Abstract
DC-SIGN, a C-type lectin expressed on the surface of dendritic cells (DCs), efficiently binds and transmits HIVs and simian immunodeficiency viruses to susceptible cells in trans. A DC-SIGN homologue, termed DC-SIGNR, has recently been described. Herein we show that DC-SIGNR, like DC-SIGN, can bind to multiple strains of HIV-1, HIV-2, and simian immunodeficiency virus and transmit these viruses to both T cell lines and human peripheral blood mononuclear cells. Binding of virus to DC-SIGNR was dependent on carbohydrate recognition. Immunostaining with a DC-SIGNR-specific antiserum showed that DC-SIGNR was expressed on sinusoidal endothelial cells in the liver and on endothelial cells in lymph node sinuses and placental villi. The presence of this efficient virus attachment factor on multiple endothelial cell types indicates that DC-SIGNR could play a role in the vertical transmission of primate lentiviruses, in the enabling of HIV to traverse the capillary endothelium in some organs, and in the presentation of virus to CD4-positive cells in multiple locations including lymph nodes.
A relatively large number of interactions between the HIV-1 envelope (Env) protein and cell surface molecules dictate which cells are susceptible to virus entry and the efficiency with which entry occurs. Infection of target cells by primary HIV-1 strains depends on the presence of CD4 molecules and chemokine receptors, most often CCR5 and CXCR4 (1–6). The surface density of these receptors is an important variable that affects the efficiency of virus entry (7–10). Equally important is simple attachment of virus to the cell surface. For many cell types, virus attachment is independent of the presence or absence of the CD4 receptor and is rate-limiting for virus infection (11–13). Attachment efficiency can be enhanced in vitro by including polycations in the virus inoculum or by centrifuging virus onto the cell surface (14). Infection efficiency can also be enhanced by pulsing dendritic cells (DCs) with virus before the addition of receptor-positive target cells (15–17). DCs are able to efficiently transmit bound virus to peripheral blood mononuclear cells (PBMCs) (16), resulting in a robust infection even though the DCs themselves are either not infected or infected inefficiently (16–20).
DC-SIGN, a type II membrane protein with a C-terminal C-type (i.e., calcium-dependent) lectin domain, has been shown to be largely responsible for the ability of dendritic cells to efficiently capture and present HIV-1 to receptor-positive cells (15). DC-SIGN appears to be a universal attachment factor for primate lentiviruses that can bind and transmit all HIV-1, HIV-2, and simian immunodeficiency virus (SIV) strains tested to date (15, ‖). In addition, DC-SIGN also interacts with the intercellular adhesion molecule-3 receptor expressed on T cells, thereby contributing to the close interaction between DCs and T cells required for efficient antigen presentation (22). Because dendritic cells are among the first cells encountered by HIV-1 during sexual transmission, it is possible that virus bound by DC-SIGN may be ultimately ferried to lymph nodes (a major site of viral replication) as a consequence of the normal cellular trafficking of DCs (23, 24). In fact, virus bound to DCs remains infectious for at least several days (15).
Identification of such a specific and efficient virus attachment factor raises the possibility that other attachment factors on relevant cell types may exist. We recently described (25) a homologue of DC-SIGN, termed DC-SIGNR (for DC-SIGN related), that exhibits 77% amino acid identity with DC-SIGN. In this study, we show that DC-SIGNR also functions as a universal attachment factor for primate lentiviruses that can bind and transmit multiple HIV-1, HIV-2, and SIV strains to receptor-positive cell lines and to human PBMCs. Using a specific antiserum we developed, we found that DC-SIGNR is expressed on sinusoidal endothelial cells in the liver, endothelial cells present in lymph node sinuses, and a significant proportion of capillary endothelial cells in term placenta but was not expressed at appreciable levels in peripheral blood-derived DCs. The presence of an efficient virus attachment and presentation factor in these cell types indicates that DC-SIGNR could influence vertical transmission and result in enhanced infection of receptor-positive cell types in lymph nodes.
Materials and Methods
Plasmids.
