Abstract
The Escherichia coli dnaX36 mutant displays a mutator effect, reflecting a fidelity function of the dnaX-encoded τ subunit of the DNA polymerase III (Pol III) holoenzyme. We have shown that this fidelity function (i) applies to both leading- and lagging-strand synthesis, (ii) is independent of Pol IV, and (iii) is limited by Pol II.
The mechanisms by which organisms achieve a high accuracy of DNA replication are of ongoing interest. Replication of the chromosome of the bacterium Escherichia coli is performed by the DNA polymerase III (Pol III) holoenzyme (HE). HE is composed of 17 subunits (10 distinct), with an overall composition (αɛθ)2β4τ2γδδ′χψ (27). It contains two (αɛθ) polymerase core assemblies, one for the leading strand and one for the lagging strand. The α subunit is the DNA polymerase, ɛ is the 3′ → 5′ proofreading exonuclease, and θ is an ɛ-stabilizing factor (2, 17, 34). Within HE, a central role is played by the τ subunit (τ2), which has several important functions, including connecting the two polymerases, enabling coupled leading- and lagging-strand synthesis. For the fidelity of replication, most of the focus has been on the DNA polymerase III core, notably the polymerase and the proofreading subunit (26, 28, 29). However, it is clear that overall chromosomal fidelity is not simply a function of polymerase fidelity but also involves activities of other HE subunits—as evidenced by mutator effects associated with defects in such subunits (24, 30, 32)—and the participation of accessory DNA polymerases (1, 8, 9, 12-14, 23, 35), of which E. coli has four (Pol I, II, IV, and V).
The present study is concerned with the fidelity role of the central τ subunit of HE, encoded by the dnaX gene, which also encodes the γ subunit (6). τ is the full-length product of the gene (643 amino acids), while the γ subunit is an early termination product (residues 1 to 430). Since τ and γ share the N-terminal protein sequence (domains I, II, and III), they share certain functions, such as the loading and unloading of the β-processivity clamps (27). However, the two additional domains (IV and V) that are present in τ permit it to perform certain unique functions. Specifically, domain IV contains the site of interaction with the DnaB helicase (10), which positively regulates the speed of the replication fork (4). Domain V contains the τ-α interaction site that enables HE to be dimeric (11). Domain V also controls the cycling of the lagging-strand polymerase, as it mediates the release of the Pol III core from its β processivity clamp upon completion of Okazaki fragments (20, 21). Thus, τ is an important control element within HE that can influence polymerase behavior, and this may extend to HE fidelity.
A fidelity role for the τ subunit was proposed based on observations of a distinct mutator activity for certain dnaX mutants (30). In particular, dnaX36 was informative since its defect (E601K) resides in domain V and hence only affects τ. Presumably, in the dnaX36 mutant, the τ-α interaction is altered, leading to the mutator effect. An additional series of dnaX mutators was also isolated, each carrying an amino acid substitution in domain V critical for interaction with the α subunit (15, 30, 33). Interestingly, they all share a unique mutational specificity: enhanced transversion base substitutions and (−1) frameshifts (in nonrun sequences) (30). To explain these observations, we suggested that the role of τ in fidelity is indirect. In this model, τ does not affect the intrinsic accuracy (insertion fidelity) of the α subunit but instead is involved in facilitating the subsequent (presumably error-free) processing of terminal mismatches created by Pol III.
dnaX36 mutator effects during chromosomal replication.
Our previous studies on the dnaX36 mutator effect were performed with lac mutational targets located on an F′ episome (12). While F′ replication is performed by Pol III HE, this type of replication may differ in important aspects from chromosomal replication. Presently, we have analyzed the effect of dnaX36 specifically on chromosomal DNA, using a system that also allows assessment of differential leading- and lagging-strand effects. We used a set of four different lacZ missense alleles that permit reversion to lac+ by a defined base-pair substitution (G·C → A·T, G·C → T·A, A·T → T·A, or A·T → G·C, respectively) (3). For each lac allele, two strains that differ only in the orientation of the lac operon are used. Comparison of the lac mutant frequencies for the two orientations allows assessment of differential leading- and lagging-strand effects (5). The experiments are performed with strains defective in mutHLS postreplicative mismatch repair, facilitating interpretation of mutation rates in terms of replication error rates (1, 5, 18, 22, 23).
