Abstract
Unsaturated fatty acids (UFA) are essential components of cells. In Saccharomyces cerevisiae, stearoyl-coenzyme A (CoA) desaturase 1 (OLE1) affects cell viability through the regulation of oleic (18:1) or palmitoleic (16:1) acid production. In this study, we used a targeted gene deletion approach to determine the impact of OLE1 on the emerging human pathogenic fungus Candida parapsilosis. We found that the deletion of OLE1 resulted in an auxotrophic yeast strain (designated OLE1 KO) that required unsaturated fatty acids for growth but not saturated fatty acids. Additionally, the production of UFA by OLE1 KO yeast cells was markedly reduced, suggesting that Ole1 is essential for UFA production. In contrast to wild-type C. parapsilosis, which produced pseudohyphal growth on UFA-supplemented medium agar, pseudohyphal formation in the OLE1 KO cells was severely impaired, suggesting that Ole1 regulates morphology. Furthermore, the OLE1 KO cells were hypersensitive to various stress-inducing factors, such as salts, SDS, and H2O2, especially at the physiological temperature. The results indicate that OLE1 is essential for the stress response, perhaps through the production of UFA for cell membrane biosynthesis. The OLE1 KO cells also were hypersensitive to human and fetal bovine serum, suggesting that targeting Ole1 could suppress the dissemination of yeast cells in the bloodstream. Murine-like macrophage J774.16 more efficiently killed the OLE1 KO yeasts, and significantly larger amounts of nitric oxide were detected in cocultures of macrophages and OLE1 KO cells than with wild-type or heterozygous strains. Moreover, the disruption of OLE1 significantly reduced fungal virulence in systemic murine infection. Taken together, these results demonstrate that Ole1 regulates the pathobiology of C. parapsilosis via UFA and that the OLE1 pathway is a promising antifungal target.
Candida parapsilosis is one of the leading causes of candidemia in humans (reviewed in references 24 and 27). Individuals at the greatest risk for infection with this pathogen are low-birth-weight infants and patients in intensive care units, especially if the patient is receiving lipid-based solutions or has prosthetic devices or indwelling catheters. C. parapsilosis is notorious for the nosocomial spread of disease through the hands of health care workers, and the fungus has increased resistance to echinocandins. Despite emerging as the second-most prevalent Candida species causing invasive disease globally, the molecular mechanisms for C. parapsilosis virulence are poorly understood. We have revealed recently that lipid metabolism is one of the major virulence pathways of the fungus (6, 10). The disruption of secreted lipases (LIP1/LIP2) (6) or fatty acid synthase 2 (FAS2) (10) severely reduced the ability of C. parapsilosis to grow under certain nutrient-limiting and/or stress conditions and attenuated the capacity of fungus to survive in macrophages or in a murine infection model.
In all eukaryotes, unsaturated fatty acids (UFA) are essential components of membrane and storage lipids (11, 22, 25). In Saccharomyces cerevisiae, saturated fatty acids (SFA) and UFA are synthesized from a acetyl-coenzyme A (CoA) precursor via the de novo fatty acid synthesis pathway, which involves a complex of enzymes (22). The initial reaction of fatty acid synthesis is the incorporation of acetyl-CoA with CO2 to generate malonyl-CoA, which is catalyzed by the biotin-bound enzyme acetyl-CoA carboxylase (AccI) (17). Thereafter, the elongation of the carbon chain is catalyzed by fatty acid synthases (Fas1 and Fas2) (reviewed in reference 19) and elongase 1 (Elo1) (18) to produce long-chain SFA. The resulting fatty acid products of the de novo fatty acid synthesis, such as palmitic and stearic acids, are the precursors for the subsequent desaturation reactions to produce UFA. The monounsaturated fatty acids in yeasts and other fungi are synthesized by stearoyl-CoA desaturase (Ole1), a conserved endoplasmic reticulum enzyme that introduces a double bond into saturated fatty acyl CoA substrates (8). In S. cerevisiae, there is a single OLE1 gene responsible for catalyzing the desaturation of palmitic and stearic acids to produce palmitoleic and oleic acids, respectively. The most abundant UFA in S. cerevisiae is oleic acid (18:1). Other fungi, such as Candida albicans (9), also express membrane-bound Δ12 and Δ15 desaturases that are important for the generation of other UFA, such as linoleic (18:2) and linolenic (18:3) acids.
The balance between SFA and UFA is critical for membrane fluidity, impacting the function and integrity of the various membrane systems of the cell. Thus, fatty acid production is subjected to highly restrictive regulation at the transcriptional and posttranslational levels. S. cerevisiae Ole1 is the target of the endoplasmic reticulum-bound transcription factors Spt23p and Mga2p (4, 12). The overexpression or depletion of OLE1 affects the cell membrane fluidity in the pathogenic yeast forms of Histoplasma capsulatum (14) and C. albicans (7), respectively. The deletion of S. cerevisiae OLE1 leads to a UFA auxotrophic mutant (21), and fatty acids produced by Ole1 are essential for mitochondrial movement and inheritance (20). C. albicans OLE1 conditional depletion mutants are auxotrophic to exogenous fatty acids, as they grew only in the presence of oleic acid (7). The filamentation of C. albicans is dependent on the amount of OLE1 expression, which suggests that Ole1 is the sensor of membrane fluidity and regulates cell morphology. Importantly, reducing OLE1 expression by the conditional inhibition of the gene can affect the virulence of C. albicans in murine infection (28). As noted above, we recently disrupted C. parapsilosis FAS2 to demonstrate that the de novo fatty acid biosynthesis pathway is essential for virulence in C. parapsilosis (10). As Ole1 is downstream of Fas2 in S. cerevisiae and probably in C. albicans, we characterized the function of C. parapsilosis Ole1 to further strengthen our understanding of the fatty acid biosynthesis pathway and to explore its potential as an antifungal target.
MATERIALS AND METHODS
Strains and culture conditions.
C. parapsilosis strains were maintained at −80°C in 35% glycerol. If not otherwise mentioned, the strains were grown in either YPD (1% yeast extract, 2% Bacto peptone, 2% glucose) or YPDPO (YPD plus 1% Tween 80 and 0.01% palmitoleic and oleic acids). Strains generated and used in this study are listed in Table 1.
TABLE 1.
C. parapsilosis strains generated and used in this study
| Name (resistance status) | Genotype | Reference |
|---|---|---|
| WT | GA1 (wild type) | 6 |
| FAS2 KO (Nous) | ΔCpfas2/ΔCpfas2::FRT | 10 |
| FAS2 HET (Nous) | CpFAS2/ΔCpfas2::FRT | 10 |
| FAS2 RE (Nous) | CpFAS2/ΔCpfas2::FRT | 10 |
| OLE1 HETr (Nour) | CpOLE1/ΔCpole1::SAT1-FLIP | This study |
| OLE1 HET (Nous) | CpOLE1/ΔCpole1::FRT | This study |
| OLE1 KOr (Nour) | ΔCpole1/ΔCpole1::SAT1-FLIP | This study |
| OLE1 KO (Nous) | ΔCpole1/ΔCpole1::FRT | This study |
| OLE1 RE (Nous) | CpOLE1/ΔCpole1::FRT | This study |
Generation of disruption construct pSFS2Ole1.