Production and characterization of DC-SIGN wt and DC-SIGN with a C-terminal AU-1 tag has been described.∥ For detection by immunostaining before the development of specific antiserum, a C-terminal AU-1 antigenic tag was also added to DC-SIGNR by PCR mutagenesis. All constructs were cloned into pcDNA3 (Invitrogen) by using the unique BamHI and EcoRI restriction sites.
Production of DC-SIGNR Antiserum.
To produce antiserum specific for DC-SIGNR, a peptide (DPTTSGIRLFPRDFQ) corresponding to a unique sequence in the cytoplasmic domain of DC-SIGNR was covalently coupled to keyhole limpet hemocyanin. Anti-peptide antiserum was raised in a chicken by three s.c. immunizations with this antigen. Immune IgY was purified from the eggs and affinity-purified against the peptide coupled to N-hydroxysuccinimide-activated Sepharose beads. Bound antibodies were eluted with 100 mM glycine (pH 2.5), and fractions containing the antibodies were pooled and dialyzed against PBS.
Virus Binding and Transfer Assays.
Both assays were performed essentially as described.∥ In brief, 293T cells were seeded in T-25 flasks and transiently transfected with DC-SIGNR and DC-SIGN expression vectors or with the pcDNA3 control plasmid. One day after transfection, the cells were detached from the flasks and seeded into 96-well plates. On the following day, the cells were incubated with p24-normalized virus stocks for 3 h, vigorously washed with culture medium, and lysed in 0.5% Triton X-100. The amount of bound viral antigen was quantified by using a commercially available p24 ELISA (Coulter Beckman, Miami, FL). For the virus transfer assay, the cells were washed with culture medium and cocultivated with C8166 T cells or PBMCs. When transfer of luciferase reporter viruses was investigated, the cultures were lysed 3 days after the start of the cocultivation and luciferase activity in 30 μl of lysate was determined with a commercially available kit (Promega). To quantify transfer of primary HIV isolates, supernatant was removed from the cocultures at regular intervals and the content of viral p24 antigen measured with the p24 ELISA. Transmission of pseudotyped green fluorescent protein (GFP) reporter virus was investigated via flow cytometry (FACS). Five days after infection, cells were harvested in FACS buffer (PBS containing 3% FCS and 0.05% sodium azide), stained with an anti-CD3 antibody (Sigma) that was directly conjugated to phycoerythrin, and analyzed via FACS. GFP-positive cells were found exclusively in the CD3+ population. Viruses were originally obtained from the National Institutes of Health AIDS Reagent Repository except for isolate SPL3, a primary virus strain isolated in the laboratory of Ron Collman (University of Pennsylvania), that has not otherwise been characterized.
Tissue Processing and Staining.
All tissues were obtained with the approval of the Local Research Ethics Committee. Histologically normal postmortem adult human tissues were obtained from the Department of Histopathology, Addenbrooke's Hospital, Hills Road, Cambridge, U.K., as were postmortem tissues from fetuses at 12 weeks and 36 weeks of gestation and tissue from histologically normal term placentas. Tissues were fixed in neutral-buffered 10% formalin before processing for paraffin wax embedding and sectioning. Rehydrated 5-μm paraffin serial sections were immunostained with anti-DC-SIGNR antiserum at a 1:50 dilution. For negative controls, anti-DC-SIGNR antiserum at a 1:50 dilution was used in the presence of a 20-fold molar excess of the peptide DPTTSGIRLFPRDFQ. Further serial sections were immunostained with rabbit polyclonal anti-Von Willebrand factor (Dako) at a 1:200 dilution. Sections were preblocked in 0.5% hydrogen peroxide in Tris-buffered saline (TBS) for 30 min and then in TBS/10% BSA/10% normal serum (donkey serum for anti-DC-SIGNR or goat serum for anti-Von Willebrand factor) for 2 h before the addition of antibody in TBS/10% BSA. After overnight incubation at 4°C in primary antibody, sections were washed with TBS and placed in secondary biotinylated antibody in TBS/10% BSA for 2 h. After washing in TBS, the StreptABC/HRP kit (Dako) was used to form avidin-biotin–horseradish peroxidase complexes. Slides were developed with diaminobenzidine (Sigma) and counterstained with Harris's hematoxylin (Dako). Slides were dehydrated in xylene and mounted in DPX (BDH Laboratory Supplies, Poole, U.K.).
