27. Array assay was not performed for sample N27.
Figure 3.
Real-time PCR results to determine sensitivity of cDNA arrays. (A) Representative real-time PCR curves for a set of dilutions of mdm-2 primers with N27 normal blood. Threshold level is indicated. Cycle times exceeding the threshold (CT) for a set of dilutions were used to generate a standard curve. (B) Standard curve generated for mdm-2 dilution series with N27 normal blood RNA. (C) Identical standard curves were generated when maspin plasmid was measured alone or in the presence of normal blood cDNA. (D) Amplification plots used to determine the expression level of maspin in blood sample N21.
Assessment of Array Sensitivity.
Numbers of disseminated solid tumor cells present in the blood have been previously quantitated by using individual and double markers by a variety of immunology and PCR-based techniques. Sensitivity of 1 in a million cells is generally considered necessary for such measurements. Maximum reported levels of tumor cells in the blood of breast cancer patients exceed 3,000 cells per ml (5, 6). Averages (SD > 100%) reported for different stages of disease include organ-confined breast cancer, 0.8 cells per ml; invasive breast cancer spread to lymph nodes only, 2.4 cells per ml; and metastatic breast cancer, 6 cells per ml (6).
To determine whether our cDNA arrays were sufficiently sensitive to detect disseminated tumor cells in blood, their sensitivity limit was assessed by quantitative real-time PCR of maspin. Maspin expression in the blood of healthy volunteers was at the limit of array detection. Low, but measurable maspin levels were detected by arrays in 33% of healthy volunteers (7 of 21), whereas the others had undetectable levels (Fig. 1 and data not shown). Maspin expression was measured by real-time PCR in three normal blood samples: N12, N21, and N27.
The absolute level of maspin transcripts was determined by comparison with a standard curve generated from dilutions of quantitated maspin plasmid (Fig. 3C). Identical standard curves were generated if maspin plasmid was measured alone or added to normal blood cDNA (Fig. 3C). Measurements of maspin transcripts (Fig. 3D) in blood were normalized to the amount of cDNA in reverse transcription reactions, which was quantitated by two methods: OD260 and fluorescence of the single-strand DNA dye OliGreen. Results were adjusted to account for the use of double-stranded plasmid in standard curves and for the size of maspin cDNA (3 kb), which is longer than an average cellular transcript (2 kb). Maspin was quantitated in cDNA preparations, and its level relative to total cDNA was assumed to be equivalent in cDNA and RNA.
In blood sample N21, a 2-μl reverse transcription reaction contained 2.4 × 10−4 pg maspin cDNA and 3.6 × 104 pg total cDNA. Hence, maspin transcripts represented 1 in 2.3 × 108 transcripts (e.g., 2.2 × 10−9 maspin messages/total cellular messages). Because a typical mammalian cell has 3.6 × 105 RNAs in its cytoplasm, this corresponds to an in vivo level of 1.6 × 10−3 copies per white blood cell. Maspin expression was similar in N27, which was not tested by arrays, and 2-fold lower in N12, which had an undetectable level of maspin by arrays. Hence the limit of detection for our arrays was approximately 1 in 2 × 108 transcripts. This is two orders of magnitude less sensitive than PCR, which has a limit of approximately 1 in 1010 messages. However, it is three orders of magnitude better than oligonucleotide microarrays with a limit of 1 in 3 × 105 messages (22). The enhanced sensitivity of our arrays can be accounted for by the use of 32P rather than fluorescence labeling, membranes rather than glass, and long cDNA tags rather than oligonucleotides.
The detection limit of 1 in 2 × 108 can be used to calculate whether arrays are capable of detecting low numbers of disseminated tumor cells in blood. If one assumes tumor cells and white cells express similar total amounts of RNA and that the marker used is abundant in tumor cells (e.g., 1,000 copies per cell), then it can be calculated that the arrays can detect as few as 1 in 106 cells (e.g., 5 tumor cells per ml of blood). This level of sensitivity is sufficient for the detection of tumor cells in blood (5, 6) and hence the assay is predicted to be a useful screening method.
Discussion
The objective of this study was to test our two-step approach of DD and high-sensitivity cDNA arrays for its ability to identify panels of expression markers that can detect disseminated solid tumor cells in the blood. We have previously used DD to identify solid tumor cell markers from blood (23, 24). Here, we demonstrate the ability of the two-step approach to identify numerous genes that detect and characterize cancer cells in the blood. These studies represent an initial phase in the development of a potentially versatile and powerful clinical assay system that can accurately detect and characterize circulating tumor cells based on an expression signature.