The pSFS2Ole1 plasmid was used to disrupt the entire open reading frames of the OLE1 genes, consisting of 1,461 nucleotides. A 473-bp upstream fragment of the OLE1 gene was amplified from genomic DNA with the use of a primer pair (C.pOLE1upF, CGGGGTACCACTCAATTGATCACCCACAGA [KpnI site underlined]; C.pOLE1upR, CCGCTCGAGGGTAGCGGAAATTATGGTTG [XhoI]). The PCR fragment was ligated into pGEMT and then transferred to pSFS2 (15) by being cloned into KpnI and XhoI sites to yield the pSFS2upOle1 plasmid. The cloning of a 477-bp fragment downstream of the OLE1 gene was performed similarly. The 447-bp downstream region was amplified by PCR from genomic DNA with the use of a primer pair (C.pOLE1doF, TCCCCGCGGTACGATTGTTATGGCAGTGG [SacII]; C.pOLE1doR, CCCGAGCTCCGAAGGTAGGTATGAGTGGG [SacI]) and then cloned into pSFS2upOle1 within the SacI and SacII sites to generate the pSFS2Ole1 disruption plasmid.
Candida parapsilosis transformation and generation of CpOle1 deletion mutants.
C. parapsilosis wild-type (WT) cells were transformed by electroporation as previously described (6), with minor modifications. Briefly, a single colony was inoculated in 50 ml YPD medium overnight with shaking at 150 rpm and 30°C. The cells were collected and centrifuged at 1,000 × g for 5 min. The cells were suspended in 45 ml TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 7.5) containing 100 mM lithium acetate and incubated for 45 min at 30°C with gentle shaking. After the addition of 0.45 ml of 1 M DTT (dithiothreitol) and an additional 15 min of shaking, the cells were washed three times with ice-cold water and once with 1 M sorbitol. Finally, the cells were diluted in 150 μl of 1 M sorbitol and kept on ice. For a single transformation, 40 μl of the cell suspension was used with 10 μl of DNA (10 μg).
To generate the heterozygous strain (OLE1 HET), the clinical C. parapsilosis isolate (WT) was transformed with 10 μg of ethanol-precipitated DNA of plasmid pSFS2Ole1, which was digested overnight with KpnI. Transformants were analyzed by Southern blotting. To eliminate the nourseothricin (Nou) selection marker cassette, the mutant strain was grown in YNB (yeast nitrogen base without glucose) medium with 2% maltose overnight. Nou-sensitive colonies were selected with a low concentration of the selection marker (20 to 25 μg/ml) in YPD medium as previously described (6). A Nou-sensitive strain subsequently was used to generate the homozygous disruptants (OLE1 KO). The transformation procedures as described for the OLE1 HET strain were repeated with the pSFS2Ole1 plasmid, except that transformants were plated in YPDPO with 200 μg/ml Nou. To identify the OLE1 KO strains, primary transformants were picked and plated on YPD and YPDPO. The yeast strains that grew on YPDPO but not on YPD were selected. Transformants were analyzed by Southern blotting. The induction in maltose YNB supplemented with fatty acid (YNBPO without glucose) medium was performed as described above to eliminate the nourseothricin selection marker cassette. Cells then were cultivated in YPD with or without fatty acids.
Generation of construct for reconstituted OLE1 gene.
As described in Results, the OLE1 KO strain is auxotrophic for UFA. Hence, the OLE1 gene could be employed as a selection marker for the mutant strain. We cloned the entire OLE1 gene, including the endogenous promoter and terminator sequence, and transformed it into the OLE1 KO strain to generate the reconstituted strain (OLE1 RE), which harbors one copy of the OLE1 gene. A 2,235-bp fragment, including the entire OLE1 gene, was amplified by PCR with the use of the primer pair Ole1ApaI (GGGGGCCCGATGTCAAACTCCCTTCCTGA) and Ole1XhoI (CCGCTCGAGCCCACTGCCATAACAATCG). The PCR product was precipitated and transformed into the OLE1 KO strain. Transformants were regenerated and analyzed by Southern blotting for the presence of the OLE1 gene in the native locus. Transformants were grown in YPD without fatty acids.
Southern blot analysis.
Total DNA was isolated and digested with appropriate enzymes. DNA then was separated on 0.8% agarose gels and transferred to nylon membranes (Amersham). The membranes were hybridized with digoxigenin-11-dUTP-labeled DNA probe that was amplified by PCR with the use of CpOLE1upF/CpOLE1upR primers. The detection and visualization of DNA was performed according to the manufacturer's instructions (DIG DNA labeling and detection kit; Roche).
Growth assays.
The growth rates of WT C. parapsilosis and the constructed mutants were analyzed in liquid YPD, YPDPO, and YNB plus 50 mM glucose, as well as in YNBPO (YNB plus 50 mM glucose, 1% Tween 80, and 0.01% palmitoleic and oleic acids), with all media at pH ∼6.5. Single colonies from the WT, HET, and RE strains were inoculated in 2.5 ml YPD medium, whereas the KO strain was inoculated in 2.5 ml YPDPO medium. These cultures were incubated overnight in an orbital shaker set at 150 rpm and 30°C. The yeast cells were washed three times with sterile phosphate-buffered saline (PBS) and counted using a hemacytometer. The experimental media then were inoculated with 5 × 106 cells/ml in 24-well plates. The cell density (optical density at 600 nm [OD600]) was read by a microtiter reader (Labsystem Multiskan MS) at the indicated times. The growth of OLE1 KO strains also was analyzed in fatty acid-containing medium that included YPD plus 0.001 to 0.01% (wt/vol) oleic acid (18:1), palmitoleic acid (16:1), a mixture of palmitoleic and oleic acids, or 0.001 to 0.1% (vol/vol) Tween 80. The growth of the OLE1 KO strain in YPD was used as a negative control. The growth rates at different times were measured by the OD600. The assays were performed in triplicate and repeated twice.
The growth of the WT and OLE1 KO strains in human serum (HS) and fetal bovine serum (FBS) also was analyzed. The WT and KO yeast cells were cultured overnight in YPD and YPDPO, respectively. Yeast cells in log-phase growth were washed, diluted to 5 × 106 cells/ml, inoculated in 20% human serum or FBS with or without heat inactivation, and incubated at 30°C with rotary shaking at 150 rpm. Aliquots were obtained at different times of growth and plated on YPD agar for the WT and on YPDPO for the KO to determine numbers of CFU. Yeast cell viability also was assessed by fluorescence microscopy. Yeast cells grown in 20% serum for 24 h were costained with 10 μM SytoxGreen and 20 μg/ml propidium iodine (PI) in PBS at room temperature for 30 min. The yeast cells then were observed with an Axiovert 200 M inverted microscope using a 63× objective and green and red fluorescence channels.
The susceptibility of yeast cells to SDS, NaCl, KCl, and H2O2 on YPDPO (pH 6.5) was tested. Overnight cultures in YPD and YPDPO broth were diluted with PBS to an OD600 of 0.1, transferred to 24-well plates, and serially diluted 1:10. Aliquots (2.5 μl) of diluted suspensions were spotted onto YPDPO solid medium containing stress-inducing factors. Plates were incubated at 30 and 37°C for 3 days.
