Abstract
The response of single DNA molecules to externally applied forces and torques was directly measured using an angular optical trap. Upon overwinding, DNA buckled abruptly as revealed by a sharp extension drop followed by a torque plateau. When the DNA was held at the buckling transition, its extension hopped rapidly between two distinct states. Furthermore, the initial plectonemic loop absorbed approximately twice as much extension as was absorbed into the plectoneme upon each additional turn. The observed extension change after buckling and the postbuckling torque support a recent DNA elasticity model.
The bending and torsional properties of DNA influence numerous cellular processes, notably DNA compaction, replication, transcription, and protein-DNA binding. DNA elasticity regulates how proteins bend and twist DNA upon binding and how translocating molecular motors exert torque and force on their DNA substrates. Single molecule techniques have proven to be powerful approaches for the investigation of the response of DNA to mechanical stress; individual DNA molecules can be stretched and twisted under physiologically relevant conditions. To date the stretching and bending elasticities of DNA have been well characterized through measurements of the force-extension relation of DNA [1,2]. However, somewhat less is known regarding the torsional elasticity of DNA, at least in part due to difficulties in making direct torque measurements. The most prevalent method to twist DNA is to use magnetic tweezers to rotate a magnetic bead via rotation of a magnetic field [3,4]. Twisting DNA can also be achieved by rotation of a micropipette cantilever [5]. These approaches have provided many important insights into DNA torsional properties even without torque detection. A recent and novel technique directly measured torque in DNA via viscous drag force on a small bead attached to the side of a DNA molecule [6]. This approach requires taut DNA to minimize writhe and thus is more suited for measurements under high force ( > 15 pN). More recently, an angular optical trap that we developed has permitted simultaneous and direct measurements of force and torque for concurrent observation of the tensile and torsional behaviors of DNA over broad ranges of forces and torques [7,8]. In addition, its wider bandwidth is well suited for detection of highly kinetic processes. Previously we showed that nanofabricated quartz cylinders are ideally suited as handles for angular trapping. During DNA supercoiling, the torque, angle, force, and extension of a DNA molecule can be simultaneously monitored at kHz rates. In this work, we have directly measured the torsional modulus of DNA in the intermediate force regime, determined basic relations regarding the dependence of torque on applied force, made the first observation of the abrupt formation of the initial plectoneme (interwound loop) in positively supercoiled DNA, and monitored previously unseen dynamics of this plectoneme formation.
Our angular optical trapping instrument, described in detail previously [7,8] features precise and immediate control of the trapping beam’s linear polarization, which is used to rotate a trapped quartz cylinder about its cylindrical axis. The physical torque exerted on the cylinder is determined by direct measurement of the change in angular momentum of the transmitted beam [7,9]. The quartz cylinders are nanofabricated to ensure uniformity and thus are ideally suited for calibration and measurement reproducibility [8]. Each cylinder is also chemically functionalized on one end for specific attachment to DNA [8].
Here we carried out experiments to measure the response of DNA as it was overwound to introduce positive supercoils. The experimental procedure resembles that previously used for magnetic tweezers studies [3], but with the addition of direct torque measurement as we previously described [8]. During an experiment as shown in Fig. 1, a DNA molecule was constrained between the end of a cylinder and the surface of a microscope coverslip, and was held under a constant force [10]. Subsequently, DNA was overwound by steady rotation of the cylinder via rotation of the input laser polarization. During this time, torque, angular orientation, position, and force of the cylinder as well as the location of the coverglass were simultaneously recorded. All experiments were performed in phosphate buffered saline with 150 mM NaCl at 23.5 °C.
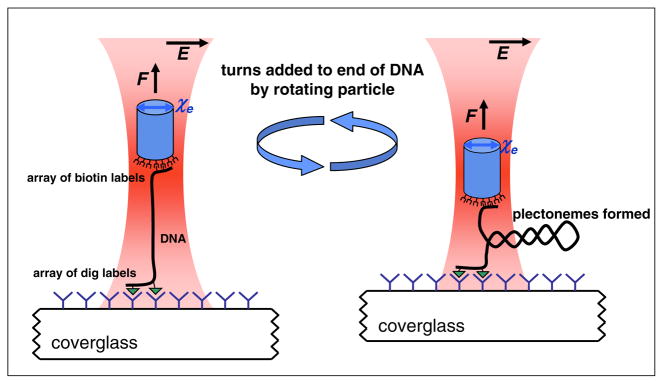
FIG. 1.
(color online). Experimental configuration. A DNA molecule was tethered to a nanofabricated birefringent quartz cylinder (extraordinary axis χe) held in an angular optical trap. Both ends of the DNA were torsionally constrained via its multiple tags: at one end via biotin-streptavidin and at the other end via digoxigenin (dig) and antidig. Force on the cylinder was applied in the axial direction and held constant by feeding back on a piezoelectric stage which displaced the coverslip. The DNA molecule was subsequently overwound by rotation of the linear polarization of the trapping laser.