In recent years, several highly sensitive methods have been developed to detect circulating cancer cells in the blood of patients with different types of malignancies. By using flow cytometry and an in vitro cell-spiking assay, Gross et al. detected 1 tumor cell in 107 peripheral blood mononuclear cells (25). The combination of immunomagnetic beads coated with antibodies against tumor antigens and flow cytometry was also sufficiently sensitive to detect cancer cells in patients' blood (6). An automated rare-event detection system by using a computerized fluorescence microscope was able to find 1 cancer cell in 106 peripheral blood cells and also allowed analysis of cell morphology and staining patterns. Immunohistochemical studies of bone marrow aspirates also show positive correlation with clinical parameters of disease progression (26). Reverse transcription–PCR-based methods are highly sensitive and have been extensively applied for the detection of tumor cells dispersed in the circulation or in regional lymph nodes. Real-time reverse transcription–PCR has also been successfully used to detect mammary carcinoma cells in the peripheral blood of breast cancer patients (4). All of these techniques are powerful methods for the detection of tumor cells; however, they require prior selection of useful markers.
Our two-step combination of DD and high-sensitivity arrays is significant in that it represents a means of screening for blood-disseminated tumor cell markers. To test the arrays, we had two specific objectives: first, to determine whether the arrays were capable of detecting disseminated tumor cells by measuring their sensitivity; second, to perform a practical test by using the arrays to screen expression levels of a set of candidate markers in a panel of breast cancer patient and normal volunteer blood. Results show that the assay is effective in both regards.
The markers we identified could have been expressed by disseminated tumor cells themselves or could represent a response of blood cells to the presence of cancer in the body. Expression changes of leukocytes may assist in detecting cancer; however, our major emphasis is on the identification of markers expressed by disseminated tumor cells. Several lines of evidence indicate that disseminated tumor cells themselves expressed our 12 markers. First, our primary DD screen selected only markers that were expressed by breast cells. Second, our arrays were sensitive enough (1 in 2 × 108 transcripts or 5 tumor cells per ml of blood) to detect tumor cells in the blood (5, 6). Third, the markers we identified were generally expressed at very low or undetectable levels in healthy individuals.
The identities of the individual markers included in cluster I are of interest. Several of these genes have been previously described as useful markers for tumor cells in the blood. Their inclusion among the cluster I genes confirms the significance of our results. In particular, maspin is a breast-specific serine-protease inhibitor that is down-regulated during the process of tumorigenesis, inhibits breast tumor cell invasion and motility (27), and inhibits angiogenesis (28). Hence, detection of this gene in the blood may indicate nonmetastatic disease. Maspin has been described as a useful marker for PCR-based detection of breast cancer cells in the blood (29, 30), although other studies have reported unsuitably high levels of expression in the blood of healthy individuals (31). Further, rates of maspin detection in blood increased after chemotherapy, presumably as a result of therapy-induced tumor cell mobilization (30, 31).
Other previously identified tumor markers found in cluster I include mdm-2, CD44, gro-alpha, and HSIX-1. mdm-2 is a cellular oncoprotein that binds to p53 protein and abrogates its growth-suppressing function (32). mdm-2 is overexpressed in 30–70% of breast tumors (33, 34), and its levels in tumor tissue are correlated with patient survival (35). CD44 RNA and protein have also been previously described as useful blood markers for breast cancer (36). Malignant breast tissue often overproduces nine or more alternatively spliced large molecular variants of CD44, whereas control tissue samples only occasionally produce minimal quantities of one or two small variants in addition to the standard product. Gro-alpha, originally identified as a melanocyte growth stimulatory cytokine, is overexpressed in colon cancers (37) and induces migration of breast tumor cells (38). HSIX1 is a homeobox transcription factor that is elevated in breast and other cancers (39). Further testing of hundreds of blood samples will be required to confirm the statistical significance of each of the cluster I genes as informative components of an expression-based blood assay.
The array assay resulted in a false positive rate of 13% (2 of 15). Three false positives initially resulted among the first blood samples drawn from 15 healthy volunteers (19%, 3 of 15). However, follow-up testing of additional blood samples from one of these women corrected one of the false positives. In the cases of these false positives, it is possible that the cluster I markers detected disseminated nonmalignant breast epithelial cells, which may have been in the circulation because of inflammation or other normal mammary processes. Further testing will be necessary to determine the reason for the elevated marker responses in healthy women.
