Abstract
Tetherin, also known as BST-2/CD317/HM1.24, is an antiviral cellular protein that inhibits the release of HIV-1 particles from infected cells. HIV-1 viral protein U (Vpu) is a specific antagonist of human tetherin that might contribute to the high virulence of HIV-1. In this study, we show that three amino acid residues (I34, L37, and L41) in the transmembrane (TM) domain of human tetherin are critical for the interaction with Vpu by using a live cell-based assay. We also found that the conservation of an additional amino acid at position 45 and two residues downstream of position 22, which are absent from monkey tetherins, are required for the antagonism by Vpu. Moreover, computer-assisted structural modeling and mutagenesis studies suggest that an alignment of these four amino acid residues (I34, L37, L41, and T45) on the same helical face in the TM domain is crucial for the Vpu-mediated antagonism of human tetherin. These results contribute to the molecular understanding of human tetherin-specific antagonism by HIV-1 Vpu.
Intrinsic immune molecules have been discovered in mammalian cells that restrict retrovirus replication (64). Tetherin, also known as BST-2/CD317/HM1.24, is a host factor that recently has been identified as a potent inhibitor against the release of HIV-1 particles (46, 67). The retained particles then are internalized into the endosomes/lysosomes and presumably degraded (4). HIV-1 is able to elude the tetherin-driven defense mechanism by expressing viral protein U (Vpu), which is expressed in a unique lineage of primate lentiviruses (64). Vpu is an 81-amino-acid (aa) type 1 integral membrane protein encoded together with the env gene from a bicistronic mRNA, and it promotes the release of HIV-1 particles by specifically antagonizing tetherin (7, 63, 65). It has been shown that Vpu inhibits the cell surface expression of tetherin by its relocalization to the trans-Golgi network (TGN) and the recycling endosomes (12), and/or by targeting it for the proteosomal and/or lysosomal degradation in a beta-transducin repeat-containing protein (β-TrCP)-dependent manner (10, 41). β-TrCP is a component of an E3 ubiquitin ligase complex that also is involved in the Vpu-induced proteasomal degradation of CD4 via the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway (64, 65, 70). An amino acid alignment of representative primate lentiviral Vpus shows that they are highly variable and have functional differences (38, 57). Only Vpu from HIV-1 pandemic group M and nonpandemic group N strains, but not from HIV-1 group O and SIVcpz, can counteract human tetherin (hu-tetherin) activity (57). These studies suggested that the evolution of a fully functional anti-tetherin protein, such as the highly adapted Vpu of the group M strains, is an important prerequisite for the ongoing spread of HIV-1 in the human population. The anti-tetherin function of HIV-1 group M Vpu is highly specific to tetherin from human, chimpanzee, and gorilla (57). Vpu does not counteract tetherin homologues from different species such as mouse, rhesus macaques, and African green monkey (agm) (17, 43). Until now, research on the interplay between hu-tetherin and Vpu has been limited mainly to the amino acid residues within the TM that appear to be important for the interaction and susceptibility of tetherin to Vpu (10, 18, 43, 54). However, whether these reported amino acid residues also play a role in the Vpu association, and whether there are other determinants of the interaction, remain to be elucidated. Thus, we set out to understand the parameters of the hu-tetherin-Vpu interaction.
Using bimolecular fluorescence complementation (BiFC) (31) and site-directed mutagenesis, we first identified three amino acid residues (I34, L37, and L41) within the TM region of human tetherin (hu-tetherin) that are crucial for its interaction with Vpu in live cells. The alteration of any one of these amino acid residues leads to a significantly reduced interaction with Vpu and also a loss of Vpu response in the virus release assay. Furthermore, structural and functional roles of these amino acid were addressed with molecular dynamics (MD) simulations of the TM domain of hu- and agm-tetherins in a lipid bilayer environment. To our knowledge, this is the first report showing the amino acid residues in hu-tetherin essential for interaction with Vpu in live cells, and we found that the proper positioning of these residues is regulated by the presence of two upstream amino acid residues in the TM helix.
MATERIALS AND METHODS
Plasmids.
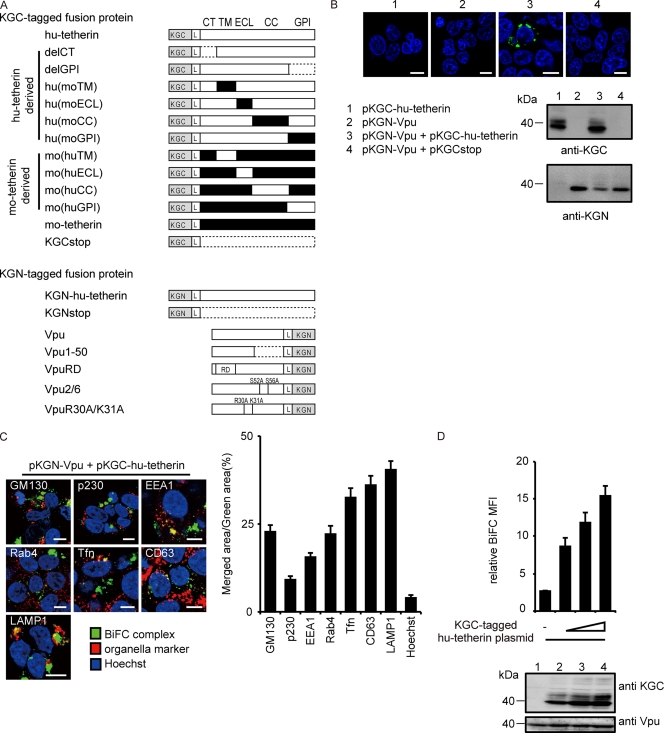
The human, mouse, and agm tetherin cDNA was amplified by reverse transcription-PCR (RT-PCR) from human peripheral blood lymphocyte (PBL), NIH 3T3, or COS-7 cells, respectively, and subcloned into Kusabira green (KG) fragment-encoding plasmids phmKGC-MC and phmKGN-MC (MBL). From pcDNA-Vphu, expressing a codon-optimized and Rev-independent HIV-1NL4-3 Vpu protein (47), an orf fragment of Vphu also was inserted into phmKGN-MN. A KGN-hu-tetherin fragment and KGN-Vphu were subcloned into CSII-CDF-GATEWAY-IRES-H2Kk (30). The mutated hu-, mo-, and agm-tetherin DNA were created by overlap extension PCR. All constructs were confirmed by DNA sequencing. A list of fusion proteins of tetherin and Vpu is summarized in Fig. 1A.
FIG. 1.
Interaction of tetherin and Vpu via TM domain. (A) Schematic diagram of KGN or KGC fusion protein used in this study. KGN- or KGC-peptide tag was fused in frame to the N terminus of hu-tetherin, mo-tetherin, or mutant proteins. KGN-tag was fused in frame to the C terminus of Vpu or its mutant proteins. The L indicates a linker sequence inserted between KG fragments and proteins of interest. Human, white; mouse, black; the broken line indicates deletion residues. (B) Tetherin-Vpu complex detected by BiFC. HEK 293 cells were transfected with the indicated DNA and examined by confocal microscopy (upper) and immunoblot analysis (lower). (C) BiFC-expressing cells were stained with markers specific for each organelle and imaged by confocal microscopy. Bar, 10 μm. The ratios of the merged area between BiFC (green) and organelles (red) were quantified (right columns) for more than 100 cells. (D) Quantitative BiFC assay. The KGN-Vpu-expressing cells were transfected with pKGC-hu-tetherin (0, 100, 200, or 400 ng) or a control [pKGCstop, 400 ng] and analyzed by flow cytometry (upper) and immunoblot analysis (lower). Relative MFI values are defined as the MFI of tetherin plasmid-transfected cells minus the MFI of untransfected cells, and results represent the means from three independent experiments plus standard deviations.
Cell culture, transfection, and transduction.
HEK 293 cells or its derivatives were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. Cells were transfected by calcium phosphate DNA precipitation or Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Stably KGN-Vpu or KGC-hu-tetherin-expressing HEK 293 was generated by a lentiviral vector as described before (30). The amount of plasmid DNA for transfection was normalized to 1.4 μg per well by pcDNA3.1. Recombinant human alpha interferon (IFN-α) was purchased from Prospec.
Confocal microscopy.
HEK 293 cells were transfected by an individual or selected pairs of BiFC constructs (0.5 μg of each plasmid). At 24 h posttransfection, cells were stained with Hoechst 33342 (Invitrogen) and fixed with 2% paraformaldehyde, followed by treatment with 0.05% saponin. Cells were sequentially incubated with monoclonal antibodies (MAbs) against GM130, p230, EEA1, Rab4, CD63, or LAMP1 (BD Transduction), followed by incubation with a goat anti-mouse IgG MAb (Molecular probes). The recycling endosomes were visualized as previously reported (12). Cells were analyzed as described before (71). All of the images were taken under similar experimental conditions (i.e., exposure time, magnification, and intensification), and image processing was the same for all of the images shown in the figure.