Killing assays and phagocytosis with macrophage-like cells.
The macrophage-like cell line J774.16 (3) was used to study the intracellular fate of the C. parapsilosis strains. Macrophages were cultured in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal calf serum (FCS) and were plated at 5 × 105 cells per well in 24-well plates. Cocultures of the J774.16 cells and C. parapsilosis were performed according to our described protocol (6). Briefly, C. parapsilosis cells were grown overnight, washed three times in PBS, counted using a hematocytometer, and suspended in DMEM (without serum). The cells then were coincubated with the macrophage monolayer at an effector/target ratio of 15:1. The cocultures were incubated at 37°C for 2 and 4 h. The wells then were washed three times with PBS to remove nonadherent Candida cells. Yeast cells were liberated from macrophages by forcibly disrupting the macrophages through pipetting in H20 for 2 min. The yeast cells were collected, counted, and serially diluted prior to being plated. Cells were plated in YPD (YPDPO for the OLE1 KO strain) agar. Phagocytosis of yeast cells was performed similarly. The intracellular yeast cells were counted after 1 h of interaction with macrophages. The phagocytic index was the ratio of the number of intracellular yeast cells to the number of macrophages counted.
The viability of yeast cells also was assessed by microscopy. Cocultures in 8-chamber glass slides were washed twice with Hank's balanced salt solution (HBSS) and stained with 0.01% acridine orange (Sigma-Aldrich) and then stained with 0.05% crystal violet (Sigma-Aldrich) dissolved in 0.15 M NaCl. The slides were stained for 45 s and washed twice with HBSS. Finally, the slides were rinsed three times with PBS. Pictures were taken with an Axiovert 200 M inverted microscope. The objective used was 20× in red, green, and phase channels. Experiments were performed in triplicate and repeated twice.
To measure nitric oxide (NO) production by macrophages challenged with yeast cells, phagocytosis assays were performed as described above. Macrophages with Candida cells in DMEM were disrupted through pipetting, and NO levels were measured from 50 μl of the suspension using the Griess reagent system (Promega). Macrophages without yeast cells were used as the control. Superoxide measurements were performed as described previously (26). Experiments were performed in triplicate and repeated twice.
Fatty acid analysis.
Fatty acid species from yeast cells grown at log phase were extracted as described by Schneiter and Daum (16), with modifications. Briefly, yeast cells from 2.5 ml overnight cultures grown in YPD and YPDPO were collected by centrifugation at 0.8 × g, washed twice with distilled water, and suspended in 1 ml of cold methanol spiked with 10 μg of heptadecanoic acid (17:0) (Sigma, St. Louis, MO) as an internal standard. The yeast cells were disrupted by being vortexed with 0.5-mm-diameter glass beads. Fatty acids were extracted with chloroform-methanol (2:1) with vigorous shaking for 1 h at room temperature. The organic phase was collected in a glass tube and dried under nitrogen gas. Fatty acid profiles were determined by a gas chromatograph as described by Stukey et al. (21).
Murine infection models.
A/J mice (female, 6 to 8 weeks of age; obtained from the National Cancer Institute) were inoculated intraperitoneally or intravenously with 3 × 107 WT or mutant yeast cells in 100 or 30 μl PBS, respectively. Animal experiments were performed according to the guidelines published by the Institute of Laboratory Animal Resources of National Research Council. Animal care for this study was approved by the institutional Animal Care and Use Committee of the Albert Einstein College of Medicine under protocol number 20080604. CFU numbers were determined from the liver, kidneys, and spleen 3 and 5 days after infection by plating tissue homogenates on YPD agar. For histological examinations, kidneys of A/J mice infected intravenously with WT and KO yeast cells were removed at day 5 after infection and fixed with formalin. The kidneys were embedded in paraffin, sectioned, and stained with periodic acid-Schiff (PAS) stain. Renal sections were examined using an Olympus AX70 microscope (Olympus America Inc.) with a 40× objective.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism version 5.02 for Windows (GraphPad Software, San Diego, CA). The significant differences between sets of data were determined by the Newman-Keuls test or analysis of variance (ANOVA) according to the data.
RESULTS
Deletion of OLE1.
To characterize the role of Ole1, the two alleles of the gene were deleted from WT C. parapsilosis. WT yeast cells (Fig. 1, lane 1) were transformed with the plasmid pSFS2Ole1 by electroporation, and more than 30 primary colonies were produced. Eight colonies were randomly analyzed by Southern blotting, which showed that one of the OLE1 alleles had been replaced by the SAT1 flipper cassette (Fig. 1, lane 2) in each transformant. To delete the remaining OLE1 gene and reuse the disruption construct, the SAT1 flipper cassette containing the selection marker was eliminated in the selected mutant to generate the sensitive heterozygous mutant. This strain subsequently was used for the second transformation with the same plasmid, pSFS2Ole1, to generate homozygous mutant strains harboring the SAT1 flipper cassette. Although initial screens on YPD failed to generate any colonies, plating onto YPD supplemented with 1% Tween 80 and 0.01% palmitoleic and oleic acids (YPDPO) led to the identification of homozygous disruptants. We also observed that with the presence of fatty acids, yeast cells were more resistant to the selection marker, requiring Nou concentrations of 200 μg/ml for selection. In two independent transformations, we identified more than 30 mutants that grew on YPDPO but not YPD. Southern blot analysis revealed that the remaining native OLE1 gene in the heterozygous mutant strains was disrupted, resulting in homozygous mutant strains (Fig. 1, lane 3). The SAT1 cassette then was eliminated to yield homozygous non-Nou-resistant, disrupted strains (Fig. 1, lane 4). These complete disruptants were used to reintroduce the entire OLE1 open reading frame, including approximately 400-bp fragments from upstream and downstream regions, by transformation. Twenty transformants were obtained by screening for the ability to grow on YPD medium and analyzed by Southern blot analysis to confirm the presence of one copy of the OLE1 gene (Fig. 1, lane 5). Successful reconstituted mutants contained the OLE1 gene in the same locus as that shown for the WT.
FIG. 1.