Results
DC-SIGNR Protein Is Expressed on Endothelial Cells in Placenta, Liver, and Lymph Nodes.
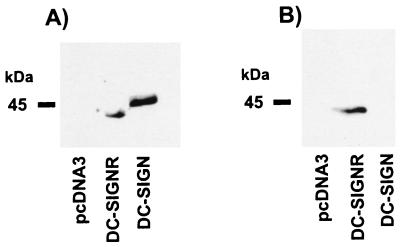
DC-SIGN and DC-SIGNR share 77% amino acid identity, with the greatest areas of homology being in the C-type lectin domain and the neck region (25). The C- and N-terminal regions of this type II membrane protein are considerably more divergent from the DC-SIGN sequence. To monitor the tissue expression of DC-SIGNR, we affinity-purified an anti-peptide antiserum raised against a unique sequence (DPTTSGIRLFPRDFQ) in the cytoplasmic domain of DC-SIGNR. The resulting antiserum recognized DC-SIGNR but not DC-SIGN by Western blot (Fig. 1).
Figure 1.
DC-SIGNR specific antiserum. Human 293T cells transiently expressing AU1-tagged versions of DC-SIGN, DC-SIGNR, or pcDNA3 vector alone were lysed in nonionic detergent. Aliquots of the resulting lysates were analyzed by SDS/PAGE and Western blotting. Blots were probed with a mAb directed against the AU1-antigenic tag (A) or with an antiserum raised against a peptide based on a unique sequence in the cytoplasmic domain of DC-SIGNR (B).
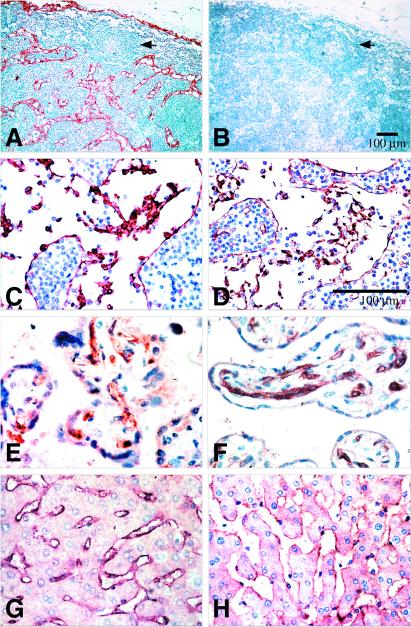
DC-SIGNR expression was demonstrated on the endothelium lining lymph node sinuses, on the endothelium of approximately half of the capillaries in term placenta, and on the endothelium lining the sinusoids of the liver (Fig. 2). This immunohistochemical staining was abrogated in the presence of specific peptide (data not shown). The endothelial identity of these cells was confirmed by immunostaining serial sections with Von Willebrand factor (factor VIII-related antigen) (Fig. 2 and ref. 26). Similar DC-SIGNR immunostaining was present in 36-week fetal liver (data not shown). No evidence of expression could be found on endothelium in multiple other tissues including lung, spleen, thymus, kidney, heart, pons, medulla, and midbrain (data not shown). In addition, no evidence of protein expression could be found on cells of the myeloid lineage, nor have we detected DC-SIGNR expression in peripheral blood-derived DCs.
Figure 2.
Expression of DC-SIGNR. Formalin-fixed paraffin-embedded sections of human tissues were immunostained with anti-DC-SIGNR chicken serum or anti-Von Willebrand factor rabbit serum. (A and C) Normal lymph node: the endothelial cells lining the sinuses are stained strongly with anti-DC-SIGNR. (B and D) Normal lymph node: an identical pattern of staining with anti-Von Willebrand factor is observed in the lymph node sinusoids. Anti-Von Willebrand factor also stained high endothelial vessels that did not stain with anti-DC-SIGNR. (E) Villi from normal term placenta: the endothelium of approximately half of the capillaries is stained with anti-DC-SIGNR antibody. (F) Villi from normal term placenta: all capillaries are stained with anti-Von Willebrand factor. (G) Normal liver: anti-DC-SIGNR antibody stained all sinusoids. (H) Normal liver: anti-Von Willebrand factor confirmed the identity of the sinusoidal lining cells as endothelium.
