Novel role and mechanism of protein inhibitor of activated STAT1 in spatial learning
PIAS1 plays a critical role in spatial learning in rats by regulating STAT1 sumoylation and DNA-binding activity.
Keywords: phosphorylation, PIAS1, spatial learning, STAT1, sumoylation
Abstract
By using differential display PCR, we have previously identified 98 cDNA fragments from rat dorsal hippocampus, which are expressed differentially between the fast learners and slow learners from water-maze learning task. One cDNA fragment, which showed a higher expression level in fast learners, encodes the rat protein inhibitor of activated STAT1 (pias1) gene. Spatial training induced a significant increase in PIAS1 expression in rat hippocampus. Transient transfection of the wild-type (WT) PIAS1 plasmid to CA1 neurons facilitated, whereas transfection of PIAS1 siRNA impaired spatial learning in rats. Meanwhile, PIAS1WT increased STAT1 sumoylation, decreased STAT1 DNA binding and decreased STAT1 phosphorylation at Tyr-701 associated with spatial learning facilitation. But PIAS1 siRNA transfection produced an opposite effect on these measures associated with spatial learning impairment. Further, transfection of STAT1 sumoylation mutant impaired spatial acquisition, whereas transfection of STAT1 phosphorylation mutant blocked the impairing effect of PIAS1 siRNA on spatial learning. In this study, we first demonstrate the role of PIAS1 in spatial learning. Both posttranslational modifications (increased sumoylation and decreased phosphorylation) mediate the effect of PIAS1 on spatial learning facilitation.
Introduction
It is well known that long-term memory formation requires de novo RNA and protein synthesis. Experiments from rats showed that inhibition of mRNA and protein synthesis impairs long-term memory formation in various learning tasks (Davis and Squire, 1984; Matthies, 1989; Lee et al, 1992). This evidence suggests that neuronal activities associated with learning result in the expression of various genes, and the protein products of these genes have an important role in memory formation. Different strategies have been used to identify these genes in different animals in the past. For example, by using two-dimensional gel analysis, Castellucci et al (1988) have identified several candidate proteins that are related to the process of long-term sensitization in Aplysia. Screening in Drosophila mutant by using the inducible transgenic method has yielded approximately 10 genes that are associated with olfactory learning and memory (Tully, 1996). By using microarray analysis, Cavallaro et al (2002) have identified 140 genes in the hippocampus that are associated with water-maze learning in rats. Similar microarray analysis also identified 50 genes that are differentially expressed between superior learners and impaired learners from water-maze learning in aged rats (Burger et al, 2007). By using differential display polymerase chain reaction (DD-PCR), we have earlier identified the integrin-associated protein gene that is associated with memory formation of one-way inhibitory avoidance learning in rats (Huang et al, 1998). More recently, by using the same method, we have identified 98 cDNA fragments from rat hippocampal CA1 area, which are differentially expressed between fast learners and slow learners from water-maze learning task in rats, and one of these cDNA fragments encodes the serum- and glucocorticoid-inducible kinase (sgk) gene (Tsai et al, 2002). Further studies demonstrate that sgk expression has a critical role in spatial memory formation and long-term potentiation in rats (Tsai et al, 2002; Ma et al, 2006; Tai et al, 2009). Moreover, sgk expression was increased after eyeblink conditioning in mice (Park et al, 2006). These studies demonstrate the importance of mRNA and protein synthesis in learning and memory formation. In addition to the sgk gene, we have identified other genes that are also associated with spatial learning in our previous report (Tsai et al, 2002). In this study, we focused on the role of another gene identified previously and examined the molecular mechanism of this gene involved in spatial learning in rats.
Results
Identification of the protein inhibitor of activated STAT1 (pias1) gene by DD–PCR
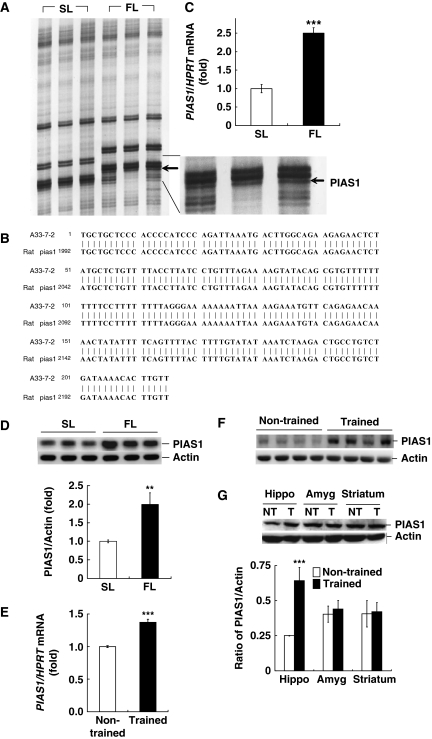
By using DD–PCR, 98 cDNA fragments were differentially expressed between fast learners and slow learners from water maze learning from our previous study (Tsai et al, 2002). When the primer set H-A33 (5′-end primer sequence as 5′-AAGCTTGCTGCTC-3′) and H-T11A (3′-end primer sequence as 5′-AAGCTTTTTTTTTTTA-3′) was used, one identified cDNA fragment that was 215 bp in length showed 100% sequence homology to the 3′-end region of the rat pias1 gene (Figure 1B;data accession number for PIAS1: NM_001106829). The expression level of this gene is much higher in the dorsal hippocampus of fast learners than slow learners (Figure 1A).
Figure 1.
Identification of the pias1 gene, and PIAS1 expression is increased after spatial training. (A) DD–PCR of hippocampal RNA associated with water maze learning in rats. FL, fast learners; SL, slow learner. The lower right panel is the magnification of the portion marked by solid lines. (B) Alignment of the sequence of A33-7-2 (the arbitrary primers used) with rat pias1. The numbers correspond to the cDNA sequences. Vertical lines indicate identity. (C) Analysis of pias1 mRNA level in FL and SL by Q–PCR. (D) Analysis of PIAS1 protein level in FL and SL by western blot. (E) Analysis of pias1 mRNA level in trained and non-trained (swimming control) animals. (F) Representative gel pattern showing PIAS1 protein level in CA1 area from trained and non-trained animals. (G) Representative gel pattern and statistics for PIAS1 protein level in CA1 area, amygdala and striatum from trained and non-trained animals. N=6–7 each group. Data are mean±s.e.m. **P<0.01 and ***P<0.001 from Student's t-test.
Spatial training increases PIAS1 expression in rat hippocampus
Quantitative real-time PCR (Q–PCR) was first carried out to confirm the results obtained from DD–PCR. Results revealed that there is a higher pias1 mRNA level in CA1 area of fast learners (n=7) than slow learners (n=7; t1,12=13.77, P<0.001; Figure 1C). Tissues from the same animals were also subjected to western blot determination of PIAS1 protein expression. Results revealed that the fast learners also have a higher PIAS1 protein level than slow learners (t1,12=4.11, P<0.01; Figure 1D). Next, we examined whether spatial training induces PIAS1 expression. Animals were randomly divided into the trained group and the non-trained group (n=7 each group). Animals in the trained group were subjected to water-maze learning with four trials in a session and two sessions in all. One session was given in the morning and the other session given in the afternoon. This is the behavioural paradigm that we used to obtain the pias1 cDNA fragment (Tsai et al, 2002). Animals in the non-trained group swam for the same period of time for each trial as the trained group (take the mean latency value for each trial), except that the visual cues and the platform were removed. Therefore, the spatial relationship between these two cannot be established. Animals in both groups were killed at the end of training and their hippocampal CA1 tissues were dissected out for pias1 mRNA and protein determination. Results from Q–PCR revealed that spatial training increased pias1 mRNA level in the CA1 area (t1,12=7.32, P<0.001; Figure 1E). Further analysis indicated that spatial training also increased PIAS1 protein level in the CA1 area (t1,12=4.62, P<0.001), but this change was not observed in the amygdala and striatum, areas not associated with spatial learning (t1,12=0.13 and 0.46, both P>0.05; Figure 1F and G).