Figure 2 depicts representative single traces of torque and extension as functions of number of turns added to the DNA at three different applied forces. As DNA was overwound at 1 turn/s, torque increased linearly while the extension remained approximately constant. This continued until the DNA buckled to form a plectoneme, indicated by a sudden decrease in extension. The buckling transition arises when the free energy of the extended DNA becomes larger than that of the initial plectonemic structure within the DNA. Beyond this transition, with additional twist the plectonemic region was extended continuously while the end-to-end extension decreased linearly and the torque plateaued at a constant value. Note that the torque and the extension slope after buckling were strongly sensitive to applied tension in the DNA. Additional experiments were carried out to verify that the data were taken under quasiequilibrium conditions [11]; when the supercoiled DNA was relaxed at the same rotation rate, the data were essentially indistinguishable.
FIG. 2.
(color online). Examples of torque and extension versus turn number. DNA molecules of 2.2 kbp in length were overwound under a constant force. Data were collected at 2 kHz and averaged with a sliding window of 1.5 s for torque and 0.05 s for extension. The torque signal had more Brownian noise relative to signal and was subjected to more filtering. As a result, higher frequency features in the torque signal, especially those near the buckling transition, might have been obscured. DNA buckling, locations indicated by dashed lines, was dependent on the applied force. (a) Torque versus number of turns. (b) The corresponding extension versus number of turns.
One of the most significant features of the overwinding data in Fig. 2(b) is the pronounced sharp drop in extension at the buckling transition, corresponding to the formation of the initial plectoneme. Interestingly, such an abrupt transition was absent in previous magnetic tweezers measurements where instead a smooth and gradual transition was observed [3]. The angular trapping method allowed detection of the abrupt transition, likely due to higher bandwidth and increased spatial resolution together with the use of shorter DNA tethers. As shown in Fig. 3(a), the magnitude of the extension drop observed at the buckling transition was dependent on the applied force, following a power law of ~F−0.5. In contrast, the extension decrease per turn after the transition followed a power law of ~F−0.4, as observed in previous magnetic tweezers studies [12]. Furthermore, the first loop of the plectoneme was able to absorb approximately twice as much extension as was absorbed into the plectoneme upon each additional single turn. These two distinct regimes of extension change versus force are depicted in Fig. 3(a). In addition, three different DNA templates were used for these experiments: a 2.2 kbp DNA, a 4.2 kbp DNA containing the 2.2 kbp sequence, and a 2.2 kbp DNA with a sequence entirely different from the first two. The extension changes were found to be the same for all three DNA templates, indicating that they are neither length nor sequence dependent within the limits of our measurements.
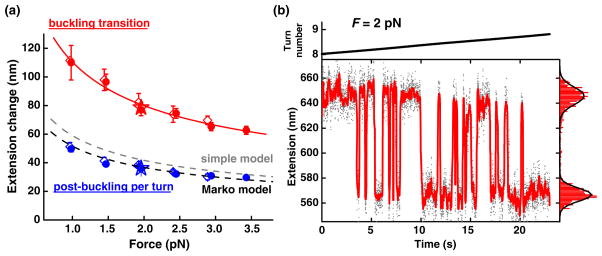
FIG. 3.
(color online). Extension change at the buckling transition. (a) The extension change versus force. The extension change for the initial plectoneme loop (red symbols) was well fit by a power law of F−0.5 (solid red) and was much larger than the extension change per turn for subsequent plectoneme growth (blue symbols). Three different DNA templates (2.2 kbp ●, N = 119; 4.2 kbp ◇, N = 35; another 2.2 kbp ★, N = 4) all exhibited the same trend. The two dashed lines show predictions by a simple model and a fit to the Marko model, respectively. Error bars are standard errors of the means. (b) A 2.2 kbp molecule was held at constant tension (2 pN) and overwound extremely slowly (0:04 turn/s) through its buckling transition. Data were taken at 2 kHz, low pass filtered to 400 Hz (solid dots), and then median filtered to 20 Hz (red curve). An extension histogram of the median-filtered data is shown on the right and was well fit by the sum of two Gaussians (black curve). The DNA was observed to rapidly fluctuate between two distinct states, separated by 79 nm, corresponding to pre- and postformation of the first plectonemic loop.
Data in Fig. 2 suggest that the energy barrier between the extended and plectonemic states is low enough for transitions between the two states to occur at an observable rate. To test this idea, a DNA tether was overwound extremely slowly through the buckling transition [Fig. 3(b)]. The extension of the DNA was observed to fluctuate between two discrete states, corresponding to pre- and postbuckling. The rates of fluctuation were highly sensitive to twist and we estimate that they were on the order of ~10 Hz near the rotational midpoint of the transition. The two states were separated by 79 nm, in good agreement with the extension drop observed at the same force in the rapid winding experiment in Fig. 3(a).