Immunoblot analysis.
At 24 h posttransfection, cells were lysed and the lysates were processed as described before (56). The membrane was incubated with anti-KGN, anti KGC MAb (MBL), anti-Flag MAb (Sigma), anti-Vpu, or anti-p24 antibody (40, 56), and then horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Santa Cruz Biotechnology). The immune complex was visualized using an enhanced chemiluminescence system (LAS 4000) according to the instructions provided by the manufacturer.
Flow cytometry.
HEK 293 cells were transfected either individually or in selected pairs of BiFC construct (0.5 μg of each plasmid) 24 h posttransfection, harvested, and analyzed as described previously (71).
Tetherin activity and Vpu sensitivity assays.
HEK 293 cells were transfected using Lipofectamine 2000 (Invitrogen) with 1 μg of pNL4-3 (wild type [WT]) or pNL4-3/Udel (Vpu deletion mutant) HIV-1 infectious plasmids (34), KGC-tagged tetherin, and its derivative plasmids. Cells and supernatants were harvested 24 h after transfection. The supernatants were filtered and infectious virion yields were measured using TZM-bl indicator cells, as described before (56), and given in relative light units (RLU).
Statistical analysis.
The Mann-Whitney's U test and Student's t test were used to determine statistical significance.
Construction of initial model for MD simulation of helix model of the tetherin TM domain in a lipid bilayer environment.
To perform MD simulations (29), we first prepared seven initial models roughly mimicking helical structures of the tetherin TM domains (hu-tetherin, agm-tetherin, agm-LL, agm-LL/I45T, hu-T45I, hu-delGI, and hu-delGI/T45I) under a lipid bilayer environment (28, 61). First, we constructed a helix peptide (D15 to C53 in hu-tetherin) in the range from the six amino acids upstream of the TM domain (K21 to K47 in hu-tetherin) to its six amino acids downstream in each tetherin, using the LEaP module of AMBER9 (35, 51). In addition, acetyl- and N-methyl groups were connected to the N- and C-terminal ends of the peptide, respectively. We also prepared a bilayer membrane structure, including about 120 palmitoyl-oleyl-sn-phosphatidyl-cholines (POPC), using VMD 1.8.7 (22). POPC is the largest component in the cellular membrane of the mammalian cell (5, 68). Finally, the helical peptide was perpendicularly inserted into the membrane, and about 10,000 water molecules were placed on both sides of the membrane surface using the LEaP module. In this study, we applied the TIP3P water model (27) as the water molecule. The AMBER ff99SB force field (20) was applied to energy and force calculations for peptide and water molecules, and the gaff (69) was used for POPC (26). The atom types and charges of each atom in POPC were automatically assigned using the Antechamber module of AMBER9 (35, 51).
MD simulations of the tetherin TM domain in a lipid bilayer environment.
Prior to MD simulations of each of seven helix models, we performed the energy minimization of the initial model and heated it until 36.85°C (310 K). The energy minimization was achieved by 1,000 steps of the steepest-descent method and by the subsequent 1,000 steps of the conjugated gradient method to release steric clashes. The minimized model subsequently was heated using Langevin dynamics (39, 50) with a collision frequency of 5.0 ps−1 (10−12 s) with the NVT ensemble for 0.1 ns (10−9 s). Preliminary MD simulation then was performed at 36.85°C under 1 atm pressure with the NPT ensemble for 0.5 s. Temperature was controlled using Langevin dynamics (39, 50) with a collision frequency of 1.0 ps−1. Until this step, to achieve equilibration for the membrane prior to that for peptide, the helical structure of the peptide was constrained. The distance of the hydrogen bond pair between main chains of the helical peptide was forced to be less than 3.0 Å with a weak constraint by a harmonic potential of 2.5 kcal/mol/Å2. We further performed 4.5 ns of MD simulations without constraints of hydrogen bonds in the peptides.
Throughout the simulations, we also applied the following conditions. In the models there are many highly charged atoms, like phosphate groups in lipids. The highly charged nature of phosphate groups tends to prevent simulations from leading to stable structures having a nanosecond lifetime (16, 23, 44, 48). To avoid the unexpected effects, the additional weak forces were applied between hetero-atoms (N, O, and S atoms) in the peptide side chain of extracellular loop (ECL) (D15 to K21 or K47 to E53 in hu-tetherin) and phosphorus atoms in lipids to weaken the electrostatic interactions of highly charged phosphate atoms, using a harmonic potential of 2.5 kcal/mol/Å2 with an equilibration distance of 7.0 Å when the distance between them was less than 7.0 Å. In addition, the periodic boundary condition was applied to avoid edge effects on the boundary face. The particle mesh Ewald (PME) method (8, 9, 15) is used for calculating the electrostatic energy of a periodic box. The cutoff distance of the nonbonded energy term was set to 12.0 Å. The movement of bonds involving hydrogen atoms was constrained by the SHAKE algorithm (55). The time step was set to 2.0 fs (10−15 s). The simulations were achieved with the PMEMD module of AMBER9 (35, 51).
Lastly, we evaluated the equilibration of the model. After 2.0 ns of MD simulation, each membrane thickness and root mean squared distance (RMSD) from a heated structure got close to a certain value (see Fig. S3 in the supplemental material). Furthermore, to validate the equilibration of peptide conformation at the C-terminal half of the TM domain (G27 to E51 in hu-tetherin), 3,000 snapshots were derived from 2.0 to 5.0 ns of MD simulation and were grouped into five clusters by the Bayesian algorithm (60) using the AMBER tool 1.2 in each model. Since the representative structure in the largest cluster is similar to the representative structures in the other four clusters (RMSD < 2.0 Å), we considered that conformations of the C-terminal domain were well equilibrated (see Table S1 in the supplemental material). In this study, among the 3,000 snapshots in each model, we selected a representative structure using the Bayesian clustering algorithm (60) to analyze its structural feature.
Comparison of 3D models of the tetherin TM domains.
We compared three-dimensional (3D) structures of the hu-tetherin TM models to those of other TM variants by following the procedure of PyMOL version 0.99 rc6 (http://www.pymol.org/) (Schrödinger LLC). We first fitted models to the hu-tetherin model using the coordinates of main-chain atoms (N, Cα, and C) around C-terminal TM domains (G27 to E51 in hu-tetherin), because agm-tetherin, hu-delGI, and hu-delGI/T45I have two amino acid deletions in the N-terminal TM domains. The RMSD values for individual amino acid residues were calculated using the coordinates of the main-chain atoms in the fitted models.
RESULTS
Visualization of tetherin-Vpu heterodimer complexes in live cells.
To detect tetherin-Vpu complexes in live cells, we used the BiFC technique (32) by the ectopic expression of two fragmented monomeric KG fluorescent proteins. This technique is based on the formation of a fluorescent complex from N- and C-terminal nonfluorescent fragments, KGN and KGC, which are brought together by association through interacting partner proteins fused to the fragments, thus allowing the specific visualization of interactive complexes in live cells (66). Preceding reports have indicated that a peptide tagged to the N terminus of human tetherin (hu-tetherin) or the C terminus of Vpu does not interfere with their function and localization. Therefore, we constructed the plasmid encoding KGC fused to the N-terminal end of hu-tetherin (KGC-hu-tetherin), designated pKGC-hu-tetherin. The plasmid encoding KGN was fused to the C-terminal end of codon optimized-Vpu (KGN-Vpu), designated pKGN-Vpu. The level of HIV-1 release inhibition by pKGC-hu-tetherin (data not shown) and its cell surface expression levels (Table 1) were equivalent to that of untagged hu-tetherin. In addition, pKGN-Vpu and untagged Vpu-expressing plasmid displayed similar levels of anti-tetherin activity (data not shown). We detected specific and significant fluorescence in cells coexpressing KGC-hu-tetherin and KGN-Vpu but not in those expressing either KGC-hu-tetherin or KGN-Vpu alone or KGN-Vpu and KGCstop (Fig. 1B). Immunoblot analysis confirmed the expression and proper size of the individual proteins (KGC-hu-tetherin and KGN-Vpu) (Fig. 1A and B). KGCstop and KGNstop, which are mutants with a termination codon inserted at a site upstream of the tetherin open reading frame (ORF), were used as negative controls. In the fluorescence-positive cells, little signal was seen at the plasma membrane, perhaps due to the downregulation of tetherin expression from the cell surface. By staining with antibodies or reagents specific for individual organelles within the cell, we detected BiFC signals from tetherin-Vpu complexes within organelles in which tetherin localization has been reported previously (42); however, distribution frequencies were different (Fig. 1C and data not shown). In fact, the previous study showed that little hu-tetherin was seen in vesicular compartments positive for the late endosome marker (LAMP-2), while in our study the complexes appear to localize predominantly in CD63+ and LAMP-1+ late endosomes (Fig. 1C, right columns) (42). These results indicate that the BiFC assay is a useful and sensitive approach to determine Vpu-tetherin interactions.
TABLE 1.