Overexpression of PIAS1 enhances spatial learning
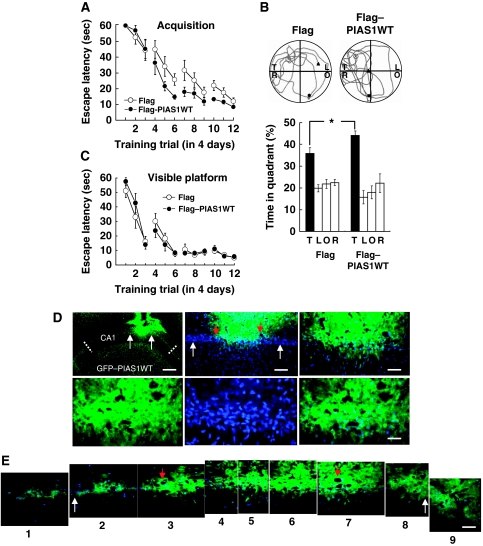
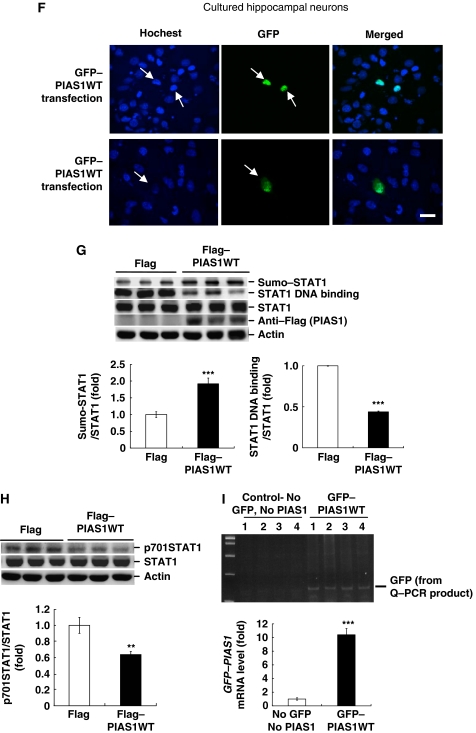
Results from the above experiments demonstrate a positive correlation between the expression of PIAS1 and spatial learning. In this study, we examined whether overexpression of PIAS1 enhances spatial learning. Animals were randomly divided into two groups to receive Flag-vector transfection (n=10) or Flag-PIAS1 wild-type (WT) transfection (n=8) to CA1 area and subjected to water-maze learning. Results revealed that PIAS1WT transfection enhanced acquisition performance (F1,16=7.07, P<0.05; Figure 2A). By analysing their probe trial performance, we found that PIAS1WT-transfected animals spent more time in the target quadrant than control animals (t1,16=2.48, P<0.05; Figure 2B). But their acquisition performance under visible platform learning was not different (F1,16=0.05, P>0.05; Figure 2C), indicating that their visual and motor functions were not altered. What they have acquired is the spatial relationship between the visual cues and the location of the platform. To confirm the transfection and expression of PIAS1WT plasmid in CA1 area, GFP-tagged PIAS1WT plasmid was transfected to the CA1 area and DAPI was added to the tissue sessions. Immunohistochemistry was carried out for visualization of the fluorescence for GFP and DAPI. Results revealed apparent immunofluorescence for GFP (green) and DAPI (blue) and their co-localization in CA1 area (Figure 2D and E). The transfected area is approximately 24% of total CA1 area viewed from a single plane (Figure 2D, upper left panel). Images at a higher magnification showed the area of GFP–PIAS1WT transfection from the most left to the most right tissue sessions (Figure 2E). Serial tissue sessions from slide 2 to slide 8 in Figure 2E represent the transfected CA1 area indicated by two white arrows in the upper left and upper middle panels of Figure 2D, which is around 620 μm in length. To estimate the transfection efficiency, we have counted the number of GFP-positive cells over that of DAPI-positive cells in CA1 area from slide 2 to slide 8 in Figure 2E. They are 57/71, 69/77, 42/43, 71/75, 110/116, 111/118 and 51/56 in order, which yields the averaged transfection efficiency approximates 92%. The expression pattern of GFP–PIAS1 in adjacent tissue sessions rostral and caudal to the present tissue session is shown in Supplementary Figure 1. These immunohistochemistry results indicated that PIAS1 seems to be expressed in both the nucleus and cytosol area of CA1 cells. In some cells, PIAS1 localization to the boundary of the nucleus is clearly seen (distinct green fluorescence dots in Figure 2E). To further examine the subcellular distribution of PIAS1 in hippocampal neurons, we have transfected the GFP–PIAS1WT plasmid to cultured hippocampal neurons and used Hochest staining for nucleus staining. Results revealed that PIAS1 is localized to the nucleus in all the neurons examined (Figure 2F, upper panels). But in approximately 10% of the neurons examined, PIAS1 is also distributed in the cytosol area at a much weaker intensity (Figure 2F, lower middle panel).
Figure 2-.
Overexpression of PIAS1 facilitates spatial learning, increases STAT1 sumoylation, decreases STAT1 DNA binding and decreased STAT1 phosphorylation at Tyr-701. PIAS1WT plasmid or Flag vector was transfected to rat CA1 area and animals were subjected to (A) water-maze learning and (B) probe trail test. T, target quadrant; L, left quadrant; O, opposite quadrant; R, right quadrant; •, start point; ▴, end point (C). The same transfection was made to different groups of rats and they were subjected to visible platform learning. (D) Immunohistochemistry showing the area of GFP–PIAS1WT transfection and the expression of PIAS1 in CA1 cell layer at different magnifications. Cells that show both green fluorescence (GFP) and blue fluorescence (DAPI) are cells successfully transfected with the plasmid. Dotted lines indicate the CA1 area. White arrows indicate the area of transfection and red arrows are markers for visualization of enlarged photos in (E). Scale bars equal 400 μm for the upper left panel, 100 μm for the upper middle panel, 50 μm for the upper right panel and 25 μm for the lower panels. (E) Enlargement of photos in upper panels of (D). The white arrows in slide 2 and slide 8 correspond to the two white arrows seen in the upper panels in (D). The red arrows (in slide 3 and slide 7) match with that seen in the upper middle panel in (D). Scale bar equals 25 μm.
Figure 2-.
(F) Immunostaining of Hochest and GFP in cultured hippocampal neurons after GFP–PIAS1WT transfection. Cells that show both green fluorescence (GFP) and blue fluorescence (Hochest) are cells successfully transfected with the plasmid. Scale bar equals 25 μm. (G) Representative gel pattern and statistics for STAT1 sumoylation, STAT1 DNA binding and anti-Flag (verification of transfection) after Flag–PIAS1WT transfection and probe trial test. (H) Representative gel pattern and statistics for STAT1 phosphorylation at Tyr-701 after Flag–PIAS1WT transfection and probe trial test. N=8–10 each group. (I) DNA gel electrophoresis showing the GFP fragment (129 bp) from Q–PCR analysis after GFP–PIAS1WT transfection to rat CA1 area. Control animals received PEI (the transfection reagent) injection only. The primers used for Q–PCR analysis of GFP–PIAS1 mRNA level were designed within the sequence of GFP (upper panel). Quantitative analysis and statistics showing the effect of GFP–PIAS1WT transfection on GFP–PIAS1 mRNA expression (lower panel). N=6 each group. Data are mean±s.e.m. *P<0.05, **P<0.01 and ***P<0.001 from Student's t-test.