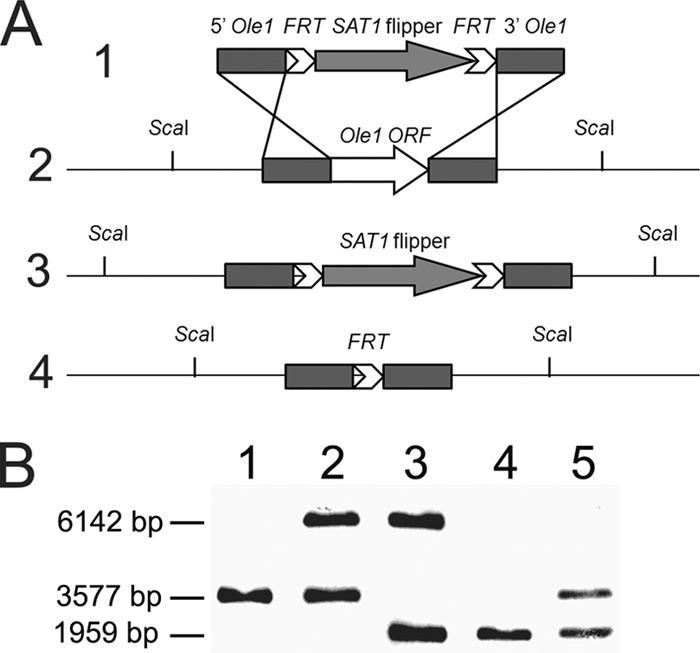
Disruption of OLE1 genes in C. parapsilosis. (A) Schematic representation of the disruption construct (1) and the genotype of the WT (2) with the OLE1 locus, disrupted OLE1 locus with the SAT1 cassette (3), and disrupted locus without the SAT1 cassette (4). (B) Southern blot analysis of the WT strain (lane 1), heterozygous resistant strain (lane 2), homozygous resistant strain (lane 3), homozygous nonresistant strain (lane 4), and reconstituted nonresistant strain (lane 5). The Southern blot probe was PCR amplified from the upstream fragment of plasmid pSFS2Ole1.
Growth dependence of OLE1 KO on UFA.
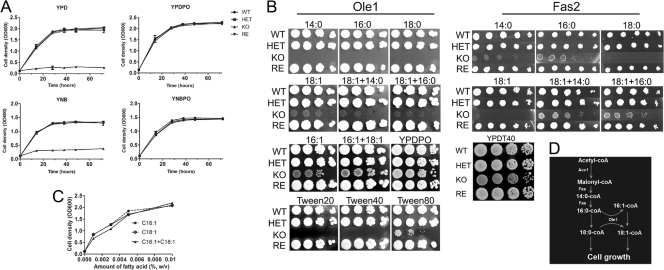
The disruption of OLE1 led to auxotrophic yeast cells that grew only in media supplemented with UFA (Fig. 2 A), thus we tested the growth dependence of the OLE1 KO strain in YPD supplemented with different fatty acids. We showed that the OLE1 KO yeast cells were unable to grow in the SFA myristic (14:0), palmitic (16:0), or stearic acids (18:0), but the yeast cells were able to grow in media supplemented with the UFA palmitoleic (16:1) or oleic acid (18:1) (Fig. 2B). The OLE1 KO strain exhibited better growth in media supplemented with palmitoleic acid than oleic acid. The disruptant also grew better in a mixture of palmitoleic and oleic acids. Combinations of palmitic with oleic acids did not enhance yeast cell growth. Since oleic acid is the predominant fatty acid in the yeast cell (10), these results suggest that palmitoleic acid could be elongated to produce oleic acid by an uncharacterized elongase. We also tested the growth of the OLE1 KO strain in different concentrations of UFA and found that growth correlated with fatty acid concentrations (Fig. 2C). Further, since combining SFA with UFA did not enhance OLE1 KO growth, the data suggest that Ole1 is the sole enzyme for producing UFA in C. parapsilosis. In comparison, our fatty acid synthase (FAS2) deletion mutant required certain SFA, such as 14:0 or 16:0, but not UFA (Fig. 2B) (10). Since UFA are essential for growth and the addition of SFA, such as 14:0 or 16:0, rescues the growth of the FAS2 deletion mutant, we further conclude that Ole1 has a role downstream from Fas2 in the de novo fatty acid biosynthesis of C. parapsilosis (Fig. 2D).
FIG. 2.
Growth dependence of OLE1 KO on unsaturated fatty acids. (A) Growth rates of wild-type (WT), heterozygous (HET), homozygous (KO), and reconstituted (RE) mutant strains. Yeast cell growth was compared in YPD, YPDPO (YPD plus fatty acids), YNB, and YNBPO (YNB plus fatty acids) broth. The growth of the Candida cells was measured by cell density at OD600. Experiments were repeated twice with four replicates for each medium, with similar results. (B) Spot growth assays of OLE1 KO and Fas2 KO on YPD supplemented with fatty acids or Tween 20, 40, or 80. A series of 10× dilutions of yeast cells were spotted onto agar. Plates were incubated at 30°C for 3 days, and images were digitally captured. Experiments were repeated twice with similar results. (C) The growth of the mutant yeast cells was dependent upon the amount of unsaturated fatty acids. Yeast cell strains were grown in YPD supplemented with 0 to 0.01% (wt/vol) palmitoleic acid (16:1), oleic (18:1) acid, or a mixture of these fatty acids. Yeast growth was determined by cell density after 24 h at 30°C with shaking. The results are the means from two independent experiments with triplicates. (D) A model of fatty acid biosynthesis generated from the assays. Fas is the Fas1 and Fas2 complex, which is required to synthesize SFA from malonyl-CoA. Ole1 is essential for UFA production from SFA precursors.
Exogenous fatty acids can overcome the growth inhibition of OLE1 KO in standard media. To test whether lipids also could be used by the OLE1 KO strain, we examined the growth of the mutant yeast cells in YPD supplemented with Tween 20, 40, or 80. We observed that the OLE1 KO strain did not grow in media supplemented with Tween 20 or 40, but it grew in media with Tween 80 (Fig. 2A). Interestingly, we observed that the OLE1 KO strain in Tween 80 medium started growing after 2 days of incubation at 30°C, suggesting that Tween 80 was digested during this time to release oleic acid. This is consistent with the OLE1 KO strain growing in the presence of oleic acid from Tween 80 but not with SFA lauric and stearic acids released from Tween 20 and 40, respectively.
Ole1 regulates production of UFA.
The OLE1 KO strain required UFA supplementation for growth. To further investigate the role of Ole1 in UFA production, we determined the fatty acid profile of the OLE1 KO and WT strains grown in YPD and YPDPO (Table 2). In the WT, we found that the addition of UFA resulted in only a slight increase in the overall quantity of UFA. The addition of UFA could negatively regulate the expression of OLE1, resulting in the accumulation of SFA. As a result, the ratio of SFA/UFA was increased about 2-fold. The deletion of OLE1 resulted in decreased levels of UFA, with a reduction in the production of palmitoleic, oleic, and linoleic acids from 0.3 to 0.1%, 10.4 to 3.4%, and 2.2 to 0.7%, respectively. Significantly, we found that the amount of stearic acid, the precursor to generate oleic acid, was elevated from 45.8% in the WT to 53.5% in the OLE1 KO strain. As a consequence of OLE1 deletion, the ratio of SFA to UFA of the mutant yeast cells was 23.08, whereas it was 6.76 for the WT grown in YPDPO. The data suggest that Ole1 regulates UFA production and the deletion of OLE1 causes an imbalance in the fatty acid composition of C. parapsilosis.
TABLE 2.