DC-SIGNR Binds HIV-1 Isolates.
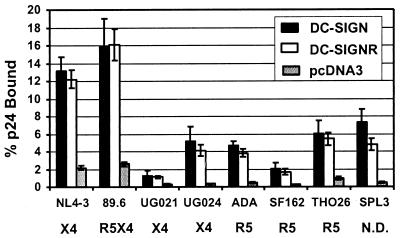
The presence of DC-SIGNR on endothelial cells, particularly in the lymph node and placenta, places this molecule in a position where it could impact vertical transmission of HIV and present virus to CD4/coreceptor-positive cell types that come into contact with these surfaces. To determine whether DC-SIGNR supports binding of R5, X4, and R5X4 HIV-1 virus strains that use CCR5, CXCR4, or either receptor in conjunction with CD4 to infect cells, respectively (27), human 293T cells were transiently transfected with DC-SIGNR or DC-SIGN expression vectors. By using versions of DC-SIGN and DC-SIGNR that possessed an AU1 antigenic tag on the C terminus of the proteins, we were able to determine that both proteins were expressed at similar levels on the surface of 293T cells, as judged by FACS analysis (∥ and data not shown). Cells expressing DC-SIGN or DC-SIGNR were incubated with p24-normalized virus stocks and extensively washed, and the bound antigen was quantified via ELISA. All virus isolates tested bound to DC-SIGNR and DC-SIGN expressing cells more efficiently than to pcDNA3-transfected control cells (Fig. 3). The laboratory-adapted NL4–3 strain and the 89.6 R5X4 viral clone bound to DC-SIGNR and DC-SIGN transfected cells with the highest efficiencies. However, those viruses also exhibited the most efficient binding to pcDNA3-transfected control cells. Thus, like DC-SIGN, DC-SIGNR functions as an efficient HIV-1 attachment factor.
Figure 3.
Binding of HIV to DC-SIGNR transfected cells. 293T cells were transiently transfected with the indicated expression plasmids. The cells were incubated with p24-normalized virus stocks, vigorously washed, and lysed in 0.5% Triton X-100, and then the content of bound p24 was quantified by ELISA. The tropism of each virus is indicated (R5, X4, or R5X4). All viruses are clade B except for the UG021 and UG024 isolates, which are clade D. Data are presented as the percent of recovered p24 antigen. The mean values ± SEM of four experiments are shown. N.D., not done.
DC-SIGNR Transmits HIV-1, HIV-2, and SIV to T Cell Lines and PBMCs with Different Efficiencies.
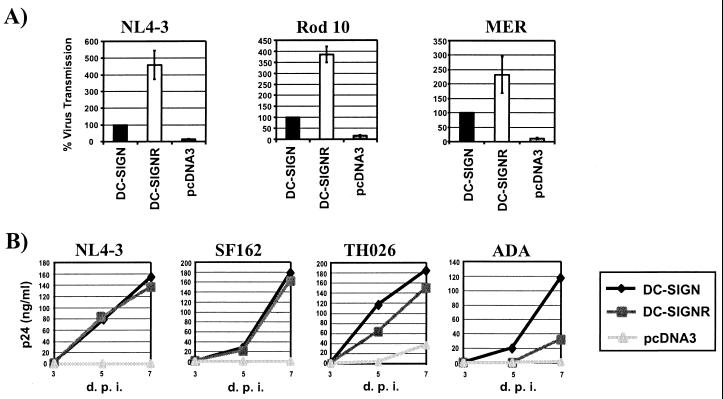
Primate lentiviruses bound to DC-SIGN can be efficiently transmitted to receptor-positive T cell lines and to primary T cells (15, ∥). We investigated the ability of DC-SIGNR to transmit virus to C8166 T cells. DC-SIGN and DC-SIGNR were expressed in 293T cells, the cells incubated with replication-competent luciferase-reporter viruses, vigorously washed, and cocultivated with C8166 cells. Three days after the start of the coculture, the cells were lysed and the luciferase activity in the lysates was quantified. DC-SIGNR-expressing cells transmitted HIV-1 NL4–3 and HIV-2 Rod10 about 4-fold and SIVmac239 MER Env about 2.5-fold more efficiently than DC-SIGN-transfected cells (Figure 4A). However, when both proteins were cotransfected, the transmission efficiencies obtained ranged between those observed for DC-SIGN and DC-SIGNR, indicating that the proteins did not function cooperatively.