Surface expression of tetherin and its derivatives used in this studya
| Name of N-terminally KGC-tagged fusion protein | MFI of tetherin |
|---|---|
| Untagged hu-tetherin | 107.6 ± 12.7 |
| hu-tetherin | 117.6 ± 16.2 |
| KGCstop | 13.1 ± 6.8 |
| mo-tetherin | ND |
| mo(huTM) | ND |
| mo(huECL) | ND |
| mo(huCC) | 123.3 ± 6.3 |
| mo(huGPI) | ND |
| hu(muTM) | 115.3 ± 4.7 |
| hu(muECL) | 138.4 ± 15.0 |
| hu(muCC) | ND |
| hu(muGPI) | 127.9 ± 9.8 |
| LLL(22-24)AAA | 116.3 ± 14.6 |
| GIG(25-27)AAA | 138.6 ± 27.2 |
| ILV(28-30)AAA | 159.4 ± 17.7 |
| LLI(31-33AAA | 160.2 ± 19.0 |
| IVI(34-36)AAA | 142.1 ± 17.9 |
| LGV(37-39)AAA | 161.0 ± 14.1 |
| PLI(40-42)AAA | 113.9 ± 12.3 |
| IFT(43-45)AAA | 149.8 ± 8.5 |
| IKA(46-48)AAA | 117.9 ± 2.0 |
| I34A | 155.9 ± 5.9 |
| V35A | 135.1 ± 34.5 |
| I36A | 157.1 ± 18.8 |
| L37A | 141.6 ± 26.8 |
| G38A | 169.1 ± 23.6 |
| V39A | 164.5 ± 14.5 |
| P40A | 165.9 ± 4.1 |
| L41A | 140.1 ± 9.8 |
| I42A | 143.6 ± 29.1 |
Values indicate the means from three independent experiments plus standard deviations. ND, not detected
Quantitative BiFC assay for tetherin and Vpu interaction.
To quantitatively measure tetherin-Vpu heterodimer complexes, we generated a HEK 293 cell line stably expressing KGN-Vpu (Fig. 1D, bottom). The mean fluorescence intensity (MFI) of the reconstituted KG fluorophore, measured by flow cytometry, is directly proportional to the number of interactive complexes in the cells (32). To examine the sensitivity of the assay with respect to hu-tetherin and Vpu complexes, the KGN-Vpu cells were transfected with various amounts of KGC-hu-tetherin plasmid DNA (Fig. 1D). The intensity of the KG BiFC signal corresponded well with the level of KGC-tetherin expression detected by an anti-KGC antibody 24 h after transfection (Fig. 1D, lanes 2 to 4, compare to the corresponding upper columns). These results demonstrate that our tetherin-Vpu BiFC assay provides sufficient sensitivity for measuring hu-tetherin-Vpu complexes.
Interaction of the TM region of hu-tetherin with Vpu.
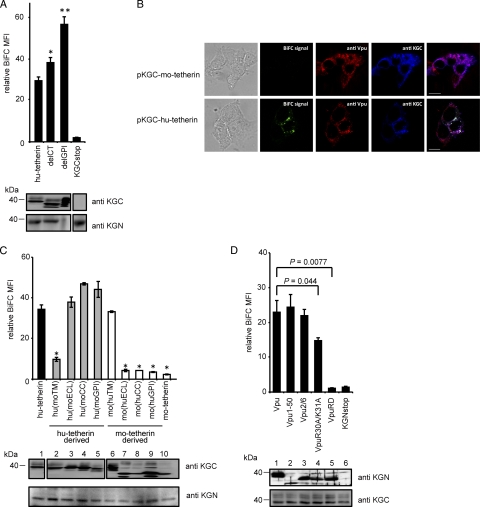
Tetherin has a unique topology: the amino-terminal cytoplasmic tail (CT) is followed by a single TM region, extracellular loop (ECL), coiled-coil domain (CC), and glycosyl phosphatidylinositol membrane anchor (GPI) at the C terminus (36). To map the domain of hu-tetherin that mediates interaction with Vpu, we first prepared KGC-tagged tetherin mutants deleted in the CT or GPI anchor (delCT and delGPI) (Fig. 1A), transfected them into the KGN-Vpu-expressing cells, and examined the resulting BiFC signals. We found that delCT and delGPI mutants produced slightly enhanced BiFC signals compared to those of WT tetherin (Fig. 2A), perhaps due to an increase in the expression level in cells (Fig. 2A, bottom) or a change in the protein localization. These data reveal that the CT and GPI regions in tetherin are not required for Vpu interaction. To investigate specific characteristics of the hu-tetherin-Vpu complex under similar topological conditions, we next used KGC-tagged mouse tetherin (KGC-mo-tetherin) (Fig. 1A). Since mo-tetherin displays resistance to Vpu counteraction (17), less interaction between Vpu and mo-tetherin was expected. In fact, the coexpression of KGN-Vpu and KGC-mo-tetherin gave no detectable fluorophore (Fig. 2B and C, lane 10), while the coexpression of KGN-tagged mo-tetherin and KGC-mo-tetherin yielded a signal (data not shown), indicating mo-tetherin homodimer formation can produce a strong fluorophore. To further verify BiFC signals, we examined the subcellular localization of Vpu and tetherin together with BiFC signals. The results showed that hu-tetherin colocalized with Vpu as well as BiFC signals (Fig. 2B, lower), whereas mo-tetherin colocalized with Vpu but did not produce BiFC florescence (Fig. 2B, upper). This indicated that BiFC signals are the results of specific interactions. These data suggest that specific region(s) of hu-tetherin participate in the interaction with Vpu. Thus, we next prepared a KGC-tagged hu- or mo-tetherin-based chimera with alterations of the TM, ECL, CC, and GPI from mo-tetherin or hu-tetherin (Fig. 1A). The subcellular localization and cell surface expression of the chimeric tetherins were detected by anti-KGC antibody or quantified using an anti-hu-tetherin antibody that reacts with the ECL domain. The anti-hu-tetherin antibody does not react with the mouse ECL domain; thus, we were only able to quantify the cell surface expression of chimera carrying the human ECL domain. The cell surface expression and localization of these chimera were similar to those of WT hu-tetherin (Table 1 and data not shown). Furthermore, we examined the total expression of the mutants by immunoblot analysis using an anti-KGC antibody. The expression of all chimeras was comparable (Fig. 2C). The mo-tetherin-based chimeric proteins migrate as multiple bands on immunoblot analysis, probably due to their differential N glycosylation (1, 17). Hu-tetherin-based chimeras with ECL, CC, or GPI domains of mo-tetherin but carrying a TM domain of hu-tetherin displayed BiFC signals comparable to that of WT hu-tetherin (Fig. 2C). In contrast, the hu-tetherin-based chimera with the reciprocal TM of mo-tetherin showed a significant reduction in BiFC signal (Fig. 2C). Conversely, the mo-tetherin-based chimera with the reciprocal TM of hu-tetherin displayed significant BiFC signals similar to those of parental hu-tetherin (Fig. 2C), indicating that the TM of hu-tetherin is necessary and sufficient for the interaction between mo-tetherin-based mutants and Vpu. Mo-tetherin-based chimera mutants replaced with the ECL, CC, or GPI of hu-tetherin showed a weak signal (Fig. 2C), suggesting that the other regions are less important in the Vpu interaction.
FIG. 2.
Interaction of tetherin and Vpu via TM domains. (A) The KGN-Vpu-expressing cells were transfected with plasmid encoding KGC-tagged hu-tetherin or deletion mutants of CT or GPI (delCT and delGPI). The expression of KGC-tetherin fusion protein was detected using an anti-KGC antibody (bottom). Relative MFI is defined as the MFI of pKGC-tetherin-transfected or its mutant-transfected cells minus the MFI of untransfected cells, and results represent the means from three independent experiments plus standard deviations. The amount of protein applied for the delGPI Western blotting was 10 times less than that for WT hu-tetherin to avoid the overexposure of the protein on polyvinylidene difluoride membrane. Statistical significance (Student's t test) in the MFI of human tetherin and mutants are represented as follows: **, P < 0.01; *, P < 0.05. (B) The KGN-Vpu-expressing HEK 293 cells were transfected with pKGC-mo-tetherin (upper) or pKGC-hu-tetherin (lower). Cells were double stained with anti-Vpu antibody and anti-KGC antibody and imaged by confocal microscopy. (C) Interaction of tetherin TM with Vpu. The KGN-Vpu-expressing cells were transfected with KGC-hu-tetherin, KGC-mo-tetherin, or chimera mutant DNA containing reciprocal exchanges of the TM, ECL, CC, or GPI between the hu- and mo-tetherin proteins and analyzed by flow cytometry. Statistical significance (Student's t test) in the MFI of hu-tetherin and chimeras are represented as follows: *, P < 0.01; standard deviation for mo(huCC) was 0.0061. Relative MFI is defined as the MFI of pKGC-tetherin-transfected or its mutant-transfected cells minus the MFI of untransfected cells. (D) Interaction of the Vpu TM with tetherin. KGC-tetherin-expressing cells were transfected with KGN-Vpu or its mutant DNA and analyzed. Statistical significances (Student's t test) in the BiFC signals are given. Relative MFI values are defined as the MFI of pKGN-Vpu or its mutant-transfected cells minus the MFI of untransfected cells. The results represent the means from three independent experiments (A, C, and D). Expression of the KGC or KGN fusion or Vpu protein was detected by immunoblot analysis with individual antibodies (A, C, and D).