Fatty acid profiles (%) of the wild-type and OLE1 KO strains
| Strain (medium) | Fatty acida |
SFAb | UFA | SFA/UFA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 14:0 | 16:0 | 16:1 | 18:0 | 18:1 | 18:2 | 20:0 | ||||
| WT (YPD) | 1.5 | 39.4 | 0.1 | 36.0 | 7.8 | 15.2 | 0.1 | 76.9 | 23.1 | 3.4 |
| WT (YPDPO) | 2.9 | 38.3 | 0.3 | 45.8 | 10.4 | 2.2 | 0.1 | 87.1 | 12.9 | 6.8 |
| OLE1 (YPDPO) | 2.8 | 39.5 | 0.1 | 53.5 | 3.4 | 0.7 | 0.1 | 95.8 | 4.2 | 23.1 |
14:0, myristic acid; 16:0, palmitic acid; 16:1, palmitoleic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid; 20:0, eicosanoic acid.
SFA (saturated fatty acids) include 14:0, 16:0, 18:0, and 20:0; UFA (unsaturated fatty acids) include 16:1, 18:1, and 18:2.
Ole1 is essential for membrane stress response.
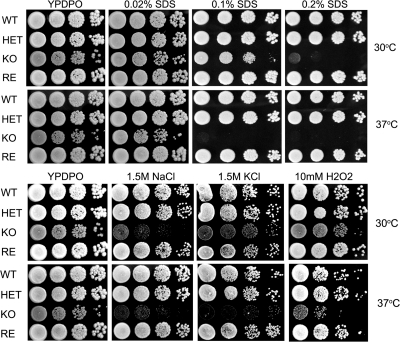
Since the deletion of OLE1 leads to the reduced production of UFA and impacts the balance of fatty acids in the cells, we tested whether the deletion enhanced the susceptibility of the OLE1 KO yeast cells to stress-inducing factors, including SDS, salts, and H2O2. Figure 3 demonstrates that the OLE1 KO yeast cells were hypersensitive to the stress-inducing factors tested. Further, increasing the temperature from 30 to 37°C increased the susceptibility of the yeast cells under the conditions assessed. The results indicate that Ole1 is essential for cell survival under stress conditions, especially at physiological temperatures. This also suggests that UFA produced by Ole1 are important for responses to cell membrane stress.
FIG. 3.
Hypersensitivity of OLE1 KO yeast cells to stress-inducing factors. A series of 10× dilutions of yeast cells were spotted on YPDPO in the presence of the indicated stress-inducing factor. The OLE1 KO yeast cells were more susceptible to SDS, NaCl, KCl, and H2O2 than the wild type. The enhanced susceptibility of the OLE1 KO cells was more profound at 37°C. The experiments were repeated with similar results.
Ole1 regulates invasive growth on solid plates.
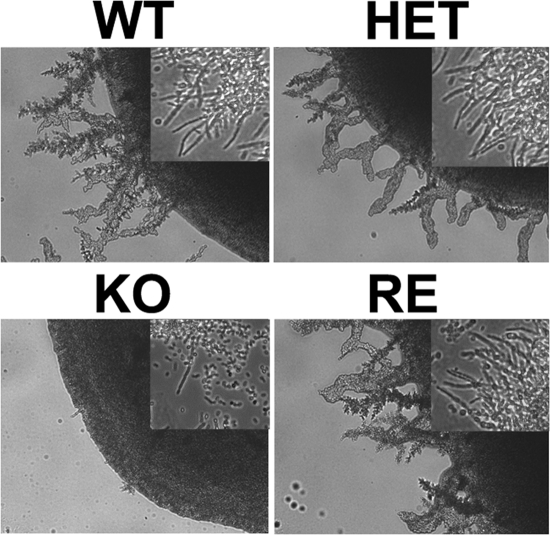
C. parapsilosis typically grows as a yeast, but it also can produce pseudohyphae. Interestingly, the conditions required to induce pseudohyphal growth in C. parapsilosis are poorly described. The filamentation of the WT does not occur on YPD (data not shown). We found that the WT strain was able to induce pseudohyphae, resulting in invasive growth on YDP agar supplemented with 1% Tween 80 and 0.01% palmitoleic and oleic acids (Fig. 4). In contrast, the OLE1 KO strain displayed limited invasive growth in the UFA solid media due to poor filamentation (Fig. 4). The filamentation patterns of the heterozygote and reconstituted mutants were similar to that of the WT. Thus, the data indicate that UFA are an important inducing factor for filamentation in C. parapsilosis and demonstrates that Ole1 regulates invasive growth and the filamentation of C. parapsilosis through UFA production.
FIG. 4.
Reduced invasive growth of OLE1 KO yeast cells. Yeast cells were plated on YPDPO agar and incubated at room temperature for 10 days. Filaments were present around the colonies of WT, HET, and RE strains but not the OLE1 KO strain (10× objective). Sections from the edges of the colonies observed on glass slides showed the pseudohyphae of the yeast cells (insets, 20× objective). The experiment was repeated with similar results.
OLE1 KO yeast cells are hypersensitive to serum.
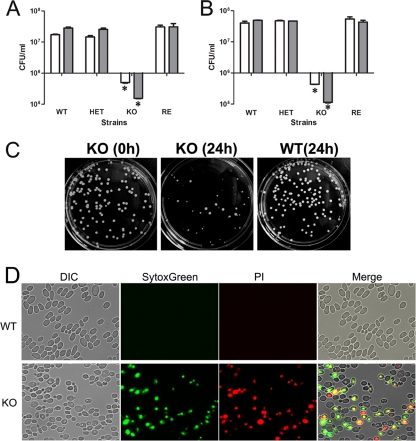
We previously reported that the inhibition of C. parapsilosis Fas2, which we demonstrated plays a role upstream of Ole1, promotes serum hypersensitivity. To examine whether the inhibition of Ole1 similarly impacts susceptibility to serum, we grew the WT and mutant yeast cells in human and fetal bovine sera. Whereas the WT, HET, or RE strains propagated well in 20% sera, we found that the number of the OLE1 KO yeast cells was reduced approximately 20, 40, 70, and 95% after 8, 12, 24, and 48 h of serum incubation, respectively (Fig. 5 and data not shown). Additionally, the colony morphology of OLE1 KO was affected in these media (Fig. 5). The deletion of OLE1 could change yeast cell membrane permeability, as suggested previously (7). To test membrane permeability, we used SytoxGreen, a high-affinity nucleic acid stain that only penetrates cells with compromised plasma membranes and will not cross the membranes of live cells. We also found that 65 to 70% of the OLE1 KO yeast cells exposed to 20% fetal bovine serum exhibited green fluorescence, indicating that SytoxGreen was able to penetrate. To verify that the SytoxGreen staining of the mutant yeast cells correlated with cell viability, we costained the serum-treated yeast cells with SytoxGreen and propidium iodine (PI). We observed that 98% of fluorescent yeast cells stained with both SytoxGreen and PI, while 2% of fluorescent yeast cells were only green. Hence, at 24 h, most mutant yeast cells with compromised cell membranes were killed by serum treatment. The results suggest that the deletion of OLE1 makes the yeast cell membrane penetrable by SytoxGreen and enhances susceptibility to microbicidal serum factors. Taken together with our previous study on Fas2 (10), we now have demonstrated that the inhibition of either gene in the de novo fatty acid biosynthesis pathway enhances serum susceptibility.
FIG. 5.