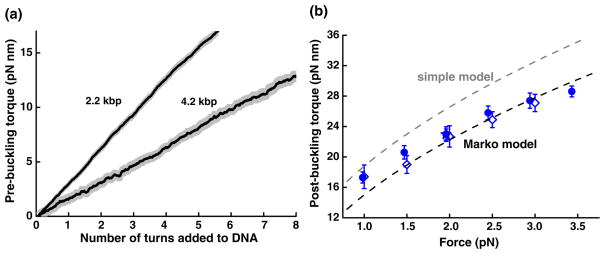
Measurements like those shown in Fig. 2(a) allowed direct determination of the torsional modulus of DNA. We plotted torque-turn relations by pooling the torque data taken at various forces prior to buckling for either the 2.2 or 4.2 kbp tethers [Fig. 4(a)]. Prior to buckling, DNA may be modeled as a simple elastic torsional rod. As twist is applied to the DNA, the restoring torque τ will increase linearly with the twist angle, as given by , where L0 is the contour length of the rod with 1 bp corresponding to 0.34 nm, n is the number of turns added, and C is the torsional modulus. The slopes of the measured torque-turn relations yielded a torsional modulus of C = 90 ± 3 nm kBT (88 ± 4 nm kBT) for the 2.2 kbp (4.2 kbp) DNA, corresponding to an intrinsic torsional modulus of ~100 nm kBT[13]. Previous studies, which have employed techniques such as DNA cyclization [14], fluorescence polarization anisotropy [15], or magnetic tweezers [16], have reported values ranging from 50 to 120 nm kBT for various temperature and buffer conditions.
FIG. 4.
(color online). Direct measurements of torque. (a) Torque prior to the buckling. Torque-turn relations prior to buckling were pooled from data for both the 2.2 kbp DNA (121 traces) and 4.2 kbp DNA (65 traces). Individual traces are shown as gray lines and resulted in a gray region when plotted together. Each solid curve indicates the average of all traces of a given length DNA. Torsional modulus for each DNA length (see text for values) was obtained by the slope of a linear fit to the average curve. (b) Torque after buckling. The torque after buckling at each force was pooled from data from the three DNA templates (blue symbols: 2.2 kbp ●, N = 119; 4.2 kbp ◇, N = 35; another 2.2 kbp ★, N = 4). The two dashed lines show predictions by the simple model and a fit to the Marko model, respectively.
These measurements also allow direct determination of the postbuckling torque. Figure 4(b) summarizes data for postbuckling torque versus force for all three DNA templates. The postbuckling torque increased with force and was independent of DNA length and sequence.
A number of models exist to explain DNA properties’ postbuckling. A simple model treats DNA as an elastic rod and assumes that a plectoneme consists of circular loops [12], predicting an extension change per turn after buckling of and a post-buckling torque given by , where Lp is the persistence length of the DNA, kBT is the thermal energy, and F is the applied force. We carried out force-extension measurements similar to those described before [1] and determined Lp = 43 ± 3 nm under our experimental conditions. The predicted postbuckling extension change per turn and post-buckling torque versus force, shown Figs. 3(a) and 4(b), respectively, are greater than measurements by as much as 25%.
Several more elaborate models exist to describe DNA supercoiling analytically [17–19]. In particular, an elegant recent theoretical work by John Marko [17] employed a detailed statistical mechanics analysis to incorporate an effective torsional flexibility of the plectonemic state (plectonemic rigidity) [20] and a force-dependent torsional flexibility of the extended state [13]. This model, which we refer to here as the Marko model, provides closed form expressions for both the extension change per turn and the post-buckling torque [21]. All parameters in the model were experimentally determined in this work, except for the plectonemic rigidity. We therefore performed a global fit of our measurements to the model using the plectonemic rigidity as the single fit parameter. The resulting best fit for the extension change per turn was in excellent agreement with the measurements [Fig. 3(a)] and the resulting best fit for the post-buckling torque agreed with the measurements to within 15% [Fig. 4(b)]. In addition, the best fit value for the plectonemic rigidity was 26 nm, within the range of 21–27 nm as previously estimated [20].
We are not aware of any analytical models suitable for prediction of the observed extension change and dynamics at the buckling transition. Mechanical rod models should be extendable to explain DNA supercoiling. Goyal et al. formulated a nonlinear dynamic rod model which shows an abrupt buckling followed by subsequent formation of a plectoneme in a macroscopic rod [22], a prediction that bears much resemblance to our measurements.