PIAS1 increases STAT1 sumoylation, decreases STAT1 DNA binding and decreases STAT1 phosphorylation at Tyr-701 associated with spatial learning facilitation
In this experiment, we studied the mechanism underlying PIAS1-mediated spatial learning facilitation. PIAS1 is known as the inhibitor of signal transducers and activators of transcription 1 (STAT1) and it inhibits the DNA-binding activity of STAT1 (Liu et al, 1998). It is also known that STAT proteins have to be first phosphorylated at the tyrosine residue and dimerize before they translocate to the nucleus to regulate gene expression (Ihle, 2001). We therefore examined whether PIAS1 may affect STAT1 phosphorylation associated with spatial learning. In addition, PIAS1 is known as a SUMO E3 ligase that promotes the sumoylation of STAT1 (Ungureanu et al, 2003). Further, PIAS1 sumoylation of STAT1 decreases the DNA-binding activity of STAT1 and negatively regulates STAT1-mediated gene transcription (Ungureanu et al, 2005; Song et al, 2006). Therefore, we also examined whether PIAS1 may alter STAT1 sumoylation and STAT1 DNA-binding activity associated with spatial learning. Animals transfected with PIAS1WT plasmid were killed after the probe trial test and their CA1 tissues were dissected out and subjected to western blot analysis of STAT1 sumoylation, STAT1 DNA binding and phospho (p)-Y701 STAT1 level. Results showed that PIAS1WT transfection increased STAT1 sumoylation (t1,16=5.17, P<0.001) and decreased STAT1 DNA binding (t1,16=9.04, P<0.001; Figure 2G). PIAS1WT transfection was further confirmed by western blot using the anti-Flag antibody (Figure 2G). Moreover, PIAS1WT transfection decreased the level of STAT1 phosphorylation at Tyr-701 without altering the STAT1 protein level (t1,16=3.75, P<0.01; Figure 2H). Additional experiment was carried out to examine whether GFP–PIAS1WT transfection actually increases the transcription of pias1. GFP–PIAS1WT fusion plasmid was transfected to the CA1 area in separate animals (n=6). Control animals received transfection reagent (PEI) injection only (n=6). At 48 h after transfection, their CA1 tissues were subjected to Q–PCR analysis of GFP–PIAS1 mRNA level with primers designed within the sequence of GFP (Supplementary Table 1). The Q–PCR product was further subjected to DNA gel electrophoresis for visualization of the GFP signal. Results revealed an apparent GFP band (129 bp in length) in GFP–PIAS1WT-transfected animals, but not in control animals. Further analysis indicated that GFP–PIAS1WT transfection increased GFP–PIAS1 mRNA level in the CA1 area (t1,10=11.03, P<0.001; Figure 2I).
PIAS1 siRNA decreases STAT1 sumoylation, increases STAT1 DNA binding and increases STAT1 phosphorylation at Tyr-701 associated with spatial learning impairment
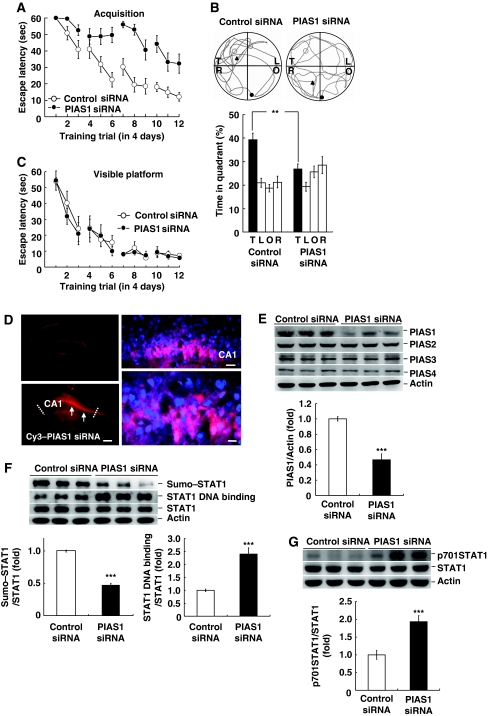
In this experiment, we further examined the role of PIAS1 in spatial learning by knocking down the expression of PIAS1. Animals were randomly divided into two groups to receive control siRNA (without Cy3) or Cy3–PIAS1 siRNA transfection to CA1 area (n=9 each group) and subjected to water-maze learning. Results showed that PIAS1 siRNA transfection impaired acquisition performance in rats (F1,16=29.4, P<0.001; Figure 3A). Further probe trial analysis indicated that PIAS1 siRNA transfection decreased the time that animals spent in the target quadrant (t1,16=12.68, P<0.01; Figure 3B). But their acquisition performance under visible platform learning was not different (F1,16=0.03, P>0.05; Figure 3C). Immunohistochemistry against Cy3 indicated that PIAS1 siRNA was indeed transfected to CA1 neurons (Figure 3D). Similar to that of PIAS1 plasmid transfection, the averaged area of PIAS1 siRNA transfection is approximately 25% of total CA1 area viewed from a single plane as indicated by arrows (Figure 3D, lower left panel). To assess the transfection efficiency, tissue sessions were added with DAPI and visualized under confocal microscope. Cells showed double staining of red fluorescence (Cy3) and blue fluorescence (DAPI) are cells successfully transfected with Cy3–PIAS1 siRNA (Figure 3D, right panels). The number of Cy3-positive neurons is 46 and the number of DAPI-positive neurons is 51. The estimated transfection efficiency is approximately 90% (Figure 3D, lower right). Further western blot analysis showed a decrease in PIAS1 protein level in PIAS1 siRNA-transfected animals (t1,16=7.88, P<0.001) (Figure 3E). But PIAS1 siRNA transfection did not affect the expression of PIAS2, PIAS3 and PIAS4 (Figure 3E). We next examined whether PIAS1 siRNA alters STAT1 sumoylation, STAT1 DNA binding and STAT1 phosphorylation associated with spatial learning. Results from western blot indicated that PIAS1 siRNA transfection decreased the level of STAT1 sumoylation (t1,16=17.95, P<0.001) and increased the level of STAT1 DNA binding (t1,16=4.49, P<0.001; Figure 3F). Meanwhile, PIAS1 siRNA increased the level of STAT1 phosphorylation at Tyr-701 without altering the STAT1 protein level (t1,16=4.58, P<0.001; Figure 3G).
Figure 3.
Transfection of PIAS1 siRNA impairs spatial learning, decreases STAT1 sumoylation, increases STAT1 DNA binding and increases STAT1 phosphorylation at Tyr-701. PIAS1 siRNA or control siRNA was transfected to CA1 area and rats were subjected to (A) water-maze learning and (B) probe trial test. (C) The same transfection was made to different groups of rats and they were subjected to visible platform learning. (D) Immunohistochemical staining against Cy3 and DAPI showing PIAS1 siRNA transfection to CA1 area at different magnifications. A control image (control siRNA without Cy3) is shown in the upper left panel and Cy3 image is shown in the lower left panel. The dotted lines indicate the CA1 area and arrows indicate the area of transfection. Scale bar=360 μm (lower left panel). Cells show both red fluorescence (Cy3) and blue fluorescence (DAPI) are cells successfully transfected with PIAS1 siRNA (right panels). Scale bar=40 μm for upper right panel and scale bar=10 μm for lower right panel. (E) Representative gel pattern for protein levels of PIAS1, PIAS2, PIAS3, PIAS4 and the statistics for PIAS1 expression after PIAS1 siRNA transfection to CA1 area and probe trial test. (F) Representative gel pattern and statistics for STAT1 sumoylation and STAT1 DNA binding after PIAS1 siRNA transfection to CA1 area and probe trial test. (G) Representative gel pattern and statistics for STAT1 phosphorylation at Tyr-701 after PIAS1 siRNA transfection to CA1 area and probe trial test. N=9 each group. Data are expressed as in Figure 2. **P<0.01 and ***P<0.001 from Student's t-test.