Table 1 shows the results of two independent experiments. “L” and “R” indicate the strand in which the underlying mutational event is assumed to take place (see footnote a) (5). The dnaX36 mutator effect is 1.6- to 4.0-fold for the lac G·C → A·T transition depending on the orientation and experiment, 10- to 35-fold for the G·C → A·T transversion, 1.5- to 4.7-fold for the A·T → T·A transversion, and 1.2- to 4.3-fold for the A·T → G·C transition. Overall, we conclude that the dnaX36 mutator effect is readily observable on the E. coli chromosome and that both strands are affected, although not to the same extent.
TABLE 1.
Mutability of dnaX36 strains as function of chromosomal lac orientationa
| lac reversion | Expt | lac orientation (strand) | No. of lac+ mutants per 108 cells |
Mutator effect (fold) | |
|---|---|---|---|---|---|
| dnaX+ | dnaX36 | ||||
| G·C → A·T | 1 | L (leading) | 68 | 106 | 1.6 |
| R (lagging) | 20 | 50 | 2.5 | ||
| 2 | L (leading) | 73 | 215 | 2.9 | |
| R (lagging) | 19 | 76 | 4.0 | ||
| G·C → T·A | 1 | L (leading) | 1.1 | 11 | 10 |
| R (lagging) | 0.6 | 16 | 27 | ||
| 2 | L (leading) | 0.7 | 8.0 | 11 | |
| R (lagging) | 0.4 | 14 | 35 | ||
| A·T → T·A | 1 | L (leading) | 1.1 | 2.2 | 2.0 |
| R (lagging) | 0.7 | 3.3 | 4.7 | ||
| 2 | L (leading) | 1.3 | 2.0 | 1.5 | |
| R (lagging) | 0.7 | 2.7 | 3.9 | ||
| A·T → G·C | 1 | L (lagging) | 3.0 | 13 | 4.3 |
| R (leading) | 25 | 29 | 1.2 | ||
| 2 | L (lagging) | 5.6 | 14 | 2.5 | |
| R (leading) | 20 | 25 | 1.3 | ||
Each mutant frequency entry is based on 30 independent cultures comprising three independent lacZ chromosomal integrants for each orientation. The strains are the lac-containing derivatives of MC4100 described previously (5), into which the dnaX36 allele was introduced by P1 transduction using linkage with the zba-2321::mini-Tn10cam transposon (12). All strains are also mismatch repair deficient (mutL). The assignment of leading- or lagging-strand events is based on the lac orientation (L or R) and the assumed mispairings that underlie each of the indicated base-pair substitutions, i.e., G·T, T·C, T·T, and T·G for the G·C → A·T, G·C → T·A, A·T → T·A, and A·T → G·C substitutions, respectively, as previously described (5).
Consistent with previous observations (1, 5, 18, 22, 23), the mutant frequencies for each of the lac alleles in control (dnaX+) strains are consistently higher for leading-strand replication than for lagging-strand replication. This persistent bias is the basis for our contention that on the E. coli chromosome, lagging-strand replication is more accurate. The mechanism underlying the higher fidelity for lagging-strand replication is not known but may be related to the more efficient editing of polymerase errors in this strand (1, 5).
For the dnaX36 strain, it is clear that in each case the mutator effect is stronger for the lagging-strand events. For example, for the lac G·C → T·A allele, the mutator effect is 10- or 11-fold on the leading strand and 27- or 35-fold on the lagging strand. In fact, for this allele, the strand bias is inverted: the lagging strand now mutates at a higher level than the leading strand. Likewise, an inversion of the strand bias is seen for the lac A·T → T·A allele. For the two transitions, G·C → A·T and A·T → G·C, no inversion is seen, but the difference between the two strands is significantly diminished. Two conclusions are drawn from these experiments. First, in the absence of proper τ function, fidelity suffers in both strands, indicating τ promotes high fidelity in both strands. Second, the fidelity role of τ is quantitatively more important in the lagging strand than in the leading strand.
Role of accessory DNA polymerases.