Hypersensitivity of OLE1 KO yeast cells to serum. The growth of the wild-type (WT), heterozygous (HET), reconstituted (RE), and homozygous (KO) mutant strains in 20% human (A) or fetal bovine (B) serum diluted in PBS. Error bars indicate standard deviations. *, P < 0.001 (ANOVA). (D) Effect of serum on the colony morphology of the OLE1 KO cells. The WT and OLE1 KO yeast cells were precultured in YPDPO liquid medium overnight, and aliquots of 5 × 106 yeast cells were transferred to 20% FBS at 30°C. Yeast cells were plated on YPDPO agar either prior to or after 24 h of incubation in FBS medium. OLE1 KO yeast cells exhibited heterogeneous colony sizes after 3 days of incubation on agar at 30°C. (C) WT and OLE1 KO strains were subjected to SytoxGreen and PI to characterize live and dead cells after 24 h of exposure to 20% human serum. Dead cells exhibited bright red fluorescence. All experiments were repeated twice with triplicates, and all results were similar to the data shown.
Ole1 regulates fungal virulence.
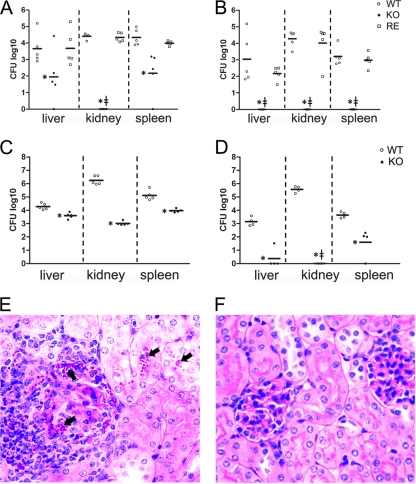
To examine whether the OLE1 KO strain was attenuated in vivo, we challenged AJ mice with the WT or mutant yeast cells intraperitoneally. Fungal burdens were examined 3 and 5 days after infection (Fig. 6 A and B). At both times, the OLE1 KO strain displayed significant reductions in CFU in the liver, spleen, and kidneys. Interestingly, despite C. parapsilosis typically displaying the highest CFU/g tissue in kidneys, no detectable yeast cells were found in the kidneys of mice infected with the OLE1 KO strain at days 3 and 5 after infection. Moreover, mice challenged with the OLE1 KO strain cleared the fungus from the assessed tissues by day 5 after infection. The WT and reconstituted strains produced similar fungal burdens. Furthermore, the fungal burdens in the kidneys, spleens, or livers of mice intravenously infected with OLE1 KO were significantly reduced compared to those of mice intravenously challenged with WT yeast cells (Fig. 6C and D). Notably, yeast cells were histologically detected in the kidneys of WT-infected mice but not in mice infected with the OLE1 KO strain (Fig. 6E). Furthermore, we also observed multifocal, widespread granulomatous and pyogranulomatous inflammation with intralesional yeast in the kidneys infected with the WT. Thus, the deletion of OLE1 severely impairs the capacity of the fungus to survive in systemic infections.
FIG. 6.
Reduced virulence of OLE1 KO. Intraperitoneal infection of A/J mice with wild-type (WT), homozygous (KO), and reconstituted (RE) yeasts (A and B). Intravenous infection of A/J mice with WT and KO yeasts (C and D). (A and C) CFU from kidneys, spleens, and livers 3 days after intraperitoneal and intravenous infection, respectively. (B and D) CFU 5 days after infection. Each symbol represents 1 mouse. *, P ≤ 0.01, (Newman-Keuls). 127, no detectable CFU of KO mutants. (E and F) Representative histological sections of kidney 5 days after intravenous infection with WT and KO cells, respectively. Arrows indicate aggregates of yeast cells in WT-infected tissues.
OLE1 KO yeast cells are susceptible to macrophages.
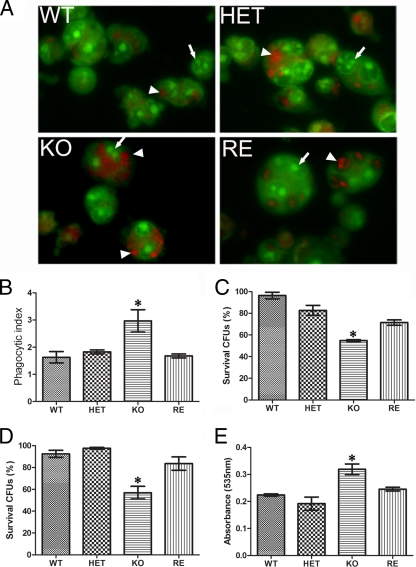
To better understand the mechanism of the reduced virulence and explore functions of macrophages in the fungal killing of OLE1 KO, we examined the phagocytosis and killing of the yeast cells with J774.16 cells. The deletion of the Ole1 genes significantly altered the phagocytosis of C. parapsilosis by macrophages. The phagocytosis of OLE1 KO yeast cells was increased by approximately 70% compared to that of the WT (Fig. 7). We also found that the intracellular survival of the OLE1 KO yeast cells was significantly reduced compared to that of WT or heterozygous cells. The intracellular survival of the OLE1 KO strain after 2 h of coculture was 55%, whereas it was 83% for the HET strain, 71.4% for the RE strain, and 97 to 99% for the WT strain (Fig. 7). We observed that prolonged inoculation time (after 4 h) did not significantly increase the reduction of the Ole1 strain (survival of 56%, whereas the survival rate for the others was 55%).
FIG. 7.
Reduced survival of OLE1 KO yeast cells exposed to macrophages. (A) Intracellular viability of yeast cells as assessed by the acridine and crystal violet staining of the yeast cells in the macrophages. The green yeast cells (arrows) are alive, whereas the orange-red cells (arrowheads) are dead. Pictures are the merge of the red, green, and phase channels at 20×. (B) Phagocytosis of the wild-type (WT), heterozygous (HET), homozygous (KO), and reconstituted (RE) mutant strains with murine-like macrophage (J774.16). The phagocytosis of each C. parapsilosis strain was assessed by counting the number of phagocytosed yeast cells in more than 500 macrophages. The phagocytosis index was the ratio of the yeast cells to macrophages. Experiments were repeated twice with triplicates, and similar results were documented. (C and D) Survival of yeast cells as determined by CFU after 2 and 4 h of coculture of yeast cells with J774.16, respectively. Experiments were repeated twice with four replicates. (E) Nitric oxide production from macrophages challenged with OLE1 KO yeast cells after 2 h. Experiments were repeated twice with four to five replicates with similar results. The results from the experiments were averaged, and error bars indicate standard deviations. For panels B to E, P < 0.05 by ANOVA (*).
Since oxidative killing is a major mechanism for controlling Candida by phagocytes, we examined the production of NO and superoxide by macrophages cocultured with yeast cells. Macrophage NO production, but not superoxide (data not shown), was significantly greater in the presence of OLE1 KO cells than WT or heterozygous strains after 2 h (Fig. 7) but not at 4 h (data not shown). This finding suggests that reactive oxygen species such as NO are major microbicidal effectors in activated macrophages.