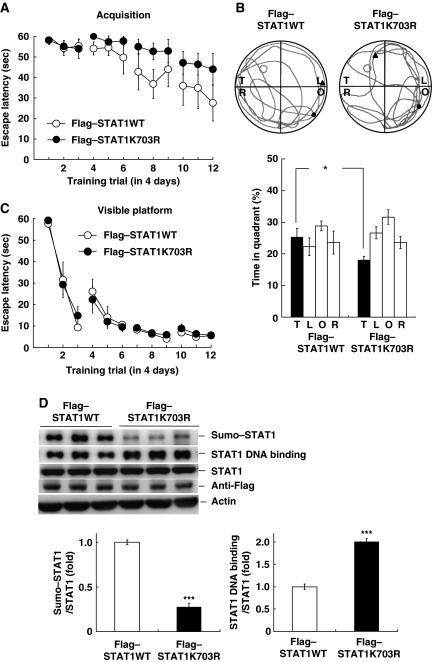
Blockade of STAT1 sumoylation impairs spatial learning
The above results showed that PIAS1 sumoylation of STAT1 is positively associated with spatial learning. If sumoylation of STAT1 has an important function in spatial learning, we expected that blockade of STAT1 sumoylation should impair spatial learning. Because STAT1 is sumoylated at Lys-703 (Ungureanu et al, 2003), we have transfected the Flag-tagged STAT1 sumoylation mutant (Flag–STAT1K703R) to CA1 area and examined its effect on spatial learning. Control animals received Flag–STAT1WT transfection (n=7 each group). Results revealed that STAT1K703R transfection impaired spatial acquisition when compared with the STAT1WT controls (t1,12=4.94, P<0.05; Figure 4A). Further probe trial analysis indicated that STAT1K703R transfection reduced the time that animals spent in the target quadrant (t1,12=4.92, P<0.05; Figure 4B). But their acquisition performance under visible platform learning was not different (F1,12=0.04, P>0.05; Figure 4C). Further western blot analysis revealed that STAT1K703R transfection decreased the level of STAT1 sumoylation (t1,12=16.39, P<0.001) and increased the level of STAT1 DNA binding (t1,12=11.45, P<0.001) compared with the STAT1WT controls without altering the STAT1 protein level (Figure 4D). STAT1WT and STAT1K703R transfections were confirmed by western blot against the Flag tag.
Figure 4.
Transfection of STAT1K703R impairs spatial learning. Flag–STAT1WT or Flag–STAT1K703R plasmid was transfected to CA1 area and rats were subjected to (A) water-maze learning and (B) probe trial test. (C) The same transfection was made to different groups of rats and animals were subjected to visible platform learning. (D) Representative gel pattern and statistics for STAT1 sumoylation, STAT1 DNA binding and anti-Flag (verification of transfection) after plasmid transfection and probe trial test. N=7 each group. Data are expressed as in Figure 2. *P<0.05 and ***P<0.001 from Student's t-test.
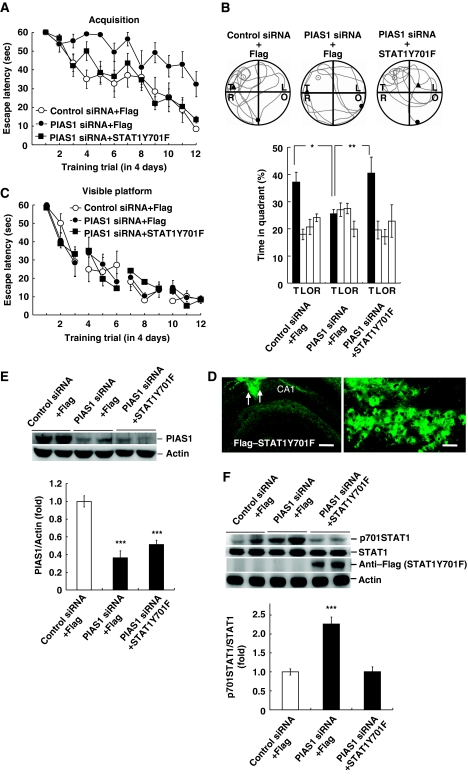
PIAS1 siRNA impairs spatial learning through increased STAT1 phosphorylation at Tyr-701
The above results revealed that PIAS1 negatively regulates STAT1 phosphorylation at Tyr-701 associated with spatial learning facilitation. But whether increased STAT1 phosphorylation at Tyr-701 actually mediates PIAS1 siRNA-induced learning impairment is not known. This issue was examined here. Animals were randomly divided into three groups to receive control siRNA (0.5 μl)+Flag-vector (0.25 μl), PIAS1 siRNA (0.5 μl)+Flag-vector (0.25 μl) and PIAS1 siRNA (0.5 μl)+STAT1Y701F (0.25 μl) transfections (n=9 each group) and subjected to water-maze learning. Results revealed that 0.5 μl PIAS1 siRNA transfection impaired acquisition performance (F2,24=8.27, P<0.01; q=5.29, P<0.01), but this effect was blocked by 0.25 μl STAT1Y701F co-transfection (q=4.66, P<0.01 comparing the PIAS1 siRNA+Flag-vector group with PIAS1 siRNA+STAT1Y701F group; Figure 5A). This result suggests that interference of PIAS1 siRNA signalling for about 50% (in terms of volume and presumably percentage of neurons) by STAT1Y701F effectively blocks the impairing effect of PIAS1 siRNA on spatial learning. Probe trial analysis indicated that animals transfected with PIAS1 siRNA spent less time in the target quadrant (F2,24=4.19, P<0.05; q=2.99, P<0.05), but this effect was also blocked by STAT1Y701F co-transfection (q=3.93, P<0.01 comparing the PIAS1 siRNA+Flag-vector group with PIAS1 siRNA+STAT1Y701F group; Figure 5B). Their acquisition performance under visible platform learning was not different (F2,24=0.02, P>0.05; Figure 5C). Because only 0.25 μl of STAT1Y701F plasmid was transfected to the CA1 area in this experiment, immunohistochemistry showed a more limited area of transfection and expression of Flag-positive cells, as indicated by arrows in Figure 5D (approximately one-third of that seen in Figure 2D). Further analysis indicated that PIAS1 siRNA transfection decreased PIAS1 protein level in both PIAS1 siRNA+Flag-vector group and PIAS1 siRNA+STAT1Y701F group (F2,24=26.76, P<0.001; q=9.91 and 7.53, respectively, when compared with the control group, both P<0.001; Figure 5E). Besides, PIAS1 siRNA transfection increased the level of STAT1 phosphorylation at Tyr-701 (F2,24=32.77, P<0.001; q=9.95, P<0.001 comparing the PIAS1 siRNA+Flag-vector group with control group), but this effect was reversed by STAT1Y701F co-transfection (q=9.27, P<0.001 comparing the PIAS1 siRNA+Flag-vector group with PIAS1 siRNA+STAT1Y701F group; Figure 5F). The STAT1 protein level remained unchanged.
Figure 5.
Transfection of STAT1Y701F blocks the impairing effect of PIAS1 siRNA on spatial learning. Flag-vector, PIAS1 siRNA or PIAS1 siRNA together with STAT1Y701F was transfected to CA1 area and rats were subjected to (A) water-maze learning and (B) probe trial test. (C) The same transfection was made to different groups of rats and they were subjected to visible platform learning. (D) Immunohistochemistry showing a smaller area of transfection for 0.25μl Flag–STAT1Y701F injection to CA1 area as indicated by arrows. The anti-Flag antibody and FITC-conjugated IgG secondary antibody were used. Scale bar=240μm for the left panel and scale bar=15μm for the right panel. (E) Representative gel pattern and statistics for PIAS1 protein level after probe trial test. (F) Representative gel pattern and statistics for STAT1 phosphorylation at Tyr-701 and anti-Flag (verification of transfection) after probe trial test. N=9 each group. Data are expressed as in Figure 2. *P<0.05, **P<0.01 and ***P<0.001 from Student's t-test.