Recent studies have shown that accessory DNA polymerases may also participate, at least occasionally, in chromosomal DNA synthesis. Obviously, DNA polymerase I plays an important role in lagging-strand replication, clearing and filling the Okazaki fragment gaps. Other DNA polymerases (Pol II, IV, and V) may occasionally gain access to the replication point and displace/replace Pol III, most obviously when the progress of Pol III HE is somehow blocked, either at DNA damage sites or, as we have proposed, at persistent terminal mismatches (1, 12, 18, 23). In the experiments shown in Fig. 1, we investigated the roles of Pol II and Pol IV. The role of Pol IV is probed by deleting the dinB gene, which encodes Pol IV, while the role of Pol II is investigated by using either a deletion allele, ΔpolB, or a proofreading (exonuclease) defective allele, polBex1 (7). The polBex1 allele (D155A, E157A) (7) is particularly useful because this allele may convert a normally error-free contribution into an error-prone contribution that may be revealed by an increase in the mutation rate (1, 7, 12, 31).
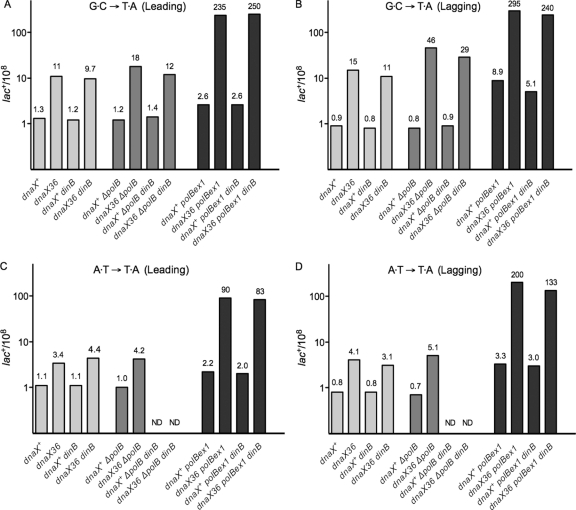
FIG. 1.
Effect of Pol IV (ΔdinB) and Pol II (ΔpolB and polBex1) on dnaX36 mutator activity. All strains are also mismatch repair deficient (mutL). Mutant frequencies were determined as described previously (1, 12). The strains used are lac-containing derivatives of MC4100 (5) into which the indicated dnaX, dinB, and polB alleles were introduced by P1 transduction as described previously (1, 5, 12). Each entry is based on at least 30 independent cultures. Average mutant frequencies with standard errors (SE) were determined using the statistical software program Prism (GraphPad). (A) Mutant frequencies for lac G·C → T·A transversions in the L (leading-strand) orientation. The mutant frequencies ± SE values were as follows: dnaX+, 1.3 ± 0.3; dnaX36, 11 ± 0.5; dnaX+ dinB, 1.2 ± 0.1; dnaX36 dinB, 9.7 ± 1; dnaX+ ΔpolB, 1.2 ± 0.2; dnaX36 ΔpolB, 18 ± 3; dnaX+ ΔpolB dinB, 1.4 ± 0.2; dnaX36 ΔpolB dinB, 12 ± 3; dnaX+ polBex1, 2.6 ± 0.3; dnaX36 polBex1, 235 ± 19; dnaX+ polBex1 dinB, 2.6 ± 0.4; dnaX36 polBex1 dinB, 250 ± 9. (B) Mutant frequencies for lac G·C → T·A transversions in the R (lagging-strand) orientation. The mutant frequencies ± SE values were as follows: dnaX+, 0.9 ± 0.2; dnaX36, 15 ± 1.2; dnaX+ dinB, 0.8 ± 0.2; dnaX36 dinB, 11 ± 0.7; dnaX+ ΔpolB, 0.8 ± 0.1; dnaX36 ΔpolB, 46 ± 6; dnaX+ ΔpolB dinB, 0.9 ± 0.2; dnaX36 ΔpolB dinB, 29 ± 5; dnaX+ polBex1, 8.9 ± 0.3; dnaX36 polBex1, 295 ± 30; dnaX+ polBex1 dinB, 5.1 ± 0.4; dnaX36 polBex1 dinB, 240 ± 13. (C) Mutant frequencies for lac A·T → T·A transversions in the L (leading-strand) orientation. The mutant frequencies ± SE values were as follows: dnaX+, 1.1 ± 0.3; dnaX36, 3.4 ± 0.6; dnaX+ dinB, 1.1 ± 0.2; dnaX36 dinB, 4.4 ± 0.5; dnaX+ ΔpolB, 1.0 ± 0.2; dnaX36 ΔpolB, 4.2 ± 0.3; dnaX+ polBex1, 2.2 ± 0.3; dnaX36 polBex1, 90 ± 7; dnaX+ polBex1 dinB, 2.0 ± 0.6; dnaX36 polBex1 dinB, 83 ± 5. ND, not done. (D) Mutant frequencies for lac A·T → T·A transversions in the R (lagging-strand) orientation. The mutant frequencies ± SE values were as follows: dnaX+, 0.8 ± 0.2; dnaX36, 4.1 ± 0.4; dnaX+ dinB, 0.8 ± 0.2; dnaX36 dinB, 3.1 ± 0.4; dnaX+ ΔpolB, 0.7 ± 0.2; dnaX36 ΔpolB, 5.1 ± 0.4; dnaX+ polBex1, 3.3 ± 0.3; dnaX36 polBex1, 200 ± 11; dnaX+ polBex1 dinB, 3.0 ± 0.4; dnaX36 polBex1 dinB, 133 ± 9. ND, not done.