The false positive rate of 13% resulting from this assay is similar to that of mammography. Although the vast majority of women screened by mammography are cancer free, up to 10% will have their mammogram interpreted as abnormal or inconclusive until further tests can be done.
We envision blood marker systems as versatile and powerful clinical screening tools for early detection and characterization of breast cancer. The assay could be part of routine screening. For example, patients at risk for breast cancer could be screened at yearly intervals in addition to mammography. Mammography typically detects 90% of breast cancers in women without symptoms.
A blood-based expression assay could be also used to detect cancers other than breast cancer. Lung, colon, prostate, and stomach cancer are additional highly prevalent cancer types that together with breast cancer account for the great majority of all cancer deaths. Inclusion of markers to detect and distinguish between these cancers would allow the assay to detect most cancers. Ovarian and pancreatic tumors, although less prevalent, lack early symptoms and patients would greatly benefit from early detection.
Important considerations for an assay to be used as a general cancer screening tool include a low rate of false positives, a robust system with a large margin of differentiation between positive and negative results, and a high rate of detection of cancers. Further clinical development may take the blood assay toward PCR methodology or antibody detection of the marker gene's protein products. Markers for numerous additional cancer types can be added to panels. Secondary assays could potentially include markers to characterize tumor cells of specific cancer types, once cancer cells have been detected and the primary site has been identified. For instance, a breast cancer panel of markers could conceivably determine estrogen receptor status, prognosis, tumor stage, grade, size, and optimum therapy.
Supplementary Material
Acknowledgments
This paper is dedicated to the late Dr. Ruth Sager, whose ideas and enthusiasm provided the groundwork for this project. This work was supported by National Institutes of Health Grant R01 CA61253-08A1 (to R. Sager and A.B.P.). E.G. is supported by FAPESP, Brazil (Grant 99/08279-5).
Abbreviation
- DD
differential display
References
- 1.Harris R. J Natl Cancer Inst Monogr. 1997;22:139–143. doi: 10.1093/jncimono/1997.22.139. [DOI] [PubMed] [Google Scholar]
- 2.Bolen J C, Rhodes L, Powell-Griner E E, Bland S D, Holtzman D. Morbid Mortal Wkly Rep. 2000;49:1–60. [PubMed] [Google Scholar]
- 3.Weidner N, Carroll P R, Flax J, Blumenfeld W, Folkman J. Am J Pathol. 1993;143:401–409. [PMC free article] [PubMed] [Google Scholar]
- 4.de Cremoux P, Extra J M, Denis M G, Pierga J Y, Bourstyn E, Nos C, Clough K B, Boudou E, Martin E C, Muller A, et al. Clin Cancer Res. 2000;6:3117–3122. [PubMed] [Google Scholar]
- 5.Kraeft S K, Sutherland R, Gravelin L, Hu G H, Ferland L H, Richardson P, Elias A, Chen L B. Clin Cancer Res. 2000;6:434–442. [PubMed] [Google Scholar]
- 6.Racila E, Euhus D, Weiss A J, Rao C, McConnell J, Terstappen L W, Uhr J W. Proc Natl Acad Sci USA. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1991;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 8.Clarke C, Titley J, Davies S, O'Hare M J. Epithelial Cell Biol. 1994;3:38–46. [PubMed] [Google Scholar]
- 9.Band V, Sager R. Proc Natl Acad Sci USA. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 11.Fournier M, Martin K J, Pardee A B. Methods in Molecular Biology: Approaches in the Investigation of Hormone Stimulated Tumors. Totowa, NJ: Humana; 2001. , in press. [Google Scholar]
- 12.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 13.Martin K J, Kritzman B M, Price L M, Koh B, Kwan C-P, Zhang X, Mackay A, O'Hare M J, Kaelin C M, Mutter G L, et al. Cancer Res. 2000;60:2232–2238. [PubMed] [Google Scholar]