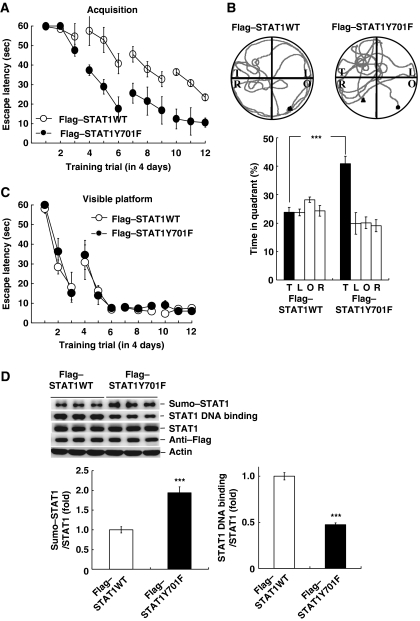
STAT1Y701F transfection enhances spatial learning
If STAT1 phosphorylation at Tyr-701 is one of the mechanisms underlying PIAS1 siRNA-mediated spatial learning impairment, it is expected that inhibition of STAT1 phosphorylation at Tyr-701 should enhance spatial learning. This issue was examined here. Animals were randomly divided into two groups to receive Flag–STAT1WT transfection or Flag–STAT1Y701F transfection (n=8 for each group) and subjected to water-maze learning. Results revealed that STAT1Y701F transfection enhanced spatial acquisition when compared with the STAT1WT controls (F1,14=18.58, P<0.001; Figure 6A). Probe trial analysis indicated that STAT1Y701F transfection increased the time that animals spent in the target quadrant (t1,14=5.54, P<0.001; Figure 6B). But the same transfection did not affect their acquisition performance under visible platform learning (F1,12=0.84, P>0.05; Figure 6C). Further western blot analyses indicated that STAT1Y701F transfection increased the level of STAT1 sumoylation (t1,14=5.37, P<0.001) and decreased the level of STAT1 DNA binding (t1,14=13.17, P<0.001) without altering the STAT1 protein level (Figure 6D). STAT1WT and STAT1Y701F transfections were confirmed by western blot against the Flag tag.
Figure 6.
Transfection of STAT1Y701F enhances spatial learning. Flag–STAT1WT or Flag–STAT1Y701F plasmid was transfected to CA1 area and rats were subjected to (A) water-maze learning and (B) probe trial test. (C) The same transfection was made to different groups of rats and they were subjected to visible platform learning. (D) Representative gel pattern and statistics for STAT1 sumoylation, STAT1 DNA binding and anti-Flag (verification of transfection) after probe trial test. N=8 each group. Data are expressed as in Figure 2. ***P<0.001 from Student's t-test.
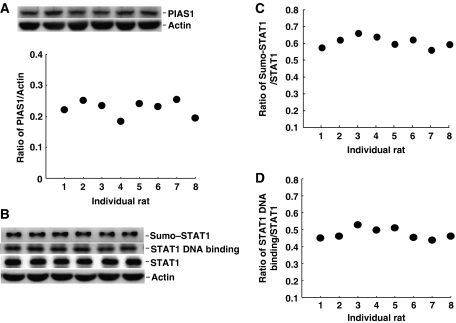
Level of PIAS1 expression, STAT1 sumoylation and STAT1 DNA binding is similar in naive animals
In our DD–PCR screening and Q–PCR analysis, we have found that the pias1 mRNA level is higher in fast learners than slow learners (Figure 1A and C). Although we have demonstrated that pias1 mRNA expression and PIAS1 protein expression are both induced after spatial training (Figure 1E and F), we like to further examine whether the fast learners may have a constitutively higher level of PIAS1 expression in the hippocampus. The level of PIAS1 expression, STAT1 sumoylation and STAT1 DNA binding in CA1 area was determined by western blot in eight randomly chosen, naive rats. Results revealed that the difference was within 38% for PIAS1 expression (Figure 7A), 18% for STAT1 sumoylation (Figure 7B and C) and 22% for STAT1 DNA binding (Figure 7B and D) among these individual animals.
Figure 7.
Level of PIAS1 expression, STAT1 sumoylation and STAT1 DNA binding is similar in CA1 area of naive rats. (A) Representative gel pattern and individual data of PIAS1 expression in randomly chosen, naive rats. (B) Representative gel pattern and (C, D) individual data of STAT1 sumoylation and STAT1 DNA binding in randomly chosen, naive rats. N=8.
Discussion
PIAS1 was initially identified as an inhibitor of STAT1 and it blocks the DNA-binding activity of STAT1 and inhibits the transcriptional activity of STAT1 in response to cytokine stimulation (Liu et al, 1998; Liao et al, 2000). PIAS1 also inhibits interferon-inducible gene expression and has an important function in innate immune responses through negative regulation of STAT1 (Liu et al, 2004). Further studies demonstrate that PIAS1 also regulates the activity of other transcription factors in addition to STAT1, such as NF-κB and Smads, and PIAS1 regulates these transcription factors through distinct mechanisms (Shuai and Liu, 2005; Sharrocks, 2006). A more recent study showed that proinflammatory stimuli could activate PIAS1 through IKKα-mediated phosphorylation of PIAS1 at Ser-90. Activated PIAS1 further inhibits the DNA-binding activity of STAT1 and NF-κB to target genes to reduce inflammation (Liu et al, 2007). PIAS1 is also a transcriptional regulator that possesses small ubiquitin-like modifier (SUMO) E3 ligase activity (Kahyo et al, 2001) and it promotes the sumoylation of several proteins including STAT1 (Johnson and Gupta, 2001; Shuai and Liu, 2005). PIAS1 is also involved in PPARγ-mediated anti-inflammation by promoting the sumoylation of PPARγ (Pascual et al, 2005). However, with the role of PIAS1 involved in inflammation and immune response been studied to a certain extent, other physiological functions that PIAS1 may regulate are barely known.
In this study, we have identified a novel role of PIAS1 by showing that overexpression of PIAS1 in CA1 neurons facilitated, whereas knockdown of PIAS1 in CA1 neurons impaired spatial learning in rats. This effect of PIAS1 is not likely due to its effect on immune response because overexpression of PIAS1 increased the expression of PIAS1, but it did not affect the expression of TNFα and IL-6, neuro-inflammatory cytokines in CNS (Arvin et al, 1996; Supplementary Figure 2). In studying the molecular mechanism underlying PIAS1-mediated spatial learning facilitation, we have found that overexpression of PIAS1 increased the sumoylation of STAT1 and decreased the DNA-binding activity of STAT1 associated with spatial learning facilitation. However, PIAS1 siRNA transfection decreased the sumoylation of STAT1 and increased the DNA-binding activity of STAT1 associated with spatial learning impairment. Further experiment with western blot confirmed that PIAS1 is mainly distributed in the nucleus. Although STAT1 is mostly distributed in the cytoplasm, STAT1 sumoylation by PIAS1 mainly takes place in the nucleus (Supplementary Figure 3). To exclude the possible off-target effect of PIAS1 siRNA used in this study, we have adopted another sequence of PIAS1 siRNA (Weber et al, 2009) and have found that the latter PIAS1 siRNA also impaired spatial learning and decreased PIAS1 expression in CA1 area (Supplementary Figure 4). These results are consistent with the reports that one important mechanism for PIAS1 regulation of STAT1 activity is sumoylation (Ungureanu et al, 2003), and that STAT1 can be SUMO-modified by PIAS1 on Lys-703 and this SUMO conjugation decreases the DNA-binding activity and transcriptional activity of STAT1 (Ungureanu et al, 2005; Song et al, 2006). The role of STAT1 sumoylation involved in spatial learning is further supported by our finding that transfection of STAT1K703R, the STAT1 sumoylation mutant, impaired water-maze performance. But our results cannot exclude the possibility that PIAS1 may also sumoylate other molecules in addition to STAT1 to facilitate spatial learning. For example, focal adhesion kinase and metabotropic glutamate receptor 8 could both be sumoylated by PIAS1 (Kadare et al, 2003; Tang et al, 2005) and both molecules are implicated in neuronal plasticity. This speculation is consistent with the idea that sumoylation is not targeted to nuclear proteins and sumoylation may have a function in regulation of certain neuronal functions, such as synaptic transmission (Scheschonka et al, 2006). To further address the issue of sumoylation involved in learning, we have transfected SUMO-1 siRNA (Meinecke et al, 2007) to rat CA1 area and have found that it did not affect spatial acquisition (Supplementary Figure 5A and B). However, when the cyclic AMP-responsive element-binding protein (CREB) sumoylation mutant (Comerford et al, 2003) was transfected to rat CA1 area, it facilitated spatial acquisition, as that of CREBWT plasmid transfection (Supplementary Figure 5C and D). These results together suggest that sumoylation of different molecules may have different effect on spatial learning, and this partially explains why SUMO-1 siRNA does not have a significant effect on spatial acquisition. In this study, we have found that rats transfected with STAT1WT plasmid seem to show a slower acquisition rate compared with the control animals (Figures 4A and 6A). Thus, we have conducted an additional experiment to assess the effect of STAT1WT transfection on spatial learning. Our preliminary results showed that STAT1WT transfection impaired spatial acquisition (Supplementary Figure 6). The role of STAT1 possibly involved in spatial learning and memory is a separate issue and is currently under investigation.