Pol IV is normally present in wild-type cells at the relatively high concentration of ∼250 molecules per cell (16). Nevertheless, studies have indicated that under those conditions, Pol IV does not affect the chromosomal error rate (19, 36). Our present results (Fig. 1) confirm this (compare dnaX+ ΔdinB results to those for dnaX+). In the dnaX36 background, the loss of Pol IV (dinB) has only some very small effects. No significant effect is seen for the G·C → T·A allele in the leading-strand (L) orientation or for both orientations of the A·T → T·A allele. An approximately 30% reduction is observed for the G·C → T·A transversion in the lagging-strand orientation (11 × 10−8 versus 15 × 10−8) (Fig. 1). This reduction, while modest, has been observed repeatedly in several experiments (data not shown) and thus likely represents a real effect. We conclude that the dnaX36 mutator effect is largely independent of the error-prone involvement of Pol IV. These results differ from those obtained with the F′ system, where a large fraction of dnaX36-mediated mutations proved Pol IV dependent (12). Presumably this reflects the fact that F′ pro lac-containing strains contain an extra copy of the dinB gene and possess some 4-fold-elevated levels of Pol IV (16).
A lack of Pol II (ΔpolB) does not affect mutagenesis in wild-type (dnaX+) cells (Fig. 1), consistent with previous reports (1), but it modestly increases the dnaX36 mutator effect for both lac alleles. The effect is most pronounced for the G·C → T·A allele: about 1.7-fold for the leading-strand orientation (18 × 10−8 for dnaX36 versus 11 × 10−8 for dnaX36 ΔpolB) and 3-fold for the lagging-strand orientation (14 × 10−8 versus 46 × 10−8) (Fig. 1). These results indicate that Pol II plays a role in preventing mutations in dnaX36 strains and that this role may be more important in lagging-strand replication. Interestingly, an additional loss of Pol IV (dnaX36 ΔpolB ΔdinB strain) leads to a reduction of the mutant frequency, although the frequency does not return to the level for the single dnaX36 strain. These results indicate that Pol II and Pol IV can compete and that, at least in the absence of Pol II, the dnaX36 mutator effect has both Pol IV-dependent and Pol IV-independent components.
The role of Pol II was further probed by using the polBex1 allele. Consistent with findings of previous studies (1), the polBex1 allele is mutagenic in a wild-type dnaX+ background. This has been interpreted to indicate that apparently Pol II, even under normal conditions, may have access to the replication fork, serving in an antimutagenic role, likely acting as a backup proofreader for Pol III (1). This editing role becomes increasingly important in the dnaX36 strain. As seen in Fig. 1, the dnaX36 polBex1 combination yields a very potent mutator activity, enhancing the effect some 20- to 22-fold for the G·C → T·A allele and 26- to 49-fold for the A·T → T·A allele (compare dnaX36 to dnaX36 polBex1). This strong mutator effect resulting from the Pol II proofreading deficiency is not dependent on the action of Pol IV, since the dnaX36 polBex1 ΔdinB triple mutant behaves essentially as does the dnaX36 polBex1 double. Thus, an important fidelity role of Pol II at the chromosomal replication fork of dnaX36 is revealed.
The τ fidelity mechanism.