DISCUSSION
We and others have shown that interference with the fatty acid biosynthesis pathway of Candida species can significantly reduce fungal growth in vitro and in vivo (5, 6, 10, 28, 29). In the current study, we applied our gene disruption method to characterize the function of Ole1 in the emerging pathogenic fungus C. parapsilosis. In contrast to C. albicans OLE1, which appears to be an essential gene, as demonstrated by an inability to produce a disruptant (7) and the impaired survival of C. albicans yeast cells subjected to the chemical or genetic repression of OLE1 (28), deletion of OLE1 in C. parapsilosis resulted in mutants auxotrophic for UFA. Since the deletion of OLE1 is not lethal in S. cerevisiae (20, 21), the regulation and function of OLE1 may reflect an evolutionary divergence in gene function. Although enzymes within the de novo fatty acid synthesis pathway are grossly conserved among yeast species, the amount and type of fatty acids produced by different fungi can vary significantly. For example, S. cerevisiae predominantly produces about 32% palmitoleic acid and 50% oleic acid (8, 22), and C. albicans produces about 7 and 34% of these fatty acids, respectively (28). The lower abundance of monounsaturated fatty acids in C. albicans could be explained by the presence of other polyunsaturated fatty acids, such as linoleic or linolenic acids (7, 9, 28). In our study, we observed that the levels of certain fatty acids in WT C. parapsilosis are different from that reported for C. albicans. For example, we found significantly higher ratios of 16:0/16:1 and 18:0/18:1 in C. parapsilosis (Table 2) (10). More than 70% of fatty acids found in C. albicans are UFA, whereas UFA comprise less than 30% in C. parapsilosis. The differential production of UFA levels in Candida strains might indicate that the regulation or function of fungal Ole1 is not uniform, which is supported by the fact that the disruption of the gene is lethal in C. albicans but not in C. parapsilosis.
C. parapsilosis OLE1 KO yeast cell growth inhibition in YPD was suppressed by the addition of exogenous UFA, which suggests that Ole1 is the key enzyme responsible for UFA production in this pathogen. Additionally, the level of UFA in the OLE1 KO strain was significantly decreased even with the addition of these fatty acids. Hence, the results suggest that (i) the incorporation of exogenous fatty acids into the OLE1 KO cells was reduced, and (ii) Ole1 is used to regulate the balance of the fatty acid pool in the WT cells by directly catalyzing UFA production. Changes in Ole1 expression lead to the imbalanced production of UFA, which impacted cell viability. This is consistent with findings for C. albicans, where the suppression of the endogenous OLE1 promoter using a Tet-off system affects the balance of SFA and UFA, leading to growth retardation in standard media (28). In yeasts, the cell membrane is the second-most outer layer to protect the cell from any stress conditions. By regulating UFA production, Ole1 appears to be essential for membrane fluidity. This notion is further supported by OLE1 overexpression in H. capsulatum, which resulted in significant changes in membrane permeability, perhaps through alterations in fatty acid composition (14). Thus, the presence of Ole1 is important for maintaining the ratio of UFA and SFA in the fatty acid pool, which is critical for maintaining membrane permeability/fluidity (7). We further found that Ole1 significantly impacted the capacity of C. parapsilosis to cope with extracellular stressors, as the disruption of OLE1 enhanced susceptibility to SDS, salts, and H2O2. Moreover, the OLE1 KO yeast cells were more susceptible to stressors at 37°C, indicating that Ole1 is involved in temperature stress responses. Taking these results together, we conclude that Ole1 produces UFA that are essential for yeast cell growth and that it regulates the capacity of C. parapsilosis to cope with diverse cellular stressors.
The clearance of Candida yeast cells is largely dependent on the appropriate function of phagocytes and is modified by the capacity of the fungus to secrete hydrolytic enzymes and maintain cellular homeostasis. We have shown that the OLE1 KO yeast cells are more avidly phagocytosed and more rapidly killed by J774.16 cells than are WT yeast. Additionally, we found that NO levels in macrophages exposed to OLE1 KO yeast cells are elevated compared to those for coculture with WT yeast, further indicating a change in the host-pathogen dynamic that benefits the host cells. Therefore, we propose that yeast cells lacking Ole1 failed to cope with stress responses generated by the macrophages and that the macrophages are able to more efficiently activate to combat the disruptant. More importantly, we demonstrated that the OLE1 KO yeast cells were significantly attenuated in murine systemic infection models. Notably, we found that the disruptant yeast cells were cleared from the kidneys of intraperitoneally infected mice within 3 days of infection and from all tissues analyzed by 5 days of infection. Similarly, OLE1 KO yeast cells were significantly cleared after intravenous challenge, as shown by CFU and histology, suggesting that Ole1 is essential for C. parapsilosis survival in a mammalian host.
The antimicrobial targeting of the fatty acid biosynthesis pathway has been examined for bacterial pathogens, although recent studies have shown that this approach is problematic, since exogenous fatty acids from host serum can overcome the inhibition of the bacterial pathway (2). We also found that the addition of exogenous fatty acids can suppress the inhibition of the C. parapsilosis fatty acid biosynthesis pathway in cultures. Indeed, we found that fatty acids extracted from human serum or bovine serum can support the growth of the OLE1 KO mutant. However, we also discovered that OLE1 KO yeast cells were hypersensitive to serum, which is similar to our findings with the deletion of FAS2 in C. parapsilosis (10). Several Candida spp., including C. parapsilosis, are human commensals, and the ability to disseminate from local residence into the bloodstream is a critical factor for systemic candidiasis. Thus, the inhibition of fatty acid biosynthesis could be an ideal way to facilitate the elimination of certain Candida spp. from the bloodstream, as we demonstrated in our intravenous infection experiments. Interestingly, the exact mechanism leading to fungal cell death presently is unclear. The inhibition of Fas2 or Ole1 could cause yeast cells to assimilate nutrients from serum such as free fatty acids or glucose to compensate for the lack of fatty acyl intermediates for fatty acid synthesis. Since the yeast cells lacking Fas2 or Ole1 cannot convert these intermediates into longer-chain fatty acids or UFA, these fatty acid precursors could accumulate and toxify the cell. For example, the inhibition of fatty acid synthase by cerulenin or RNA interference (RNAi) techniques induces the apoptosis of breast cancer cells, purportedly due to the accumulation of malony-CoA and ceramide (1, 13, 23). Alternatively, as the inhibition of Fas2 or Ole1 alters cell permeability and fluidity, the fungus could simply be more susceptible to damage from serum substances such as ions, vitamins, microbicidal peptides, or oxidative radicals. We are in the process of exploring the impact of various serum components on C. parapsilosis viability in an attempt to elucidate the component(s) of sera responsible for the enhanced killing of our C. parapsilosis mutants defective in de novo fatty acid synthesis.
Fungal Ole1 is composed of two domains (8). The N domain contains the catalytic motif for the desaturase activity, and the C domain contains the cytochrome b5 activity. In humans, the desaturase (SCD) and cytochrome b5 are two separate enzymes. The difference in enzyme structure could make fungal Ole1 an ideal antimicrobial target. Our current study provides supportive evidence for this rationale. The disruption of OLE1 or the inhibition of the protein interferes with C. parapsilosis growth, impairs filamentation, alters interactions with phagocytes, and attenuates virulence. Recently, genetic repression or chemical inhibition by small molecules of C. albicans OLE1 has been shown to impair C. albicans virulence even in mice receiving high-fat diets (28). Since interference with OLE1 also attenuated H. capsulatum virulence (14), Ole1 may be considered a potential broad-spectrum antifungal target.