In addition to increased STAT1 sumoylation by PIAS1, another mechanism underlying PIAS1-mediated spatial learning facilitation is decreased STAT1 phosphorylation by PIAS1. Our results showed that overexpression of PIAS1 decreased STAT1 phosphorylation at Tyr-701 associated with spatial learning enhancement, whereas PIAS1 siRNA increased STAT1 phosphorylation at Tyr-701 associated with spatial learning impairment. Moreover, transfection of the STAT1 phosphorylation mutant, STAT1Y701F, blocked the impairing effect of PIAS1 siRNA on spatial learning. STAT1Y701F at a higher concentration by itself enhanced spatial learning. Meanwhile, it increased the sumoylation of STAT1 and decreased the DNA-binding activity of STAT1. These results are consistent with the observation that STAT1 phosphorylation at Tyr-701 induced by interferon-β inhibits the sumoylation of STAT1 and support the idea that STAT1 phosphorylation and STAT1 sumoylation are mutually exclusive (Zimnik et al, 2009). However, although we have found that decreased STAT1 phosphorylation mediates the enhancing effect of PIAS1 on spatial learning, the molecular mechanism underlying PIAS1 inhibition of STAT1 phosphorylation is not known and needs to be further investigated. Moreover, in addition to increase in STAT1 sumoylation and decrease in STAT1 phosphorylation, there are other mechanisms that may also contribute to the enhancing effect of PIAS1 on spatial learning. For example, PIAS1 is shown as a binding partner for CREB-binding protein (CBP) in epithelial cells (Yin et al, 2005). Further, PIAS1 recruits the co-activator CBP or p300 to enhance CREB-mediated transcription (Shuai and Liu, 2005). Thus, PIAS1 may facilitate spatial learning through enhanced interaction with CBP and/or p300 to enhance CREB-mediated gene transcription. Experiments are undertaken to further investigate the molecular mechanism underlying PIAS1-mediated spatial learning facilitation and the signalling pathway in regulation of PIAS1 expression in the hippocampus.
This study showed that fast learners have a higher PIAS1 expression level than slow learners. This result raises the possibility that there may be a subgroup of rats that have a constitutively higher PIAS1 level in the hippocampus. To address this issue, we have examined the endogenous PIAS1 protein level in eight randomly chosen, naive rats. Results revealed that the PIAS1 level in hippocampal CA1 area is not very different among these individual animals (with the maximum difference approximates 38%; Figure 7A). This result suggests that PIAS1 expression is actually induced upon spatial training. When we examined hippocampal PIAS1 expression at longer intervals after training, we found that the magnitude of increase in PIAS1 expression is diminished at 3 days after training when compared with that immediately after training (Supplementary Figure 7 versus Figure 1F), and it is further diminished at 5 days after training. No apparent difference was observed at 21 days after training (Supplementary Figure 7). These results suggest that pias1 may function as an immediate early gene that is rapidly induced upon training. But it probably further activates other mechanisms to facilitate the learning process. In further addressing the issue whether PIAS1 may also facilitate memory consolidation, we have transfected PIAS1 siRNA to CA1 area in one of two equally trained groups of rats 2 days before the last training day (i.e. day 2), so that PIAS1 siRNA knocks down endogenous PIAS1 expression at the beginning stage of memory consolidation (Supplementary Figure 8). PIAS1 siRNA was transfected again at the end of training (i.e. day 4) to maintain its knockdown effect during the period of memory consolidation. Probe trial was performed 4 days after the last training day (i.e. day 8). Control animals received control siRNA transfection. Our results showed that PIAS1 siRNA transfection starts effective after 24 h (trial 9) and impairs memory consolidation (Supplementary Figure 8). This result suggests that PIAS1 may also have a function in memory consolidation. In another experiment, we have tried to examine the effect of PIAS1WT plasmid transfection on memory consolidation by measuring probe trial performance also at a longer interval after training. However, this would require repeated PIAS1 transfection for three times because the effect of plasmid transfection lasts for a shorter period of time (it significantly diminishes 72 h after transfection, as suggested by Abdallah et al (1996) and according to our previous experience) than siRNA transfection does (which lasts for 96 h in the hippocampus; Supplementary Figure 8D). Our result revealed that most of the animals transfected with PIAS1WT plasmid for three times show tissue inflammation in the CA1 area (DJC Tai, unpublished observation). Therefore, we cannot include this result and the effect of PIAS1 plasmid transfection can only be limited to spatial learning in this study. Furthermore, the effect of overexpression of PIAS1 on spatial learning (Figure 2A) is not as significant as that of knockdown of PIAS1 expression (Figure 3A). One speculation is that PIAS1 expression at a physiological level is sufficient to mediate signal transduction for spatial learning, whereas knockdown of endogenous PIAS1 expression blocks its essential signal transduction mediating spatial learning. To further extend the role of PIAS1 in memory processing, we have examined PIAS1 expression in the medial prefrontal cortex at 21 days after training, a time point that memory storage presumably takes place in this brain region (Liang et al, 1996). Results revealed that there is about 22% decrease in PIAS1 expression in trained animals compared with non-trained animals in this brain area (Supplementary Figure 9). This result suggests that PIAS1 may have a differential role in memory formation and memory storage. Alternatively, PIAS1 expression may be downregulated due to chronic inhibition of STAT1 by elevated PIAS1 level after spatial training because the promoter region of the pias1 gene may contain the STAT1-binding site based on the prediction from the Transcription Element Search System (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME). Future studies are required to elucidate the role of PIAS1 in memory storage and the regulation mechanism for PIAS1 expression. In addition, spatial training induced a nearly two-fold increase in PIAS1 protein level in the CA1 area, but the PIAS1 mRNA level was elevated for approximately 40% only (Figure 1E and G). One possible explanation for this is that translation takes place efficiently and that untranslated pias1 mRNA degrades rapidly. However, spatial training did not increase PIAS1 protein level in the amygdala and striatum. This result suggests that spatial training-induced PIAS1 expression is specific to the hippocampus and this result is consistent with the idea that the dorsal hippocampus has an important role in spatial learning and memory function (Moser et al, 1993). Moreover, the fact that fast learners have a higher PIAS1 protein level than slow learners raises the possibility that the fast learners may have a higher level of stress in the water maze, and stress may upregulate PIAS1 expression. To test this possibility, we have analysed corticosterone level in the blood of fast learners and slow learners and have found that fast learners do not have a higher corticosterone level than slow learners (Supplementary Figure 10). To further test this possibility, we have injected dexamethasone, a synthetic glucocorticoid, to CA1 area in separate animals and examined its effect on PIAS1 expression. Results revealed that dexamethasone did not apparently alter PIAS1 expression at the two doses examined (5 and 10ng; Supplementary Figure 10). Similar result was found when foot shock stress was given to the rats (Supplementary Figure 10). These results together suggest that fast learners do not have a higher level of stress than slow learners and the facilitating effect of PIAS1 on spatial learning is not due to stress.
In this study, transfection was made only to a limited area in CA1 neurons (about 620μm viewed from a single plane and 200 μm in thickness; that counts for less than 5% of total volume of the punched tissue), but significant biochemical and behavioural changes were observed. This is possibly because the protein extraction method we used is suitable for extraction of proteins in the cell, but there are presumably more fibres (such as the neuronal processes of CA1 neurons and dentate gyrus neurons and part of the corpus callosum) than cells in the punched area. When biochemical assays were performed, total amount of proteins, instead of total tissue volume, was used as a criterion. Thus, the percantage of transfected cells should be much more than 5% of total number of cells in the punched tissue. This is probably why significant biochemical changes were still observed in a limited transfection area. In another study, Han et al (2007) have found that transfection of the CREBWT plasmid to approximately 16–20% of amygdala cells successfully rescues fear memory deficit in CREB-deficient mice. Further, selective depletion of neurons overexpressing CREB after training blocks the expression of that fear memory (Han et al, 2009). These results suggest that activation of a specific subpopulation of neurons and their neuronal activity is sufficient to mediate memory process and to form memory trace. The same mechanism may take place in the hippocampus in this study. But the exact mechanism of how do these neurons communicate with other untransfected neurons to mediate the behavioural changes remains to be investigated. In addition, transfection was made to neurons near the central part of CA1 cell layer of dorsal hippocampus in this study. Whether CA1 neurons located more laterally (such as close to the midline and the boundary of CA2/CA3 layer) and ventrally contribute equally to spatial learning needs to be further examined. Future study with selective depletion of the transfected neurons in CA1 area would also help to elucidate the casual relationship between activation of these neurons and spatial learning.