The highly dynamic nature of a twisted DNA molecule at the buckling transition may have important biological consequences in vivo. The specific supercoiling density (0.00–0.10) and applied force (1.0–3.5 pN) are well within the range commonly experienced by DNA in the cell. If a DNA molecule is subject to moderate stresses, distant elements on the sequence may transiently be brought into contact, which may facilitate the binding of DNA looping proteins or transcription factors [23]. The rapid formation and loss of these transient loops could therefore greatly reduce the search time needed for a protein to find two spatially separated sequence elements on the template.
Direct measurements of DNA torsional response lays an important foundation for the understanding of many biological processes that are regulated by torque. For example, topoisomerases are known to mediate linking numbers in DNA by sensing torsional stress in the DNA [24,25]. RNA polymerases as well as other groove-tracking enzymes are expected to rotate about the DNA helical axis [26,27], and would thereby generate and move against positive torque in the downstream DNA. We anticipate major progress in these areas with the advent of a number of biophysical techniques (including the one presented here) to measure and control the rotation of microscopic particles [6,8,28,29]. The angular optical trap, with its wide bandwidth, high spatial resolution, and ability to simultaneously measure force and torque, should prove to be a valuable tool to understand these highly kinetic and mechanical processes.
Supplementary Material
Acknowledgments
We thank members of the Wang lab for critical reading of the manuscript, L. Bai, J. Jing, and D. S. Johnson for helpful advice on DNA template designs, and J. F. Marko, D. Bensimon, V. Croquette, and N. H. Dekker for helpful discussions. M. D. W. wishes to acknowledge support from NSF Grant (No. DMR-0517349), NIH Grant (No. R01 GM059849), the Keck Foundation, and the Cornell Nanobiotechnology Center.
References
- 1.Wang MD, et al. Biophys J. 1997;72:1335. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith SB, Finzi L, Bustamante C. Science. 1992;258:1122. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 3.Strick T, Allemand JF, Bensimon D, Bensimon A, Croquette V. Science. 1996;271:1835. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 4.Crut A, et al. Proc Natl Acad Sci USA. 2007;104:11 957. [Google Scholar]
- 5.Leger JF, et al. Phys Rev Lett. 1999;83:1066. [Google Scholar]
- 6.Bryant Z, et al. Nature (London) 2003;424:338. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 7.LaPorta A, Wang MD. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 8.Deufel C, et al. Nat Meth. 2007;4:223. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 9.Bishop AI, et al. Phys Rev A. 2003;68:033802. [Google Scholar]
- 10.Deufel C, Wang MD. Biophys J. 2005;90:657. doi: 10.1529/biophysj.105.065458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.See auxiliary material [21] on reversibility of overwinding DNA.
- 12.Strick T, et al. Rep Prog Phys. 2003;66:1. [Google Scholar]
- 13.Moroz JD, Nelson P. Proc Natl Acad Sci USA. 1997;94:14 418. doi: 10.1073/pnas.94.26.14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz DS, Wang JC. J Mol Biol. 1984;173:75. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- 15.Selvin P, et al. Science. 1992;255:82. doi: 10.1126/science.255.5050.1380. [DOI] [PubMed] [Google Scholar]
- 16.Strick T, Bensimon D, Croquette V. Genetica (The Hague) 1999;106:57. doi: 10.1023/a:1003772626927. [DOI] [PubMed] [Google Scholar]
- 17.Marko J. Phys Rev E. 2007;76:021926. [Google Scholar]
- 18.Bouchiat C, Mezard M. Phys Rev Lett. 1998;80:1556. [Google Scholar]
- 19.Purohit PK, Nelson PC. Phys Rev E. 2007;75:039903. [Google Scholar]
- 20.Vologodskii A, et al. J Mol Biol. 1992;227:1224. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 21.See EPAPS Document No. E-PRLTAO-100-046814 for auxiliary text on predictions by the Marko model and an auxiliary figure on reversibility of overwinding DNA. For more information on EPAPS, see http://www.aip.org/pubservs/epaps.html.
- 22.Goyal S, et al. Int J Non-Linear Mech. 2008;43:65. [Google Scholar]
- 23.Nelson P. Proc Natl Acad Sci USA. 1999;96:14 342. [Google Scholar]
- 24.Koster DA, et al. Nature (London) 2005;434:671. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 25.Strick T, Bensimon D, Croquette V. Nature (London) 2000;404:901. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 26.Harada Y, et al. Nature (London) 2001;409:113. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 27.Revyakin A, et al. Science. 2006;314:1139. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop AI, et al. Phys Rev Lett. 2004;92:198104. doi: 10.1103/PhysRevLett.92.198104. [DOI] [PubMed] [Google Scholar]
- 29.Oroszi L, et al. Phys Rev Lett. 2006;97:058301. doi: 10.1103/PhysRevLett.97.058301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.