- 14.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor-Papadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane E B, Leigh I M. J Cell Sci. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- 16.Slade M J, Smith B M, Sinnett H D, Cross N C, Coombes R C. J Clin Oncol. 1999;17:870–879. doi: 10.1200/JCO.1999.17.3.870. [DOI] [PubMed] [Google Scholar]
- 17.Grunewald K, Haun M, Urbanek M, Fiegl M, Muller-Holzner E, Gunsilius E, Dunser M, Marth C, Gastl G. Lab Invest. 2000;80:1071–1077. doi: 10.1038/labinvest.3780112. [DOI] [PubMed] [Google Scholar]
- 18.Wasserman L, Dreilinger A, Easter D, Wallace A. Mol Diagn. 1999;4:21–28. doi: 10.1016/s1084-8592(99)80046-0. [DOI] [PubMed] [Google Scholar]
- 19.Berois N, Varangot M, Aizen B, Estrugo R, Zarantonelli L, Fernandez P, Krygier G, Simonet F, Barrios E, Muse I, Osinaga E. Eur J Cancer. 2000;36:717–723. doi: 10.1016/s0959-8049(99)00338-x. [DOI] [PubMed] [Google Scholar]
- 20.Eltahir E M, Mallinson D S, Birnie G D, Hagan C, George W D, Purushotham A D. Br J Cancer. 1998;77:1203–1207. doi: 10.1038/bjc.1998.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox D R, Hinkley D V. Theoretical Statistics. London: Chapman & Hall; 1974. pp. 182–187. [Google Scholar]
- 22.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 23.Fournier M V, Guimaraes F C, Paschoal M E, Ronco L V, Carvalho M G, Pardee A B. Cancer Res. 1999;59:3748–3753. [PubMed] [Google Scholar]
- 24.Fournier M V, Carvalho M G, Pardee A B. Mol Med. 1999;5:313–319. [PMC free article] [PubMed] [Google Scholar]
- 25.Gross H J, Verwer B, Houck D, Hoffman R A, Recktenwald D. Proc Natl Acad Sci USA. 1995;92:537–541. doi: 10.1073/pnas.92.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich C R, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, et al. N Engl J Med. 2000;342:525–533. doi: 10.1056/NEJM200002243420801. [DOI] [PubMed] [Google Scholar]
- 27.Sheng S, Carey J, Seftor E A, Dias L, Hendrix M J, Sager R. Proc Natl Acad Sci USA. 1996;93:11669–11674. doi: 10.1073/pnas.93.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Volpert O, Shi Y H, Bouck N. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 29.Sabbatini R, Federico M, Morselli M, Depenni R, Cagossi K, Luppi M, Torelli G, Silingardi V. J Clin Oncol. 2000;18:1914–1920. doi: 10.1200/JCO.2000.18.9.1914. [DOI] [PubMed] [Google Scholar]
- 30.Luppi M, Morselli M, Bandieri E, Federico M, Marasca R, Barozzi P, Ferrari M G, Savarino M, Frassoldati A, Torelli G. Ann Oncol. 1996;7:619–624. doi: 10.1093/oxfordjournals.annonc.a010680. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Guerrero J A, Gilabert P B, Gonzalez E B, Sanz Alonso M A, Perez J P, Talens A S, Oraval E A, de la Rubia Comos J, Boix S B. J Hematother. 1999;8:53–61. doi: 10.1089/106161299320578. [DOI] [PubMed] [Google Scholar]
- 32.Momand J, Zambetti G P. J Cell Biochem. 1997;64:343–352. [PubMed] [Google Scholar]
- 33.Soslow R A, Carlson D L, Horenstein M G, Osborne M P. Breast Cancer Res Treat. 2000;61:161–170. doi: 10.1023/a:1006479113769. [DOI] [PubMed] [Google Scholar]
- 34.Bueso-Ramos C E, Manshouri T, Haidar M A, Yang Y, McCown P, Ordonez N, Glassman A, Sneige N, Albitar M. Breast Cancer Res Treat. 1996;37:179–188. doi: 10.1007/BF01806499. [DOI] [PubMed] [Google Scholar]
- 35.Jiang M, Shao Z M, Wu J, Lu J S, Yu L M, Yuan J D, Han Q X, Shen Z Z, Fontana J A. Int J Cancer. 1997;74:529–534. doi: 10.1002/(sici)1097-0215(19971021)74:5<529::aid-ijc9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 36.Matsumura Y, Tarin D. Lancet. 1992;340:1053–1058. doi: 10.1016/0140-6736(92)93077-z. [DOI] [PubMed] [Google Scholar]
- 37.Cuenca R E, Azizkhan R G, Haskill S. Surg Oncol. 1992;1:323–329. doi: 10.1016/0960-7404(92)90094-2. [DOI] [PubMed] [Google Scholar]
- 38.Youngs S J, Ali S A, Taub D D, Rees R C. Int J Cancer. 1997;71:257–266. doi: 10.1002/(sici)1097-0215(19970410)71:2<257::aid-ijc22>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 39.Ford H L, Kabingu E N, Bump E A, Mutter G L, Pardee A B. Proc Natl Acad Sci USA. 1998;95:12608–12513. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.