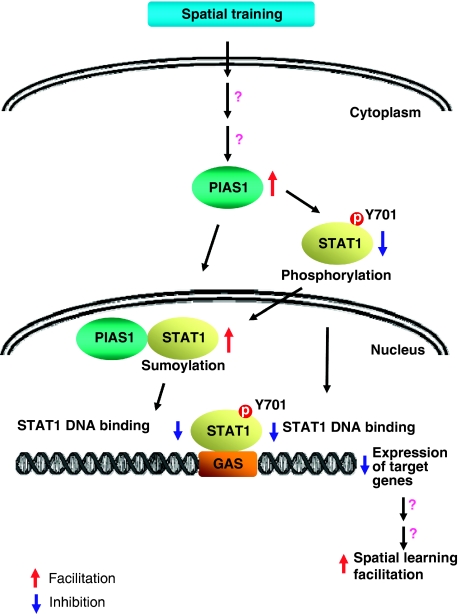
In summary, PIAS1 is well documented in the immune system, but other physiological functions that PIAS1 may also regulate are barely known. In this study, we are the first demonstrating a novel role of PIAS1 involved in spatial learning in rats. We also first demonstrate the role for sumoylation in regulation of spatial learning. Moreover, in addition to STAT1 sumoylation by PIAS1, we have identified a novel regulation mechanism in which PIAS1 decreases the phosphorylation of STAT1 at Tyr-701. Both posttranslational modifications mediate the enhancing effect of PIAS1 on spatial learning (Figure 8). This study opens the door for future investigations of the role and function of PIAS1 in the central nervous system.
Figure 8.
Schematic diagram showing the relationship among spatial training, PIAS1 expression, STAT1 sumoylation, STAT1 phosphorylation, STAT1 DNA binding and spatial learning facilitation. Spatial training upregulates PIAS1 expression in hippocampal CA1 area. PIAS1 expression leads to increased STAT1 sumoylation and decreased STAT1 phosphorylation at Tyr-701 (which further increases STAT1 sumoylation). Both events result in decreased STAT1 DNA binding activity and downregulation of STAT1-targeted gene expression. Downregulation of these gene expressions ultimately results in spatial learning facilitation.
Materials and methods
Animals
Adult male Sprague-Dawley rats (250–350 g) bred at the Animal Facility of the Institute of Biomedical Sciences, Academia Sinica in Taiwan were used. Animals were housed in a room maintained on a 12/12 h light/dark cycle (light on at 0630 hours) with food and water continuously available. Experimental procedures follow the Guidelines of Animal Use and Care of the National Institute of Health.
Water-maze learning
The water maze used was a plastic, circular pool, 2.0 m in diameter and 0.6 m in height, which was filled with water (25±2°C) to a depth of 20 cm. A circular platform (8 cm in diameter) was placed at a specific location away from the edge of the pool. The top of the platform was submerged 1.5 cm below the water surface. Water was made cloudy by adding milk powder. Distinctive, visual cues were set on the wall.
For spatial learning, animals were subjected to three trials a day with one given early in the morning, one in the early afternoon and another in the late afternoon. The training procedure lasted for 4 days and a total of 12 trials were given. For these trials, animals were placed at different starting positions spaced equally around the perimeter of the pool in a random order. Animals were given 60 s to find the platform. If an animal could not find the platform, it was guided to the platform and was allowed to stay on the platform for 20 s. The time that each animal took to reach the platform was recorded as the escape latency. A probe trial of 60 s was given on day 5 to test their memory retention. In the experiment assessing the effect of PIAS1 siRNA transfection on memory consolidation, probe trial was conducted on day 8. Animals were placed in the pool with the platform been removed and the time they spent in each quadrant (target quadrant, left quadrant, opposite quadrant and right quadrant) was recorded. For screening of the fast learners and slow learners, the criteria and procedures used were the same as that described previously (Tsai et al, 2002).
DD–PCR
DD-PCR was performed in a previous study and the cDNA fragment examined here was also obtained previously (Tsai et al, 2002). Briefly, 80 arbitrary random primers (H-AP1∼H-AP80, RNAimage Kit) were purchased from GenHunter (Nashville, TN). The reverse transcribed (RT) products of dorsal hippocampal tissues from three fast learners and slow learners were subjected to different amplification reactions by using these primers according to the procedures described previously (Tsai et al, 2002). Differentially expressed cDNA fragments were resolved from the sequencing gels and cloned into the PCR 2.1 TA vector (Invitrogen, Carlsbad, CA).
Plasmid DNA construction and DNA/polyethyleneimine (PEI) complex preparation
For construction of the Flag-tagged pias1 plasmid, full-length pias1 was cloned by amplifying the rat hippocampal pias1 cDNA with primers 5′-ATCGGGATCCCATGGCGGACAGTGCGGAAC-3′ and 5′-ATCGGAATTCTCAGTCCAACGAGATAATG-3′. The PCR product was sub-cloned between the BamHI and EcoRI sites of the mammalian expression vector pCMV-Tag2A. For construction of the GFP-tagged pias1 plasmid, full-length pias1 was sub-cloned into the pEGFP-C1 expression vector with RsrII sites. The full-length human stat1-Flag plasmid was purchased from Addgene (Cambridge, MA). The stat1Y701F mutant plasmid was generated by using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The method used for plasmid DNA transfection to brain tissues was adopted from that of a previous report (Abdallah et al, 1996). The non-viral vector transfection reagent, PEI, was used because we have previously demonstrated that PEI does not produce toxicity to hippocampal neurons (Chao et al, 2010). Before injection, plasmid DNA was diluted in 5% glucose to a stock concentration of 2.77 μg/μl. Branched PEI of 25 kDa (Sigma, St Louis, MO) was diluted to 0.1 M concentration in 5% glucose and added to the DNA solution. Immediately before injection, 0.1 M PEI was added to reach a ratio of PEI nitrogen per DNA phosphate equals to 10. The mixture was subjected to vortex for 30 s and allowed to equilibrate for 15 min.
RNA interference
The rat PIAS1 siRNA was used to knock down PIAS1expression in CA1 area. The sense and antisense sequences used were adopted from a previous study (Kawai-Kowase et al, 2005). The sequence for the sense strand is: 5′-UCCGGAUCAUUCUAGAGCUTT-3′ and that for the antisense strand is: 5′-AGCUCUAGAAUGAUCCGGATT-3′. The Silencer Negative Control number 1 siRNA (control siRNA) was used as a control. These are the siRNAs with sequences that do not target any gene product (Ambion, Austin, TX). They both were synthesized from Ambion.