Specific interaction of Vpu TM with hu-tetherin.
HIV-1 Vpu protein consists of two major elements: an N-terminal TM domain that anchors Vpu in the cellular membranes and a CT consisting of two putative α-helices separated by a conserved phosphorylation site (21). To understand which domain of Vpu interacts with hu-tetherin, we generated HEK 293 cells stably expressing KGC-tagged hu-tetherin (KGC-hu-tetherin) (Fig. 2D). Cells were transfected with plasmids encoding KGN-Vpu WT or mutants. We prepared KGN-tagged Vpu mutants by truncating its CT at residue 50 (Vpu1-50), replacing the TM region with a scrambled TM sequence (VpuRD), inserting an alanine at serine residues 52 and 56 in the cytoplasmic domain (Vpu2/6), or by inserting positively charged residues within the putative overlapping tyrosine- and dileucine-based sorting motifs with alanine (VpuR30A/K31A) (Fig. 1A) (12, 46, 58, 59). The levels of KGN-Vpu expression of these mutants were nearly equivalent (Fig. 2D, upper) and did not significantly alter KGC-hu-tetherin expression (Fig. 2D, lower). Our BiFC assay revealed that VpuRD interacted poorly with hu-tetherin, while Vpu1-50 displayed BiFC signals equivalent to those of WT Vpu (Fig. 2D, top columns), indicating that Vpu interacts with hu-tetherin through its TM domain. Moreover, VpuR30A/K31A displayed a modest decrease in BiFC signal. A previous study showed that the VpuR30A/K31A mutant displayed a partial (∼40%) defect in its TGN localization and its efficient delivery to late endosomal degradation compartments (12). This indicates that the trafficking of Vpu to the TGN is important, to some extent, for hu-tetherin interaction. Our results confirm that Vpu and hu-tetherin interact with each other via their respective TM regions.
Mapping of the Vpu interaction domain in hu-tetherin TM.
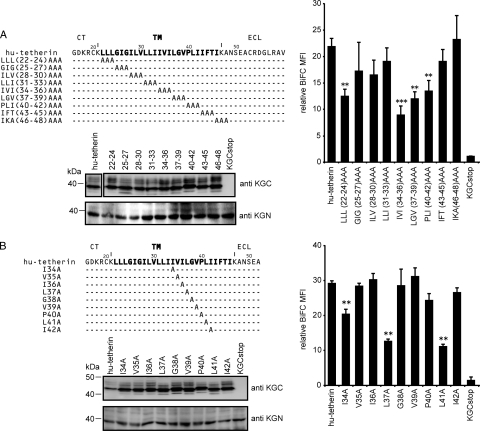
To reveal the Vpu-interacting amino acids in the TM region of tetherin, we next prepared plasmids encoding KGC-tagged hu-tetherin mutants with triple-alanine substitution in the TM region (Fig. 3A, top) and then transfected them into KGN-Vpu-expressing cells. Alanine is the substitution residue of choice, since it eliminates the side chain beyond the β-carbon and yet does not alter the main-chain conformation (as can glycine or proline), nor does it impose extreme electrostatic or steric effects (37). Levels of the cell surface expression of these mutants were nearly equivalent to or greater than that of WT hu-tetherin (Table 1). The levels of KGC expression detected by immunoblot analysis were equivalent to those of WT hu-tetherin (Fig. 3A, bottom), while LLL(22-24)AAA, IVI(34-36)AAA, LGV(37-39)AAA, and PLI(40-42)AAA showed clearly reduced BiFC signals compared to that of WT hu-tetherin (Fig. 3A). These data suggest that the residues covering 22 to 24 and 34 to 42 of hu-tetherin play a crucial role in the Vpu interaction. The reduction of Vpu binding capacity by the alteration of residues 22 to 24 in hu-tetherin might correspond to a previous observation that the deletion of 22L and 23L from hu-tetherin disrupts the interaction with Vpu (54).
FIG. 3.
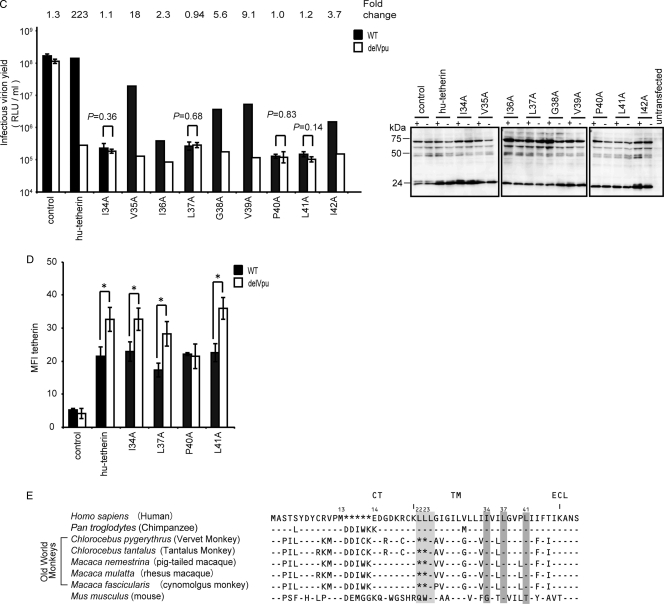
Amino acid residues of hu-tetherin TM region required for Vpu interaction and counteraction. (A and B) BiFC assay of hu-tetherin TM mutants on Vpu interaction. Schematic diagram showing the amino acid alterations made in the C-terminal TM region of hu-tetherin. Identity is indicated by dashes. The KGN-Vpu-expressing cells were transfected with a series of KGC-hu-tetherin TM mutants and analyzed by flow cytometry as described in Materials and Methods. The protein expression of KGC-fused hu-tetherin and its mutants in cell lysates were detected using the indicated antibody. Relative MFI values are defined as the MFI of pKGN-Vpu-transfected or its mutant-transfected cells minus the MFI of untransfected cells, and results represent the means from three independent experiments plus standard deviations. (C) Tetherin activity and Vpu sensitivity of hu-tetherin TM mutants. HEK 293 cells were cotransfected with WT HIV-1 infectious DNA (pNL4-3) or its vpu-deleted DNA (pNL4-3/Udel) (1 μg) with or without (control) pKGC-hu-tetherin and its derivative expression plasmids (100 ng). The expression of HIV-1 (Pr55Gag) in the cell lysates was detected using anti-p24 antibody. Results represent the means from more than three independent experiments. Statistical significance (Student's t test) is represented as P < 0.05 (**) and P < 0.01 (***) (A and B) or as numerical values (C). (D) Surface expression of tetherin and its mutants. HEK 293 cells were cotransfected with WT HIV-1 infectious DNA (pNL4-3) or its vpu-deleted DNA (pNL4-3/Udel) (1 μg) with or without (control) pKGC-hu-tetherin and its mutant expression plasmids (100 ng). The cell surface expression of tetherin was detected using anti-tetherin antibody. Statistical significance (Student's t test) is represented as P < 0.05 (*). (E) Amino acid alignment of seven primates and the mo-tetherin TM domain. Identity is indicated by dashes, and sequence gaps are indicated by an asterisk. Gray boxes indicate amino acid sequences corresponding to the mutants for which the BiFC signals were significantly decreased.
To narrow down interacting amino acid residues within the 34 to 42 region of hu-tetherin, we next prepared single-alanine-substituted mutants (Fig. 3B, top). The levels of the cell surface protein expression of the mutants were equivalent to that of WT hu-tetherin (Table 1), and their electrophoretic mobilities were consistent with the expected sizes (Fig. 3B, bottom). Among the nine single-alanine-substituted mutants, I34A, L37A, and L41A mutations produced significantly reduced BiFC signals compared to hu-tetherin or its other mutants (Fig. 3B, right columns).
I34, L37, and L41 of hu-tetherin are involved in the determination of Vpu susceptibility.