Figure 4.
DC-SIGNR mediates transmission of HIV-1, HIV-2, and SIV strains. DC-SIGNR, DC-SIGN, or pcDNA3 control plasmids were transiently transfected into 293T cells. The cells were pulsed with virus, extensively washed, and cocultivated with target cells. (A) Transmission of luciferase-reporter viruses to C8166 T cells. HIV-1 NL4–3, HIV-2 Rod10, and SIVmac239 MER Env luciferase reporter viruses were used in the transfer assay described above. Luciferase activity was determined 3 days after the start of the cocultivation. Data are the mean ± SEM of three experiments. (B) Transmission of HIV isolates to PBMCs. The transmission of primary HIV isolates and the laboratory-adapted NL4–3 virus was investigated as described above. The culture supernatants were removed as indicated (d.p.i., days postinfection), and their p24 content was measured by ELISA. Similar results were obtained in an independent experiment.
We also investigated whether DC-SIGNR was capable of transferring HIV isolates to primary cells. The virus-transfer experiment was carried out as described above, except that the transfer of six primary HIV-1 isolates of various tropisms to cultured human PBMCs was analyzed. The supernatants of the 293T/PBMC cocultures were harvested at regular intervals and assayed for their p24 content with a p24-antigen-capture ELISA. All isolates tested replicated more efficiently in PBMCs cocultured with 293T cells expressing DC-SIGN or DC-SIGNR than in PBMCs cocultured with 293T control cells. DC-SIGNR and DC-SIGN expressing cells induced comparable replication of the laboratory-adapted NL4–3 strain and the SF162 primary isolate. However, robust replication of the ADA isolate was only observed after coculture with DC-SIGN-expressing cells. This was surprising because ADA bound to DC-SIGNR and DC-SIGN equally well (Fig. 3). However, the binding assay was done under equilibrium binding conditions, and it is possible that ADA dissociates from DC-SIGNR more quickly than the other viruses tested. If so, this could have a more dramatic effect on virus transmission than on binding under saturating conditions. The THO26 (Fig. 4B), SPL-3, UG021 and UG024 isolates (data not shown) were transmitted slightly more efficiently by DC-SIGN than by DC-SIGNR. To confirm the identity of the cells that were infected, we also investigated the transmission of a replication defective GFP reporter virus pseudotyped with the NL4–3 Env. Five days after the start of the coculture, the cells were stained for CD3 and GFP-positive cells were quantified via FACS. All positive cells were in the CD3-positive gate, indicating that only PBMCs, not the DC-SIGN-positive 293T cells, were infected (data not shown). Thus, like DC-SIGN, DC-SIGNR can transmit various bound HIV-1 virus strains to receptor-positive cell lines and to PBMCs. DC-SIGNR transmitted virus more efficiently than DC-SIGN when C8166 cells were used and more variably than DC-SIGN when PBMCs were used as targets. Therefore, some virus strains may be transmitted by DC-SIGNR to primary cells more efficiently than others.
DC-SIGNR and DC-SIGN Exhibit Comparable Ligand Specificity.
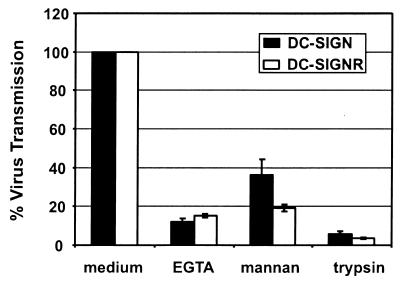
EGTA and mannan (a carbohydrate that binds to the lectin domain) block virus transmission by DC-SIGN, suggesting a critical role of the calcium-dependent lectin domain in this process (15, ∥). In agreement with this data, we found that deletion of the lectin domain in DC-SIGN prevents virus binding and transmission.∥ Moreover, washing virus-pulsed cells with trypsin/EDTA blocks transmission of virus from DC-SIGN-expressing 293T cells to CD4/coreceptor-positive cells.∥ We therefore investigated whether DC-SIGNR-mediated transmission is sensitive to the same agents. The virus transmission assay was carried out as described above, but the cells were incubated with EGTA and mannan before the addition of reporter virus or the cells were treated with trypsin/EDTA after the incubation with reporter virus. Addition of EGTA reduced virus transmission by both DC-SIGNR- and DC-SIGN-expressing cells to a comparable degree. Preincubation with mannan also reduced virus transmission, although somewhat less efficiently than did EGTA (Fig. 5). Trypsin/EDTA strongly inhibited virus transfer by DC-SIGNR and DC-SIGN, indicating that, in both cases, bound virus remained at the cell surface.