Acknowledgments
J.D.N. is supported in part by an Irma T. Hirschl/Monique Weill-Caulier Trust Research Award. A.G. is supported by EMBO Installation Grant 1813, by OTKA PD73250, and by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
Editor: G. S. Deepe, Jr.
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Bandyopadhyay, S., R. Zhan, Y. Wang, S. K. Pai, S. Hirota, S. Hosobe, Y. Takano, K. Saito, E. Furuta, M. Iiizumi, S. Mohinta, M. Watabe, C. Chalfant, and K. Watabe. 2006. Mechanism of apoptosis induced by the inhibition of fatty acid synthase in breast cancer cells. Cancer Res. 66:5934-5940. [DOI] [PubMed] [Google Scholar]
- 2.Brinster, S., G. Lamberet, B. Staels, P. Trieu-Cuot, A. Gruss, and C. Poyart. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83-86. [DOI] [PubMed] [Google Scholar]
- 3.Chan, J., Y. Xing, R. S. Magliozzo, and B. R. Bloom. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 175:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chellappa, R., P. Kandasamy, C. S. Oh, Y. Jiang, M. Vemula, and C. E. Martin. 2001. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 276:43548-43556. [DOI] [PubMed] [Google Scholar]
- 5.Gácser, A., F. Stehr, C. Kroger, L. Kredics, W. Schafer, and J. D. Nosanchuk. 2007. Lipase 8 affects the Pathogenesis of Candida albicans. Infect. Immun. 75:4710-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gácser, A., D. Trofa, W. Schafer, and J. D. Nosanchuk. 2007. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J. Clin. Investig. 117:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnamurthy, S., A. Plaine, J. Albert, T. Prasad, R. Prasad, and J. F. Ernst. 2004. Dosage-dependent functions of fatty acid desaturase Ole1p in growth and morphogenesis of Candida albicans. Microbiology 150:1991-2003. [DOI] [PubMed] [Google Scholar]
- 8.Martin, C. E., C. S. Oh, and Y. Jiang. 2007. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771:271-285. [DOI] [PubMed] [Google Scholar]
- 9.Murayama, S. Y., Y. Negishi, T. Umeyama, A. Kaneko, T. Oura, M. Niimi, K. Ubukata, and S. Kajiwara. 2006. Construction and functional analysis of fatty acid desaturase gene disruptants in Candida albicans. Microbiology 152:1551-1558. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen, L. N., D. Trofa, and J. D. Nosanchuk. 2009. Fatty acid synthase impacts the pathobiology of Candida parapsilosis in vitro and during mammalian infection. PLoS One 4:e8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen, J. 2009. Systems biology of lipid metabolism: from yeast to human. FEBS Lett. 583:3905-3913. [DOI] [PubMed] [Google Scholar]
- 12.Oh, C. S., and C. E. Martin. 2006. Candida albicans Spt23p controls the expression of the Ole1p Delta9 fatty acid desaturase and regulates unsaturated fatty acid biosynthesis. J. Biol. Chem. 281:7030-7039. [DOI] [PubMed] [Google Scholar]
- 13.Pizer, E. S., J. Thupari, W. F. Han, M. L. Pinn, F. J. Chrest, G. L. Frehywot, C. A. Townsend, and F. P. Kuhajda. 2000. Malonyl-coenzyme-A is a potential mediator of cytotoxicity induced by fatty-acid synthase inhibition in human breast cancer cells and xenografts. Cancer Res. 60:213-218. [PubMed] [Google Scholar]
- 14.Porta, A., A. Eletto, Z. Torok, S. Franceschelli, A. Glatz, L. Vigh, and B. Maresca. 2010. Changes in membrane fluid state and heat shock response cause attenuation of virulence. J. Bacteriol. 192:1999-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 16.Schneiter, R., and G. Daum. 2006. Extraction of yeast lipids. Methods Mol. Biol. 313:41-45. [DOI] [PubMed] [Google Scholar]
- 17.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Delta is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneiter, R., V. Tatzer, G. Gogg, E. Leitner, and S. D. Kohlwein. 2000. Elo1p-dependent carboxy-terminal elongation of C14:1Delta(9) to C16:1Delta(11) fatty acids in Saccharomyces cerevisiae. J. Bacteriol. 182:3655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweizer, E., and J. Hofmann. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68:501-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart, L. C., and M. P. Yaffe. 1991. A role for unsaturated fatty acids in mitochondrial movement and inheritance. J. Cell Biol. 115:1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stukey, J. E., V. M. McDonough, and C. E. Martin. 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264:16537-16544. [PubMed] [Google Scholar]
- 22.Tehlivets, O., K. Scheuringer, and S. D. Kohlwein. 2007. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 1771:255-270. [DOI] [PubMed] [Google Scholar]
- 23.Thupari, J. N., M. L. Pinn, and F. P. Kuhajda. 2001. Fatty acid synthase inhibition in human breast cancer cells leads to malonyl-CoA-induced inhibition of fatty acid oxidation and cytotoxicity. Biochem. Biophys. Res. Commun. 285:217-223. [DOI] [PubMed] [Google Scholar]
- 24.Trofa, D., A. Gacser, and J. D. Nosanchuk. 2008. Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 21:606-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turkish, A., and S. L. Sturley. 2007. Regulation of triglyceride metabolism. I. Eukaryotic neutral lipid synthesis: “many ways to skin ACAT or a DGAT.” Am. J. Physiol. Gastrointest. Liver Physiol. 292:G953-G957. [DOI] [PubMed] [Google Scholar]
- 26.Ukeda, H., S. Maeda, T. Ishii, and M. Sawamura. 1997. Spectrophotometric assay for superoxide dismutase based on tetrazolium salt 3′-1-(phenylamino)-carbonyl-3, 4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate reduction by xanthine-xanthine oxidase. Anal. Biochem. 251:206-209. [DOI] [PubMed] [Google Scholar]
- 27.van Asbeck, E. C., K. V. Clemons, and D. A. Stevens. 2009. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit. Rev. Microbiol. 35:283-309. [DOI] [PubMed] [Google Scholar]
- 28.Xu, D., S. Sillaots, J. Davison, W. Hu, B. Jiang, S. Kauffman, N. Martel, P. Ocampo, C. Oh, S. Trosok, K. Veillette, H. Wang, M. Yang, L. Zhang, J. Becker, C. E. Martin, and T. Roemer. 2009. Chemical genetic profiling and characterization of small-molecule compounds that affect the biosynthesis of unsaturated fatty acids in Candida albicans. J. Biol. Chem. 284:19754-19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, X. J., G. E. McElhaney-Feser, W. H. Bowen, M. F. Cole, S. E. Broedel, Jr., and R. L. Cihlar. 1996. Requirement for the Candida albicans FAS2 gene for infection in a rat model of oropharyngeal candidiasis. Microbiology 142:2509-2514. [DOI] [PubMed] [Google Scholar]