To examine whether the interaction between hu-tetherin and Vpu through I34, L37, and L41 residues plays a functional role in the determination of Vpu susceptibility, we tested these mutants for Vpu susceptibility and tetherin activity. HEK 293 cells were transfected with infectious WT HIV-1 DNA (pNL4-3) or its Vpu-deleted HIV-1 DNA (pNL4-3/Udel) together with WT hu-tetherin or its mutants. The amount of HIV-1 in the culture supernatants together with HIV-1 protein in cell extract was determined. Importantly, hu-tetherin mutants with I34A, L37A, P40A, and L41A mutations completely lost their Vpu susceptibility, while V35A, I36A, G38A, V39A, and I42A remained Vpu sensitive (Fig. 3C). To determine whether mutants that conferred resistance to antagonism by Vpu in virion release assays also conferred resistance to the downregulation of tetherin from the cell surface, we examined cell the surface expression of tetherin or its derivatives on these cells. In our results, the levels of the tetherin downregulation of I34A, L37A, and L41A mutants were similar to that of WT tetherin (Fig. 3D). On the other hand, only P40A displayed resistance to Vpu-induced downregulation. These results suggest that different mechanisms account for the loss of Vpu sensitivity in P40A and other mutants. Overall, these results provide direct evidence that I34, L37, and L41 of hu-tetherin are involved in the determination of Vpu susceptibility in addition to Vpu interaction. Interestingly, these amino acid residues are highly conserved among primate tetherins and are present not only in Vpu-sensitive but also in Vpu-insensitive tetherins (Fig. 3E, gray boxes 34, 37, and 41). This suggests that the presence of these residues alone is insufficient to render tetherin Vpu sensitive. Indeed, a previous report demonstrated that agm-tetherin interacted poorly with HIV-1 Vpu (54). Thus, we hypothesize that the difference of Vpu interaction efficiency between hu- and agm-tetherin is due to differences in secondary structure rather than the primary amino acid sequence of the TM domain. Furthermore, additional residues in human tetherin that are not conserved in monkey tetherins may contribute to Vpu sensitivity.
Acquisition of Vpu-interacting activity in agm-tetherin by the insertion of two amino acid residues and acquisition of Vpu susceptibility by a single amino acid mutation.
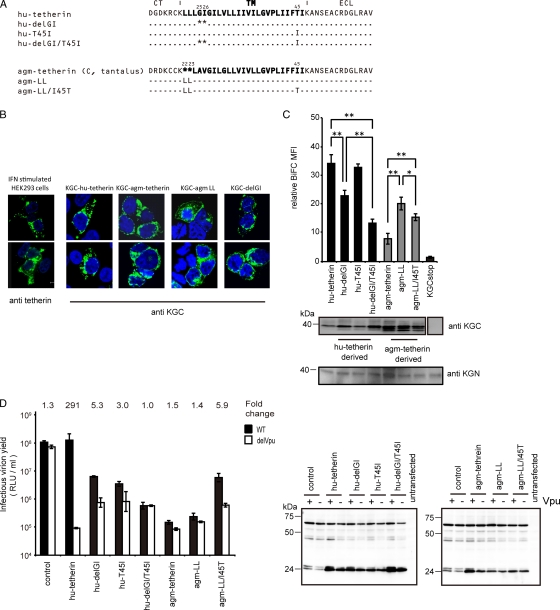
To determine additional critical amino acid residues that affect Vpu interaction and susceptibility, we carried out both loss- and gain-of-function approaches using hu- or agm-tetherin. Human and chimpanzee tetherins contain a dileucine motif upstream of the Vpu binding domain that is absent from other primate tetherins (Fig. 3E). The deletion of this motif (L22 and L23) in hu-tetherin clearly influenced its Vpu binding capacity (54). Another report demonstrated that a two-amino-acid deletion and a single-amino-acid substitution in the hu-tetherin TM (hu-delGI/T45I) can confer complete resistance to antagonism by Vpu (43). Hence, we focused on these two regions for the subsequent experiments (Fig. 4A). To confirm the lack of alteration of intracellular distribution of tetherin by deleting or inserting amino acid in its TM region, we examined the distribution of representative mutants (hu-delGI and agm-LL). The localizations of these mutants were very similar to that of hu-tetherin or agm-tetherin, respectively (Fig. 4B).
FIG. 4.
C-terminal motif in tetherin TM domain is a determinant for Vpu counteraction. (A) Amino acid alignment of hu-tetherin, agm-tetherin, and their mutants. Identity is indicated by dashes, and sequence gaps are indicated by an asterisk. (B) Subcellular localization of tetherin 2-amino acid-deletion mutant or insertion mutant. KGN-Vpu-expressing cells were transfected with plasmid encoding KGC-tagged hu-tetherin, KGC-tagged agm-tetherin, or their mutants, respectively, and cells were stained with an anti-KGC antibody. IFN-treated HEK 293 cells were stained with an anti-tetherin antibody. Cells were examined by confocal microscopy. (C) BiFC assay of hu- and agm-tetherin TM mutants on Vpu interaction. KGN-Vpu-expressing cells were transfected with plasmid encoding KGC-hu-tetherin, KGC-agm-tetherin, or the mutants and was analyzed by flow cytometry. The expression of the protein in cell lysates was detected using the indicated antibody. Relative MFI values are defined as the MFI of pKGC-tetherin or its mutant transfected cells minus the MFI of untransfected cells, and results represent the means from three independent experiments plus standard deviations. (D) Tetherin activity and Vpu sensitivity assays of hu- and agm-tetherin TM mutants. HEK 293 cells were cotransfected as described in the legend to Fig. 2B without (control) or with 100 ng KGC-hu-tetherin or agm-tetherin and the derivative DNA. The HIV-1 proteins (Pr55Gag) in the cell lysates were detected using anti-p24 antibody. Results represent the means from three independent experiments plus standard deviations.
For the loss-of-function approach for hu-tetherin, we prepared KGC-tagged hu-delGI, hu-T45I, and hu-delGI/T45I tetherin mutants (Fig. 4A). Although the levels of KGC expression of these mutants were equivalent to that of WT hu-tetherin (Fig. 4C), hu-delGI and hu-delGI/T45I displayed significantly lower-level BiFC signals than WT hu-tetherin, while hu-T45I displayed a level of BiFC signals similar to that of WT hu-tetherin (Fig. 4C). Among these hu-tetherin mutants, hu-delGI/T45I showed the lowest interaction profile with Vpu. These data indicate that by itself T45 does not play a significant role in Vpu interaction; however, it exacerbates the effect of the GI deletion (G25 and I26). From tetherin activity and Vpu susceptibility assays, we confirmed that hu-delGI remained partially sensitive to Vpu (Fig. 4D) as previously described (43).
For the gain-of-function approach we prepared two KGC-tagged agm-tetherin mutants by inserting two leucine residues (agm-LL) or by combining the insertion of two leucines with a mutation of I45T (corresponding to hu-tetherin residues) (Fig. 4A). As demonstrated in Fig. 4C, agm-LL displayed a more-than 2-fold increase of BiFC signal compared to that of WT agm-tetherin. We also tested these mutants for tetherin activity and Vpu sensitivity. Notably, an agm-LL mutant was insensitive to HIV-1 Vpu antagonism (Fig. 4D) even though it interacted with Vpu to a certain extent (Fig. 4C). Finally, the agm-LL/I45T mutant, containing the combined two-leucine insertion and I45T in the TM of agm-tetherin, clearly displayed the Vpu sensitivity (Fig. 4D). However, its BiFC signal was not increased by I45T substitution (Fig. 4C). These results suggest that T45 of hu-tetherin has an important role for Vpu counteraction against tetherin without affecting Vpu-tetherin association.
Structural study of the TM domain of hu- and agm-tetherins in a lipid bilayer environment.
To obtain structural insights into the mechanisms by which the amino acid at specific positions in the tetherin TM domain control interaction and susceptibilities to neutralization by Vpu, we constructed 3D models of the TM domains of hu-tetherin, agm-tetherin, and five tetherin mutants, including agm-LL, agm-LL/I45T, hu-delGI, hu-T45I, and hu-delGI/T45I. We constructed these models in a lipid bilayer environment by MD simulations (28, 29, 61) of the models and compared the structures. Representative structures were obtained by cluster analysis (60) of the 3,000 snapshots during 2.0 to 5.0 ns of each MD simulation for the structural comparisons. Details of the simulation methods and processes, such as the time course of structural changes during MD simulations, are described in the supplemental material (see Table S1 and Fig. S1 and S2).