Figure 5.
EGTA, mannan, and trypsin/EDTA inhibit virus transfer. The transfer of NL4–3 luciferase reporter virus was analyzed as described above, except that the 293T cells were preincubated with EGTA or with mannan or one of the three wash steps was carried out with trypsin/EDTA instead of medium.
Discussion
The discovery of DC-SIGN helps explain the mechanisms by which DCs make initial contact with resting T cells, a step that can ultimately lead to T cell activation (28). In addition, DC-SIGN appears to be largely responsible for the unique ability of DCs to present virus to cells that express the necessary viral receptors. DC-SIGN binds to multiple strains of HIV-1, HIV-2, and SIV (15, ∥) and does so in a manner that maintains the integrity and infectivity of the virus for up to several days (15). Thus, virus bound to DC-SIGN-positive DCs can take advantage of the normal cellular trafficking patterns of DCs, perhaps being ferried by DCs to lymph nodes (15), the major site of HIV replication in vivo (29).
We have found that DC-SIGNR functions similarly to DC-SIGN with regards to virus attachment and transmission, as might be expected given the high degree of homology between these molecules (25). Like DC-SIGN, DC-SIGNR appears to function as a universal attachment factor for primate lentiviruses, supporting binding and transmission of all strains of HIV-1, HIV-2, and SIV examined. However, transmission of virus strains from DC-SIGNR-expressing cells to PBMCs was more variable than when virus was transmitted via DC-SIGN-positive cells. Therefore, DC-SIGNR may be more selective than DC-SIGN with regards to the virus strains that it can efficiently bind and transmit. To accurately discern this, however, it will be important to study virus binding and transmission with multiple cell types and to correlate expression levels with function. For example, we have found that DC-SIGN function is tightly linked to its expression levels, with ≈100,000 copies of DC-SIGN per 293T cell being needed for maximal activity.∥ In our experimental systems, high levels of DC-SIGN and DC-SIGNR expression were attained. Thus, it may be important to reduce expression levels of these attachment factors to identify differences in how these molecules interact with diverse virus strains.
Most studies of HIV entry have focused on the interactions of Env, CD4, and coreceptors, and the resulting conformational changes in Env that lead to membrane fusion. The impressive abilities of DC-SIGN and DC-SIGNR to increase the efficiency of virus infection underscore the importance of virus attachment as a first step in the entry pathway, both in cis and in trans. A number of molecules can support HIV attachment to both receptor-positive and receptor-negative cells, including cell surface heparan sulfate proteoglycans and interactions between LFA (lymphocyte function-associated antigen) and intercellular adhesion molecule-1 (11, 30). In the case of DCs, DC-SIGN appears to be largely responsible for virus attachment. Thus, there are now several examples where a virus receptor on one cell can function in trans to support infection of an adjoining cell. In some cases, binding of HIV-1 to CD4-positive cells in vitro enables virus to infect CD4-negative coreceptor-positive cells in trans (31). DC-SIGN and now DC-SIGNR also support infection in trans but do not obviate the need for either CD4 or coreceptor for viral entry (15). The precise mechanisms by which DC-SIGN and DC-SIGNR transmit virus are not known beyond the obvious dependence on carbohydrate recognition. It will therefore be important to determine whether interactions between Env and these attachment factors involve specific carbohydrate or polypeptide structural motifs.