We have suggested that the dnaX36 mutator effect is most productively interpreted in terms of a defect in the processing of terminal mismatches (misinsertion errors) produced by the Pol III α-subunit (1, 12, 18, 22, 23). The current data on chromosomal DNA synthesis support this proposal. While most terminal mismatches created by the α subunit are expected to be removed by the ɛ proofreading subunit, a subset of errors may be refractory to this mechanism and require the action of the τ subunit for resolution. This mechanism operates on both leading and lagging strands. How τ promotes error removal is not yet known, but it may involve facilitating the required conformational change in α to place the mismatch in the exonuclease site. Alternatively, τ may channel the mismatch toward the exonuclease of Pol II or the exonuclease of a third Pol III core that has been proposed to be present at the replication fork (25). Obviously, in the absence of this τ function (as in the dnaX36 mutant), this mechanism may be inoperative. Pol III may be forced to eventually extend the terminal mismatch, accounting for the dnaX mutator effect. On the other hand, our data indicate that the major fate of the mismatch is editing by the exonuclease of Pol II. In fact, more than 90% of the potential mutations (those seen in the dnaX36 polBex1 mutant) appear to be edited away by Pol II. Thus, Pol II is the primary backup polymerase at the replication fork.
Acknowledgments
We thank L. Garcia-Villada and R. Kasiviswanathan of the NIEHS for their critical reading of the manuscript for this article and helpful comments.
This research was supported by project no. Z01 ES065086 of the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, and by grant 2 PO4A 061 30 (to I.J.F. and P.J.) from the Polish Ministry of Science and Higher Education.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Banach-Orlowska, M., I. J. Fijalkowska, R. M. Schaaper, and P. Jonczyk. 2005. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol. Microbiol. 58:61-70. [DOI] [PubMed] [Google Scholar]
- 2.Chikova, A. K., and R. M. Schaaper. 2005. The bacteriophage P1 hot gene product can substitute for the Escherichia coli DNA polymerase III θ subunit. J. Bacteriol. 187:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. U. S. A. 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallmann, H. G., S. Kim, A. E. Pritchard, K. J. Marians, and C. S. McHenry. 2000. Characterization of the unique C terminus of the Escherichia coli τ DnaX protein. Monomeric C-tau binds α and DnaB and can partially replace τ in reconstituted replication forks. J. Biol. Chem. 275:15512-15519. [DOI] [PubMed] [Google Scholar]
- 5.Fijalkowska, I. J., P. Jonczyk, M. M. Tkaczyk, M. Bialoskorska, and R. M. Schaaper. 1998. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 95:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flower, A. M., and C. S. McHenry. 1990. The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. U. S. A. 87:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster, P. L., G. Gudmundsson, J. M. Trimarchi, H. Cai, and M. F. Goodman. 1995. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:7951-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii, S., and R. P. Fuchs. 2007. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J. Mol. Biol. 372:883-893. [DOI] [PubMed] [Google Scholar]
- 9.Furukohri, A., M. F. Goodman, and H. Maki. 2008. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J. Biol. Chem. 283:11260-11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, D., and C. S. McHenry. 2001. τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of τ, binds the replication fork, helicase, DnaB. J. Biol. Chem. 276:4441-4446. [DOI] [PubMed] [Google Scholar]
- 11.Gao, D., and C. S. McHenry. 2001. τ binds and organizes Escherichia coli replication through distinct domains. Partial proteolysis of terminally tagged τ to determine candidate domains and to assign domain V as the α binding domain. J. Biol. Chem. 276:4433-4440. [DOI] [PubMed] [Google Scholar]