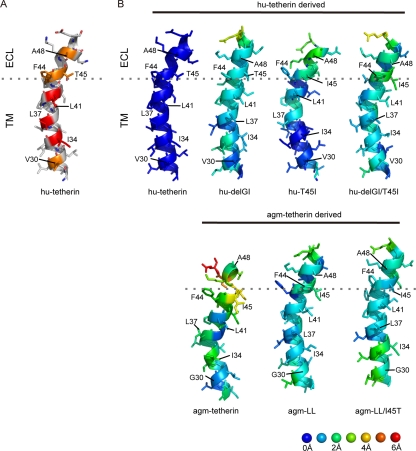
The thermodynamically stable hu-tetherin TM model shows that the TM consists of helical structures in the lipid bilayer that are consistent with its location in the plasma membrane. In the model, amino acid residues I34, L37, and L41, which we demonstrated above to be indispensable for Vpu interaction, are positioned on the same face of the helix in the lipid bilayer environment, leading to the protrusion of the hydrophobic side chains in a similar direction (Fig. 5A, amino acid residues with red color). V30, F44, T45, and A48 of hu-tetherin also were positioned on the same helical face with these amino acid residues (Fig. 5A, amino acid residues with orange color). This 3D arrangement of particular amino acids on the helical surface was significantly disordered in the TM domains of agm-tetherin; side chains of I34, L37, and L41, which correspond to the I34, L37, and L41 in hu-tetherin, respectively, protruded in different directions (Fig. 5B, agm-tetherin). When the agm-tetherin TM model was superimposed on the hu-tetherin TM model using the main-chain atoms (see Materials and Methods), more than 2.0 Å of 3D positional shifts were detected at the many main-chain atoms. This included I34, L37, F44, I45, and A48, which correspond to I34, L37, F44, T45, and A48, respectively, in hu-tetherin (Fig. 5B; also see Fig. S2 in the supplemental material). On the other hand, conformational changes from the hu-tetherin TM were detected in the smaller regions of other TM variants examined (Fig. 5B, agm-LL, agm-LL/I45T, hu-delGI, hu-T45I, and hu-delGI/T45I). More than 2.0 Å of positional shifts were observed only at I45 of agm-LL, I34 of agm-LL/I45T, A48 of hu-T45I, and F44 and I45 of hu-delGI/T45I (Fig. 5B; also see Fig. S2). hu-delGI was more similar in structure to hu-tetherin than the others (Fig. 5B; also see Fig. S2).
FIG. 5.
Structure model of tetherin TM domains. (A) Structural models of C-terminal TM domains (G27 to E51 in hu-tetherin). I34, L37, and L41, whose mutations influence interactions with Vpu, are highlighted with a red cartoon and sticks. V30, F44, T45, and A48, whose side chains are positioned in the same face with side chains of I34, L37, and L41, are highlighted with an orange cartoon and sticks. (B) Comparisons of three-dimensional structures of individual residues between hu-tetherin and others. RMSD values between hu-tetherin and other models were calculated using the coordinates of N, Cα, and C atoms in the C-terminal TM domain (G27 to E51 in hu-tetherin) in the representative structures, each of which was selected from 3,000 snapshots during 2.0 to 5.0 ns of the respective MD simulations using the Bayesian clustering algorithm. The individual residues in the structures were colored according to the RMSD values. The three-dimensional images of the structures were made with PyMOL version 0.99 rc6 (Schrödinger LLC).
Experimental data showed that I34, L37, and L41 in hu-tetherin are critical for Vpu interaction (Fig. 3B). Hence, we examined how the 3D positions of these amino acid residues are maintained. For hu-T45I, hu-delGI, and agm-LL, which have relatively high competence for Vpu interaction, positional shifts of three amino acid residues were less than 1.3 Å (Fig. 5B; also see Fig. S2). In contrast, the others showed greater positional shifts of the three amino acid residues. Positions of I34 in agm-thetherin and agm-LL/I45T shifted 1.9 to 2.4 Å, while that of L41 in hu-delGI/T45I shifted 1.6 Å. These results are consistent with their competence for Vpu interaction.
Experimental data also pointed out the crucial importance of the amino acid residue at position 45 for the susceptibility of hu-tetherin to Vpu (Fig. 4D). The hu-tetherin and two mutants (agm-LL/I45T and hu-delGI) have threonine at this position and shared relatively similar conformations at the threonine and neighboring phenylalanine (F44 in hu-tetherin); less than 1.5 Å of positional shifts was detected when these mutant TMs were superimposed on the hu-tetherin TM (Fig. 5B; also see Fig. S2). The conformational differences from the hu-tetherin TM were greater for those having the change of threonine into isoleucine (agm-tetherin, agm-LL, hu-T45I, and hu-delGI/T45I); the differences ranged from 1.7 to 3.5 Å with these mutants (Fig. 5B; also see Fig. S2).
We also analyzed the snapshot models at 5.0 ns of each MD simulation. All of these structural characteristics and differences found with the representative models were reproducible with the snapshot model at 5.0 ns of each MD simulation (see Fig. S3 in the supplemental material) where fluctuations of TM structures during MD simulations were relatively small (see Fig. S1). Our structural models support the results that the Vpu-specific interaction and susceptibility with hu-tetherin is defined by critical amino acid residues that lie in the TM (I34, L37, L41, and T45) and imply that the appropriate positioning of these amino acid residues in hu-tetherin are important for Vpu interaction and sensitivity.
DISCUSSION
The molecular basis behind the HIV-1 Vpu counteraction of hu-tetherin has not been fully elucidated. Despite the observation that the specific antagonism of hu-tetherin occurs through a Vpu-mediated pathway, the specific residues of hu-tetherin that define the association with Vpu remain unknown. In this report, we utilized a BiFC approach to characterize the interaction between Vpu and tetherin (Fig. 1). We first validated our BiFC approach by detecting the specific hu-tetherin-Vpu heterodimer complexes (Fig. 1B and D), which have been shown previously by immunoprecipitation studies (25, 54). The complementation-based method is a simple way for confirming the protein-protein interaction in live cells. As BiFC depends on the two interacting proteins to bring the complementary fluorescence protein fragments close enough to fold into stable fluorescence complex, a positive BiFC signal implies a distance of less than 15 nm between the two interacting proteins (33). Consistently with our BiFC results, hu-tetherin and Vpu interaction recently were reported using a fluorescence resonance energy transfer (FRET) assay (2), suggesting a distance of less than 6 nm between the interacting Vpu and tetherin (52). However, it should be noted that although the BiFC assay demonstrated a positive association of tetherin and Vpu, it remains unclear whether the interaction is direct or indirect. As a result, we were able to identify three amino acid residues (I34, L37, and L41) that were shown in our 3D model to align on the same TM helical surface for Vpu interaction (Fig. 5). Altering any one of these amino acid residues led to a disruption of the Vpu-hu-tetherin complex and also abrogated Vpu susceptibility (Fig. 3). Furthermore, our finding that the T45 residue of hu-tetherin had no bearing on the Vpu interaction yet significantly abated Vpu susceptibility (Fig. 4) implies that certain residues participate exclusively in the Vpu-mediated antagonism of tetherin independently of the interaction. In addition, our 3D models offer a glimpse into the possible dynamics of the viral and host molecules and allow us to visualize the arrangement of the amino acid residues in the hu-tetherin TM involved in the possible binding with Vpu.
Recently, several groups have demonstrated that the hu-tetherin TM and specific residues within it are essential for Vpu interaction and hu-tetherin counteraction (10). The studies identified hu-tetherin delGI T45I, delGI I33V I36L (43), del22/23 (54), T45I, and I26V/V30G/I36L/T45I (18) as mutants that are resistant to Vpu, but they mainly focused on hu-tetherin residues that differed from rhesus or agm-tetherin residues (10). In contrast, we conducted a comprehensive screening of the entire TM region and identified three residues (I34, L37, and L41) as critical determinants for Vpu interaction and susceptibility (Fig. 3B and C). Our model predicts that these three amino acid residues are directly involved in helix-helix interactions between the TMs of tetherin and Vpu, composing an interacting domain (Fig. 5A). This possibility is feasible considering the biochemical characteristics of hydrophobic isoleucine and leucine, which often are located at the surfaces of protein-protein interaction sites in membrane proteins, as exemplified in leucine zipper and leucine/isoleucine zipper motifs. The deletion mutants delGI and L22-23 had a significant impact on perturbing the Vpu-mediated antagonism of hu-tetherin (Fig. 4D). Our gain-of-function studies showed that the insertion of two residues (LL) in the TM of agm-tetherin was enough to restore a partial level of interaction with Vpu, and in combination with the I45T mutation, it led to a partial restoration of Vpu sensitivity (Fig. 4D). We speculate that missing residues in the helix, such as the delGI mutant and agm-tetherin, have an overall impact on the TM amino acid residue positioning, thereby affecting both interaction and sensitivity. Interestingly, mutations of L22 and L23 to alanine in our triple-alanine scan mutants disrupted Vpu interaction, whereas mutations of G25 and I26 to alanine did not have any effect (Fig. 3A). This finding leads us to believe that the deletion of LL has a more-specific effect on the Vpu interaction in conjunction with orienting the tetherin helix TM residues.
In an evolutionary aspect, two amino acids in the hu-tetherin TM might have been inserted when advanced primates evolved from primitive primates, resulting in an adaptation by HIV-1 Vpu to recognize hu-tetherin. It is unclear why these insertions at positions 22 and 23 occur in hu-tetherin, but the fact that these regions also evolved under selective pressure shows that these events might have occurred as a consequence of their role in other functional activities of tetherin, such as B cell differentiation, the inhibition of virus production, or the regulation of interferon release (6, 18, 24, 43).
In contrast to other reports (10), we could not definitively verify the importance of the mutant I36A. The mutation of I36 did not have an impact on the Vpu and hu-tetherin interaction in our BiFC assay; even though it attenuated Vpu susceptibility during virus release, the effect was not as dramatic as mutating L34, L37, L41, or T45 (Fig. 3B and C and 4C and D). Mutants I26A and V30A were not investigated further, because they did not disrupt the Vpu and tetherin BiFC fluorophore in our preliminary interaction screen (Fig. 3A).