We documented expression of DC-SIGNR on endothelial cells in human placenta, lymph node sinuses, and hepatic sinusoids but not on peripheral blood-derived DCs. Expression of DC-SIGNR in other tissues has not yet been demonstrated, although the development of additional antiserum or mAbs may reveal expression in other locations. If the virus attachment and transmission functions of DC-SIGNR require high levels of expression, as we have observed for DC-SIGN, then the mere presence of DC-SIGNR or DC-SIGN in a given cell type may not necessarily mean that an additional site at which virus can be efficiently captured and presented has been identified. Thus, quantitative measures of expression rather than just measurements of expression per se may be required.
What might the consequences of DC-SIGNR expression on endothelial cells be for HIV-1 infection? The expression of DC-SIGNR in lymph node and placenta is intriguing. Lymph nodes represent the major site of HIV replication in vivo (29), and the presence of DC-SIGNR on the surface of endothelial cells in lymph node sinuses represents an obvious mechanism by which cell-free virus can be transmitted to CD4-positive cells that come into contact with these surfaces. If DC-SIGNR binds to intercellular adhesion molecule-3 and other cell surface receptors, interactions between T cells and the endothelial cell surface may occur more frequently, increasing the likelihood of virus transmission. We have also documented DC-SIGNR expression in a significant proportion of capillaries in villi of term placenta and, in a separate study (E.J.S., J.T., and N.C., unpublished results), have found that DC-SIGN is expressed on decidual macrophages and fetal Hofbauer cells, a macrophage-like cell type in the placenta that supports HIV infection (32). The presence of these attachment factors on either side of the trophoblast layer that separates the maternal and fetal circulation could serve to concentrate virus at this site and influence vertical transmission. Expression of DC-SIGNR on hepatic sinusoidal endothelial cells is of less obvious relevance to HIV pathogenesis, although there is at least one report that these cells may be infected by HIV-1 in vitro (33). In addition, hepatic endothelial cells may also be involved in antigen presentation (21, 34), perhaps presenting opportunities to transmit virus to circulating CD4-positive cells.
In summary, DC-SIGNR joins DC-SIGN as a specific virus attachment factor that could have profound effects on the tropism and pathogenesis of HIV-1, HIV-2, and SIV. The discovery of these proteins stresses the need to reexamine virus attachment in general, not only to cells bearing CD4 and appropriate coreceptors but also to cells that frequently come into contact with these targets of virus infection. Perhaps other highly specific high-affinity virus attachment proteins will be identified. Finally, DC-SIGN and DC-SIGNR, which appear to share similar attachment mechanisms, potentially represent antiretroviral targets.
Acknowledgments
We thank Dr. D. G. D. Wight, Department of Histopathology, Addenbrooke's Hospital, Hills Road Cambridge, U.K. for advice about liver histology and Dr S. M. Rushbrook and Mrs K. Bird for assistance with immunohistochemistry. We also thank Kirsten Hiebenthal-Millow and Frank Kirchhoff for providing luciferase-reporter virus proviral genomes and Victor Holubowsky and Farida Shaheen for generation and quantification of virus stocks. This work was supported by Grant P30-AI45008 of the Viral/Cell/Molecular Core of the Penn Center for AIDS Research and by National Institutes of Health Grants R01 35383 and R01 40880 to R.W.D. This work was also supported by a Burroughs Wellcome Fund Translational Research Award to R.W.D. R.W.D. is a recipient of an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation. F.B. was supported by a fellowship from the Swiss National Science Foundation (Grant 823A-61172). E.J.S. was supported by a Medical Research Council (MRC) clinical training fellowship and also by the Sackler foundation. N.C. and L.S.M. were supported by grants from the MRC and Cancer Research Campaign. S.P. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Abbeviations: DC, dendritic cell; DC-SIGN, DC-specific, ICAM-3 grabbing, nonintegrin; DC-SIGNR, DC-SIGN related; PBMC, peripheral blood mononuclear cell; GFP, green fluorescent protein; FACS, flow cytometry.
S.P., F.B., B.L., G.J.L., M. D. Sanchez, K. Hiebenthal-Millow, J. Münch, F. Kirchhoff, and R.W.D., unpublished work.