- 12.Gawel, D., P. T. Pham, I. J. Fijalkowska, P. Jonczyk, and R. M. Schaaper. 2008. Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant. J. Bacteriol. 190:1730-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heltzel, J. M., R. W. Maul, S. K. Scouten Ponticelli, and M. D. Sutton. 2009. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc. Natl. Acad. Sci. U. S. A. 106:12664-12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Indiani, C., L. D. Langston, O. Yurieva, M. F. Goodman, and M. O'Donnell. 2009. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. U. S. A. 106:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jergic, S., K. Ozawa, N. K. Williams, X. C. Su, D. D. Scott, S. M. Hamdan, J. A. Crowther, G. Otting, and N. E. Dixon. 2007. The unstructured C-terminus of the τ subunit of Escherichia coli DNA polymerase III holoenzyme is the site of interaction with the α subunit. Nucleic Acids Res. 35:2813-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, S. R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 17.Kirby, T. W., S. Harvey, E. F. DeRose, S. Chalov, A. K. Chikova, F. W. Perrino, R. M. Schaaper, R. E. London, and L. C. Pedersen. 2006. Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex. J. Biol. Chem. 281:38466-38471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuban, W., M. Banach-Orlowska, M. Bialoskorska, A. Lipowska, R. M. Schaaper, P. Jonczyk, and I. J. Fijalkowska. 2005. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J. Bacteriol. 187:6862-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuban, W., P. Jonczyk, D. Gawel, K. Malanowska, R. M. Schaaper, and I. J. Fijalkowska. 2004. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J. Bacteriol. 186:4802-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leu, F. P., R. Georgescu, and M. O'Donnell. 2003. Mechanism of the E. coli τ processivity switch during lagging-strand synthesis. Mol. Cell 11:315-327. [DOI] [PubMed] [Google Scholar]
- 21.Lopez de Saro, F. J., R. E. Georgescu, and M. O'Donnell. 2003. A peptide switch regulates DNA polymerase processivity. Proc. Natl. Acad. Sci. U. S. A. 100:14689-14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makiela-Dzbenska, K., M. Jaszczur, M. Banach-Orlowska, P. Jonczyk, R. M. Schaaper, and I. J. Fijalkowska. 2009. Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity. Mol. Microbiol. 74:1114-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maliszewska-Tkaczyk, M., P. Jonczyk, M. Bialoskorska, R. M. Schaaper, and I. J. Fijalkowska. 2000. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proc. Natl. Acad. Sci. U. S. A. 97:12678-12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maul, R. W., L. H. Sanders, J. B. Lim, R. Benitez, and M. D. Sutton. 2007. Role of Escherichia coli DNA polymerase I in conferring viability upon the dnaN159 mutant strain. J. Bacteriol. 189:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInerney, P., A. Johnson, F. Katz, and M. O'Donnell. 2007. Characterization of a triple DNA polymerase replisome. Mol. Cell 27:527-538. [DOI] [PubMed] [Google Scholar]
- 26.Mo, J. Y., and R. M. Schaaper. 1996. Fidelity and error specificity of the alpha catalytic subunit of Escherichia coli DNA polymerase III. J. Biol. Chem. 271:18947-18953. [DOI] [PubMed] [Google Scholar]
- 27.O'Donnell, M. 2006. Replisome architecture and dynamics in Escherichia coli. J. Biol. Chem. 281:10653-10656. [DOI] [PubMed] [Google Scholar]
- 28.Pham, P. T., M. W. Olson, C. S. McHenry, and R. M. Schaaper. 1999. Mismatch extension by Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 274:3705-3710. [DOI] [PubMed] [Google Scholar]
- 29.Pham, P. T., M. W. Olson, C. S. McHenry, and R. M. Schaaper. 1998. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J. Biol. Chem. 273:23575-23584. [DOI] [PubMed] [Google Scholar]
- 30.Pham, P. T., W. Zhao, and R. M. Schaaper. 2006. Mutator mutants of Escherichia coli carrying a defect in the DNA polymerase III τ subunit. Mol. Microbiol. 59:1149-1161. [DOI] [PubMed] [Google Scholar]
- 31.Rangarajan, S., G. Gudmundsson, Z. Qiu, P. L. Foster, and M. F. Goodman. 1997. Escherichia coli DNA polymerase II catalyzes chromosomal and episomal DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 94:946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saveson, C. J., and S. T. Lovett. 1997. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 146:457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su, X. C., S. Jergic, M. A. Keniry, N. E. Dixon, and G. Otting. 2007. Solution structure of domains IVa and V of the τ subunit of Escherichia coli DNA polymerase III and interaction with the α subunit. Nucleic Acids Res. 35:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taft-Benz, S. A., and R. M. Schaaper. 2004. The θ subunit of Escherichia coli DNA polymerase III: a role in stabilizing the ɛ proofreading subunit. J. Bacteriol. 186:2774-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchida, K., A. Furukohri, Y. Shinozaki, T. Mori, D. Ogawara, S. Kanaya, T. Nohmi, H. Maki, and M. Akiyama. 2008. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol. Microbiol. 70:608-622. [DOI] [PubMed] [Google Scholar]
- 36.Wolff, E., M. Kim, K. Hu, H. Yang, and J. H. Miller. 2004. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 186:2900-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]