Interestingly, a P40A mutation also abolished susceptibility to Vpu (Fig. 3C), although McNatt et al. reported that a P40I mutation does not affect the Vpu susceptibility (43). However, this discrepancy more likely is due to an overall conformational rearrangement of the hu-tetherin TM from the replacement of proline with hydrophobic amino acid of different sizes. Proline residues generally play significant roles in maintaining the conformational structure, and changing proline to alanine can affect the overall conformation more dramatically than changing proline to isoleucine (53). In addition to P40A, I36A also displayed, to a lesser extent, Vpu counteraction (Fig. 3C). In a previous study, I36 has been shown to be one of the positively selected residues in the hu-tetherin TM sequence (18, 43). When this residue was exchanged for alanine, a loss of susceptibility to Vpu was observed (Fig. 3C). From our structural analysis of human tetherin, I36 is not likely to be involved in interaction; however, exchanging I36 for other residues can affect Vpu susceptibility.
One of the more-important observations of our study was the function of T45, which repeatedly has been reported to be essential for Vpu susceptibility by others (10, 18, 43). In our hands, a mutation of this residue did not significantly affect Vpu and hu-tetherin complex formation when measuring the interaction by BiFC; however, the mutation of T45 still affected Vpu sensitivity (Fig. 4D). Our gain-of-function studies further illustrated the importance of T45 by rendering agm-tetherin susceptible to Vpu in the agm-LL/I45T mutant, an effect that was not observed for the agm-LL mutant without the I45T substitution. This suggests that Vpu-tetherin association alone is not sufficient for acquiring antagonistic activity.
Changes in the amino acid at position 45 in hu-tetherin TM are predicted to influence the conformation of not only itself but also its immediate downstream amino acid at position 44 (Fig. 5B; also see Fig. S2 in the supplemental material). When hu-tetherin has a threonine at position 45, the amino acids at positions 44 and 45 tended to be positioned on the same surface as the functionally important amino acid, such as I34, L37, and L41 (Fig. 4B; also see Fig. S4). Concurrently with this folding property, tetherins tended to preserve the Vpu-susceptibility (Fig. 4D, hu-delGI). In contrast, when agm-tetherin has an isoleucine at position 45, the ordered 3D arrangement fluctuated, particularly at positions 44 and 45, and the tetherins tended to have severely reduced susceptibilities to Vpu activity (Fig. 4D and 5B; also see Table S1 and Fig. S2). These results indicate that the amino acid at position 45 plays crucial roles in both folding and the Vpu susceptibility of the hu-tetherin TM domain. This is consistent with biochemical characteristics of threonine; in general, threonine is rare in TMs of proteins; however, if it exists, the threonine can play very important roles in the folding and protein-protein interaction of TMs (49), such as the tight packing of helices (14), the formation of intermolecular hydrogen bonds (13, 62), and the formation of biologically functional protein complexes (3). Therefore, it is conceivable that T45 in hu-tetherin plays key roles in forming a biologically functional complex of hu-tetherin, Vpu, and undefined proteins for the Vpu-mediated degradation of tetherin. T45 may play an important role in one or more of the reported mechanisms by which Vpu antagonizes hu-tetherin: (i) by targeting hu-tetherin for proteasomal and/or lysosomal degradation via the β-TrCP-dependent ubiquitin/proteasome pathway (10); (ii) by trafficking hu-tetherin to intracellular organelles such as the TGN, Golgi complex, or ER (12); and (iii) by sequestering hu-tetherin within intracellular compartments and preventing its distribution to the plasma membrane (11, 19). Although our results indicate that T45 behaves in this manner, residues identified by Gupta et al., such as I26, V30, and I36, also may contribute similarly (18). Further structural and biochemical studies are necessary to address this issue.
In conclusion, the main advantage our modeling results provides is the visualization and positioning of the TM amino acid residues in a manner that allows for Vpu binding. To our knowledge, this is the first 3D model for the TM domain of tetherin that offers a plausible explanation for the residues involved in the Vpu-hu-tetherin interaction. Having this insight will aid in identifying the complementary motifs in Vpu that interact with the three amino acid residues found in this study. Our BiFC system also will be a useful tool for building a structure-activity relationship and elucidating the Vpu TM residues contributing to tetherin binding. Moreover, it also is conceivable to design a type of peptide decoy molecule (45) consisting of only a TM domain homologous to human tetherin that is able to sequester Vpu while permitting the retainment of HIV-1 virions by tetherin. With this in mind, and guided by structure-based predictions, the Vpu-tetherin complex could become a novel target for the future pharmacological intervention of HIV-1 dissemination.
Supplementary Material
Acknowledgments
We thank Jun Komano and Chuanyi Nie for helpful discussions of the manuscript.
This work was supported by Grants-in-Aid for Science Research of Priority Areas from the Ministry of Education, Culture, Sports and Technology of Japan, a Health and Labor Science research grant (Research on Publicly Essential Drugs and Medical Devices) and a grant for HIV/AIDS research from the Ministry of Health, Labor and Welfare of Japan. K.S., T.Y., and S.P.Y. were supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print on 10 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andrew, A. J., E. Miyagi, S. Kao, and K. Strebel. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banning, C., J. Votteler, D. Hoffmann, H. Koppensteiner, M. Warmer, R. Reimer, F. Kirchhoff, U. Schubert, J. Hauber, and M. Schindler. 2010. A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5:e9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beahm, D. L., A. Oshima, G. M. Gaietta, G. M. Hand, A. E. Smock, S. N. Zucker, M. M. Toloue, A. Chandrasekhar, B. J. Nicholson, and G. E. Sosinsky. 2006. Mutation of a conserved threonine in the third transmembrane helix of alpha- and beta-connexins creates a dominant-negative closed gap junction channel. J. Biol. Chem. 281:7994-8009. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brügger, B., B. Glass, P. Haberkant, I. Leibrecht, F. T. Wieland, and H. G. Krausslich. 2006. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 103:2641-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, W., L. Bover, M. Cho, X. Wen, S. Hanabuchi, M. Bao, D. B. Rosen, Y. H. Wang, J. L. Shaw, Q. Du, C. Li, N. Arai, Z. Yao, L. L. Lanier, and Y. J. Liu. 2009. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 206:1603-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, E. A., E. F. Terwilliger, J. G. Sodroski, and W. A. Haseltine. 1988. Identification of a protein encoded by the vpu gene of HIV-1. Nature 334:532-534. [DOI] [PubMed] [Google Scholar]
- 8.Crowley, M. F., T. A. Darden, T. E. Cheatham, and D. W. Deerfield. 1997. Adventures in improving the scaling and accuracy of a parallel molecular dynamics program. J. Supercomput. 11:255-278. [Google Scholar]
- 9.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald-an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089-10092. [Google Scholar]
- 10.Douglas, J. L., J. K. Gustin, K. Viswanathan, M. Mansouri, A. V. Moses, and K. Fruh. 2010. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6:e1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubé, M., B. B. Roy, P. Guiot-Guillain, J. Binette, J. Mercier, A. Chiasson, and E. A. Cohen. 2010. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 6:e1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubé, M., B. B. Roy, P. Guiot-Guillain, J. Mercier, J. Binette, G. Leung, and E. A. Cohen. 2009. Suppression of tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J. Virol. 83:4574-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duong, M. T., T. M. Jaszewski, K. G. Fleming, and K. R. MacKenzie. 2007. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J. Mol. Biol. 371:422-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eilers, M., S. C. Shekar, T. Shieh, S. O. Smith, and P. J. Fleming. 2000. Internal packing of helical membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 97:5796-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577-8593. [Google Scholar]
- 16.Fritsch, V., G. Ravishanker, D. L. Beveridge, and E. Westhof. 1993. Molecular dynamics simulations of poly(dA)·poly(dT): comparisons between implicit and explicit solvent representations. Biopolymers 33:1537-1552. [DOI] [PubMed] [Google Scholar]
- 17.Goffinet, C., I. Allespach, S. Homann, H. M. Tervo, A. Habermann, D. Rupp, L. Oberbremer, C. Kern, N. Tibroni, S. Welsch, J. Krijnse-Locker, G. Banting, H. G. Krausslich, O. T. Fackler, and O. T. Keppler. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285-297. [DOI] [PubMed] [Google Scholar]
- 18.Gupta, R. K., S. Hue, T. Schaller, E. Verschoor, D. Pillay, and G. J. Towers. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser, H., L. A. Lopez, S. J. Yang, J. E. Oldenburg, C. M. Exline, J. C. Guatelli, and P. M. Cannon. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornak, V., R. Abel, A. Okur, B. Strockbine, A. Roitberg, and C. Simmerling. 2006. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hout, D. R., E. R. Mulcahy, E. Pacyniak, L. M. Gomez, M. L. Gomez, and E. B. Stephens. 2004. Vpu: a multifunctional protein that enhances the pathogenesis of human immunodeficiency virus type 1. Curr. HIV Res. 2:255-270. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27-38. [DOI] [PubMed] [Google Scholar]