References
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 3.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P D, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 4.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 5.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 7.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharron M P, Pöhlmann S, Price K, Tsang M, Kirchoff F, Doms R W, Lee B. Blood. 2000;96:41–49. [PubMed] [Google Scholar]
- 9.Kuhmann S E, Platt E J, Kozak S L, Kabat D. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doms R W. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 11.Mondor I, Ugolini S, Sattentau Q J. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ugolini S, Mondor I, Parren P, Burton D, Tilley S, Klasse P J, Sattentau Q J. J Exp Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ugolini S, Mondor I, Sattentau Q J. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 14.O'Doherty U, Swiggard W J, Malim M H. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geijtenbeek T B H, Kwon D S, Torensma R, Vliet S J v, Duijnhoven G C F v, Middel J, Cornelissen I L M H A, Nottet H S L M, Kewalramani V N, Littman D R, et al. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 16.Cameron P U, Freudenthal P S, Barker J M, Gezelter S, Inaba K, Steinman R M. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 17.Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman R M. J Virol. 1998;72:2733–2737. doi: 10.1128/jvi.72.4.2733-2737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayehunie S, Garcis-Zepeda E A, Hoxie J A, Horuk R, Kupper T S, Luster A D, Ruprecht R M. Blood. 1997;90:1379–1386. [PubMed] [Google Scholar]
- 19.Canque B, Bakri Y, Camus S, Yagello M, Benjouad A, Gluckman J C. Blood. 1999;93:3866–3875. [PubMed] [Google Scholar]
- 20.Weissman D, Li Y, Ananworanich J, Zhou L J, Adelsberger J, Tedder T F, Baseler M, Fauci A S. Proc Natl Acad Sci USA. 1995;92:826–830. doi: 10.1073/pnas.92.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knolle P A, Germann T, Treichel U, Uhrig A, Schmitt E, Hegenbarth S, Lohse A W, Gerken G. J Immunol. 1999;162:1401–1407. [PubMed] [Google Scholar]
- 22.Geijtenbeek T B H, Torensma R, Vliet S J v, Duijnhoven G C F v, Adema G J, Kooyk Y v, Figdor C G. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 23.Stahl-Hennig C, Steinman R M, Tenner-Racz K, Pope M, Stolte N, Matz-Rensing K, Grobschupff G, Raschdorff B, Hunsmann G, Racz P. Science. 1999;285:1261–1265. doi: 10.1126/science.285.5431.1261. [DOI] [PubMed] [Google Scholar]
- 24.Barratt-Boyes S M, Watkins S C, Finn O J. J Immunol. 1997;158:4543–4547. [PubMed] [Google Scholar]
- 25.Soilleux E J, Barten R, Trowsdale J. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 26.Sehested M, Hou-Jensen K. Virchows Arch A Pathol Anat. 1981;391:217–225. doi: 10.1007/BF00437598. [DOI] [PubMed] [Google Scholar]
- 27.Berger E A, Doms R W, Fenyö E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. Nature (London) 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 28.Steinman R M. Cell. 2000;100:491–494. doi: 10.1016/s0092-8674(00)80684-4. [DOI] [PubMed] [Google Scholar]
- 29.Fauci A. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 30.Fortin J-F, Cantin R, Bergeron M G, Tremblay M J. Virology. 2000;268:493–503. doi: 10.1006/viro.2000.0190. [DOI] [PubMed] [Google Scholar]
- 31.Speck R F, Esser U, Penn M L, Eckstein D A, Pulliam L, Chan S Y, Goldsmith M A. Curr Biol. 1999;9:547–550. doi: 10.1016/s0960-9822(99)80241-3. [DOI] [PubMed] [Google Scholar]
- 32.Newell M L. AIDS. 1998;12:831–837. doi: 10.1097/00002030-199808000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Steffan A M, Lafon M E, Gendrault J L, Schweitzer C, Royer C, Jaeck D, Arnaud J P, Schmitt M P, Aubertin A M, Kirn A. Proc Natl Acad Sci USA. 1992;89:1582–1586. doi: 10.1073/pnas.89.5.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knolle P A, Gerken G. Immunol Rev. 2000;174:21–34. doi: 10.1034/j.1600-0528.2002.017408.x. [DOI] [PubMed] [Google Scholar]