- 23.Ibragimova, G. T., and R. C. Wade. 1998. Importance of explicit salt ions for protein stability in molecular dynamics simulation. Biophys. J. 74:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa, J., T. Kaisho, H. Tomizawa, B. O. Lee, Y. Kobune, J. Inazawa, K. Oritani, M. Itoh, T. Ochi, K. Ishihara, et al. 1995. Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST2, that may be involved in pre-B-cell growth. Genomics 26:527-534. [DOI] [PubMed] [Google Scholar]
- 25.Iwabu, Y., H. Fujita, M. Kinomoto, K. Kaneko, Y. Ishizaka, Y. Tanaka, T. Sata, and K. Tokunaga. 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J. Biol. Chem. 284:35060-35072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jójárt, B., and T. A. Martinek. 2007. Performance of the general amber force field in modeling aqueous POPC membrane bilayers. J. Comput. Chem. 28:2051-2058. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen, W. L., J. Chandrasekhar, J. Madura, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926-935. [Google Scholar]
- 28.Kandasamy, S. K., and R. G. Larson. 2006. Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers: a systematic investigation of hydrophobic mismatch. Biophys. J. 90:2326-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karplus, M., and J. A. McCammon. 2002. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9:646-652. [DOI] [PubMed] [Google Scholar]
- 30.Kawano, Y., T. Yoshida, K. Hieda, J. Aoki, H. Miyoshi, and Y. Koyanagi. 2004. A lentiviral cDNA library employing lambda recombination used to clone an inhibitor of human immunodeficiency virus type 1-induced cell death. J. Virol. 78:11352-11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerppola, T. K. 2006. Complementary methods for studies of protein interactions in living cells. Nat. Methods 3:969-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerppola, T. K. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1:1278-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerppola, T. K. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimkait, T., K. Strebel, M. D. Hoggan, M. A. Martin, and J. M. Orenstein. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kollman, P. A., I. Massova, C. Reyes, B. Kuhn, S. Huo, L. Chong, M. Lee, T. Lee, Y. Duan, W. Wang, O. Donini, P. Cieplak, J. Srinivasan, D. A. Case, and T. E. I. Cheatham. 2000. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33:889-897. [DOI] [PubMed] [Google Scholar]
- 36.Kupzig, S., V. Korolchuk, R. Rollason, A. Sugden, A. Wilde, and G. Banting. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694-709. [DOI] [PubMed] [Google Scholar]
- 37.Lefèvre, F., M. H. Remy, and J. M. Masson. 1997. Alanine-stretch scanning mutagenesis: a simple and efficient method to probe protein structure and function. Nucleic Acids Res. 25:447-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, E. S., H. S. Malik, and M. Emerman. 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84:7124-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loncharich, R. J., B. R. Brooks, and R. W. Pastor. 1992. Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-actylananyl-N′-methylamide. Biopolymers 32:523-535. [DOI] [PubMed] [Google Scholar]
- 40.Maldarelli, F., M. Y. Chen, R. L. Willey, and K. Strebel. 1993. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol. 67:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangeat, B., G. Gers-Huber, M. Lehmann, M. Zufferey, J. Luban, and V. Piguet. 2009. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 5:e1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuyama, N., T. Kuronita, R. Tanaka, T. Muto, Y. Hirota, A. Takigawa, H. Fujita, Y. Aso, J. Amano, and Y. Tanaka. 2009. HM1.24 is internalized from lipid rafts by clathrin-mediated endocytosis through interaction with alpha-adaptin. J. Biol. Chem. 284:15927-15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNatt, M. W., T. Zang, T. Hatziioannou, M. Bartlett, I. B. Fofana, W. E. Johnson, S. J. Neil, and P. D. Bieniasz. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5:e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miaskiewicz, K., R. Osman, and H. Weinstein. 1993. Molecular dynamics simulation of the hydrated d(CGCGamino acidTTCGCG)2 dodecamer. J. Am. Chem. Soc. 115:1526-1537. [Google Scholar]
- 45.Montal, M. 2009. Vpu matchmakers as a therapeutic strategy for HIV infection. PLoS Pathog. 5:e1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen, K. L., M. llano, H. Akari, E. Miyagi, E. M. Poeschla, K. Strebel, and S. Bour. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163-175. [DOI] [PubMed] [Google Scholar]
- 48.Nilsson, L. 1998. Protein nucleic acid interactions, p. 2220-2229. In P. von Ragué Schleyer, et al. (ed.), Encyclopedia of computational chemistry. John Wiley & Sons, New York, NY.
- 49.Nyholm, T. K., S. Ozdirekcan, and J. A. Killian. 2007. How protein transmembrane segments sense the lipid environment. Biochemistry 46:1457-1465. [DOI] [PubMed] [Google Scholar]
- 50.Pastor, R. W., B. R. Brooks, and A. Szabo. 1988. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 65:1409-1419. [Google Scholar]
- 51.Pearlman, D. A., D. A. Case, J. W. Caldwell, W. S. Ross, T. E. I. Cheatham, S. DeBolt, D. Ferguson, G. Seibel, and P. Kollman. 1995. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 91:1-41. [Google Scholar]
- 52.Poole, E., P. Strappe, H. P. Mok, R. Hicks, and A. M. Lever. 2005. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic 6:741-755. [DOI] [PubMed] [Google Scholar]
- 53.Ramachandran, G. N., C. Ramakrishnan, and V. Sasisekharan. 1963. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7:95-99. [DOI] [PubMed] [Google Scholar]
- 54.Rong, L., J. Zhang, J. Lu, Q. Pan, R. P. Lorgeoux, C. Aloysius, F. Guo, S. L. Liu, M. A. Wainberg, and C. Liang. 2009. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J. Virol. 83:7536-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryckaert, J.-P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327-341. [Google Scholar]
- 56.Sato, K., S. P. Yamamoto, N. Misawa, T. Yoshida, T. Miyazawa, and Y. Koyanagi. 2009. Comparative study on the effect of human BST-2/tetherin on HIV-1 release in cells of various species. Retrovirology 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauter, D., M. Schindler, A. Specht, W. N. Landford, J. Munch, K. A. Kim, J. Votteler, U. Schubert, F. Bibollet-Ruche, B. F. Keele, J. Takehisa, Y. Ogando, C. Ochsenbauer, J. C. Kappes, A. Ayouba, M. Peeters, G. H. Learn, G. Shaw, P. M. Sharp, P. Bieniasz, B. H. Hahn, T. Hatziioannou, and F. Kirchhoff. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schubert, U., S. Bour, A. V. Ferrer-Montiel, M. Montal, F. Maldarell, and K. Strebel. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70:809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schubert, U., and K. Strebel. 1994. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 68:2260-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao, J., S. W. Tanner, N. Thompson, and T. E. Cheatham. 2007. Clustering molecular dynamics trajectories. 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 3:2312-2334. [DOI] [PubMed] [Google Scholar]
- 61.Shen, L., D. Bassolino, and T. Stouch. 1997. Transmembrane helix structure, dynamics, and interactions: multi-nanosecond molecular dynamics simulations. Biophys. J. 73:3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, S. O., M. Eilers, D. Song, E. Crocker, W. Ying, M. Groesbeek, G. Metz, M. Ziliox, and S. Aimoto. 2002. Implications of threonine hydrogen bonding in the glycophorin A transmembrane helix dimer. Biophys. J. 82:2476-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strebel, K., T. Klimkait, and M. A. Martin. 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241:1221-1223. [DOI] [PubMed] [Google Scholar]
- 64.Strebel, K., J. Luban, and K. T. Jeang. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tokarev, A., M. Skasko, K. Fitzpatrick, and J. Guatelli. 2009. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res. Hum. Retrovir. 25:1197-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ueyama, T., T. Kusakabe, S. Karasawa, T. Kawasaki, A. Shimizu, J. Son, T. L. Leto, A. Miyawaki, and N. Saito. 2008. Sequential binding of cytosolic Phox complex to phagosomes through regulated adaptor proteins: evaluation using the novel monomeric Kusabira-Green system and live imaging of phagocytosis. J. Immunol. 181:629-640. [DOI] [PubMed] [Google Scholar]
- 67.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Meer, G., D. R. Voelker, and G. W. Feigenson. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, J., R. M. Wolf, J. W. Caldwell, P. A. Kollman, and D. A. Case. 2004. Development and testing of a general amber force field. J. Comput. Chem. 25:1157-1174. [DOI] [PubMed] [Google Scholar]
- 70.Willey, R. L., F. Maldarelli, M. A. Martin, and K. Strebel. 1992. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J. Virol. 66:226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida, T., H. Ebina, and Y. Koyanagi. 2009. N-linked glycan-dependent interaction of CD63 with CXCR4 at the Golgi apparatus induces downregulation of CXCR4. Microbiol. Immunol. 53:629-635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.