Intra-hippocampal gene transfection and siRNA injection
Rats were anaesthetised with pentobarbital (40 mg/kg, i.p.) and subjected to stereotaxic surgery. Two 23-gauge, stainless-steel, thin-wall cannulae were implanted bilaterally to the CA1 area of the dorsal hippocampus at the following coordinates: 3.5 mm posterior to the bregma, 2.5 mm lateral to the midline, and 3.4 mm ventral to the skull surface. After recovery from the surgery, 0.7 μl plasmid DNA complex (1.5 μg/μl) was injected to the CA1 area at a rate of 0.1 μl/min. For siRNA injection, 0.7 μl of PIAS1 siRNA (8 pmol/μl) or control siRNA was transfected to the CA1 area by using the cationic polymer transfection reagent jetSI™ 10 mM (Polyplus-Transfection, New York, NY). For the PIAS1 siRNA and STAT1Y701F co-transfection experiment, 0.5 μl of PIAS1 siRNA and 0.25 μl of STAT1Y701F plasmid was injected. The reason for using these volumes is because they together yield a total volume of 0.75 μl that is similar to the transfection volume used in all other experiments. The reason that we did not use equal volume of PIAS1 siRNA and STAT1Y701F transfection is because that we wanted to avoid a simple additive effect (one impairs learning and the other facilitates learning). Instead, we aimed to study their interaction effect. The inner diameter of the injection needle is 0.31 mm and the wall thickness of the injection needle is 0.12 mm each side. The injection needle was left in place for 5 min to limit the diffusion of injected DNA and siRNA. Spatial training started 48 h after DNA injection or 72 h after siRNA injection. Plasmid DNA or siRNA was injected again at the beginning of the second training day. For the experiment assessing the effect of PIAS1 siRNA on memory consolidation, PIAS1 siRNA was injected at the end of the second and fourth training days (two injections in all). A time span of 1 h was allowed between the second injection and spatial training.
Animals were killed after the probe trial test or after spatial training. Their brains were removed and the hippocampal tissue slices (2-mm thick; two slices in all) were dissected out by using a brain slicer. The CA1 tissue was further punched out by using a punch of 2 mm diameter.
Western blot
The CA1 tissue lysate was lysed in RIPA buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% IGEPAL CA-630, 1 mM phenymethylsulfonyl fluoride (PMSF), 20 μg/ml pepstatin A, 20 μg/ml leupeptin, 20 μg/ml aprotinin, 50 mM NaF and 1 mM Na3VO4). The lysate was resolved by 8% SDS–PAGE. The proteins resolved by SDS–PAGE were transferred to the polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA) and western blot analysis was conducted by using the following antibodies: rabbit anti-PIAS1 (Epitomics, Burlingame, CA), anti-PIAS2 (Abcam, Cambridge, UK), anti-PIAS3 (Cell Signaling, Danvers, MA), anti-PIAS4 (Cell Signaling), anti-STAT1 (Millipore), anti-p-Tyr701 STAT1 (Cell Signaling) and anti-actin (Chemicon, Temecula, CA). The secondary antibody used was HRP-conjugated goat-anti rabbit IgG antibody (Chemicon). Membrane was developed by reacting with chemiluminescence HRP substrate and exposed to the LAS-3000 image system (Fujifilm, Tokyo, Japan) for visualization of protein bands. The protein bands were quantified by using the NIH Image J Software.
Immunoprecipitation and in vitro sumoylation assay
To analyze the endogenous PIAS1 SUMO E3 ligase activity, immunoprecipitation (IP) of PIAS1 as the source of E3 was performed for in vitro sumoylation assay (for PIAS1WT transfection and PIAS1 siRNA transfection experiments). Hippocampal CA1 tissue lysate was prepared in the same way as that prepared for western blot. For IP PIAS1, the clarified lysate (0.5 mg) was immunoprecipitated with 0.5 μl of anti-PIAS1 antibody (Epitomics) at 4°C overnight. The protein A agarose beads (30 μl, 50% slurry, GE Healthcare, Barrington, IL) were added to the IP reaction product to catch the immune complex at 4°C for 3 h. The immune complex on beads were washed three times with washing buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 1 mM DTT, 50 mM β-glycerophosphate, 50 mM NaF, 10 μg/ml PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin and 4 μg/ml pepstatin and subjected to in vitro sumoylation reaction with the addition of recombinant STAT1 protein (0.5 μg) (Prospec, Rehovot, Israel). In vitro sumoylation assay was performed by using the SUMO link™ kit according to the manufacturer's instructions (Active Motif, Carlsbad, CA) and boiled in Laemmli sample buffer at 95°C for 10 min. The in vitro sumoylation product was subjected to 8% SDS–PAGE followed by transferring onto the PVDF membrane (Millipore). The membrane was immunoblotted with anti-STAT1 antibody (Millipore). For other experiments transfected with STAT1K703R, STAT1Y701F, and in naive animals, and in animals subjected to water-maze training, endogenous STAT1 sumoylation was determined. In these experiments, hippocampal CA1 tissue lysate (0.5 mg) was immunoprecipitated with anti-STAT1 antibody instead of anti-PIAS1 antibody. The remaining procedures are the same as that for carrying out the in vitro sumoylation assay.
Biotinylated oligonucleotides pull-down assay for STAT1 DNA-binding activity
DNA oligonucleotides containing two GAS elements (underlined) (5′-GAGACTCAGTTTCCCGTAAATCGTCCAGTTTCCCGTAAAGACTATGC-3′) were conjugated with a 5′ biotin on the sense strand according to the method described previously (Meyer et al, 2003). Both complementary oligonucleotides were re-suspended in the annealing buffer (10 mM Tris (pH 8.0), 50 mM NaCl and 1 mM EDTA). For annealing the sense and antisense oligonucleotides, 10 μl each of the complementary oligonucleotides together with 80 μl of the annealing buffer were mixed in a 0.5-ml microtube and the tube was placed in a heating block at 90°C. The heating block was allowed to gradually cool down to room temperature and stored on ice or at −20°C until use. For the STAT1 pull-down assay, the clarified hippocampal CA1 tissue lysate (0.4 mg) was added with 6 μl duplex oligonucleotides (100 μM) and poly dI-dC (1 μg/ml, GE Healthcare) at 4°C overnight. The streptavidin agarose beads (10 μl, Sigma) were added to the pull-down reaction product to catch the STAT1–DNA oligonucleotide complex at 4°C for 3 h. The pull-down reaction complex on beads were then washed three times with washing buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 1 mM DTT, 50 mM β-glycerophosphate, 50 mM NaF, 10 μg/ml PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin and 4 μg/ml pepstatin and boiled in Laemmli sample buffer at 95°C for 10 min. For the analysis of STAT1 DNA-binding activity, the pull-down assay product was subjected to 8% SDS–PAGE followed by transferring onto the PVDF membrane (Millipore) and immunoblotted with anti-STAT1 antibody (Millipore).
Quantitative real-time PCR and DNA gel electrophoresis
Total RNA from CA1 tissue was isolated by using the RNAspin mini kit (GE Healthcare). The cDNA was generated from total RNA with Superscript III reverse transcriptase (Invitrogen). Real-time PCR analysis was performed by using the ABI PRISM 7500 real-time PCR system with Power SYBR Green PCR Master Mix according to the instruction manual (Applied Biosystems (ABI), Foster City, CA). The primers for pias1 were designed according to a previous report (Kawai-Kowase et al, 2005). The primers for GFP were designed by using the Primer Design Program Primer3 (http://frodo.wi.mit.edu/primer3). The primer sequences for pias1 and HRPT are shown in Supplementary Table 1. The cycle threshold (Ct) value and data were analysed by using the 7500 system Sequence Detection Software (ABI). The amount of pias1 gene expression was normalized to that of HPRT gene expression. The relative expression level (in fold) was determined by using the 2−(ΔΔCt) method (Livak and Schmittgen, 2001). The PCR product from GFP was mixed with EZ-VISION DNA DYE (Amresco, Solon City, OH) and analysed by gel electrophoresis on 8% DNA PAGE in 1 × TBE buffer (90 mM Tris (pH 8.0), 90 mM boric acid and 2 mM EDTA). The gel was photographed on the gel image system. The intensity of the cDNA band was quantified by using the NIH Image J Software.
Primary hippocampal culture and plasmid transfection
Primary hippocampal culture was prepared as described previously (Yang et al, 2006). Briefly, cultured hippocampal neurons at embryonic day 18 were plated on dish at a density of 4 × 105 cells/ml and were fixed with 4% paraformaldehyde-4% sucrose for 15 min and permeabilized with 0.1% Triton X-100 for 20 min at room temperature. GFP–PIAS1WT plasmid was transfected to cultured neurons at day in vitro (DIV) 7 by using the Lipofectamine 2000 reagent (Invitrogen) and neurons were fixed 48 h later. Hochest was added to the culture medium for 3 h as a nucleus marker. Coverslips were mounted and images were taken by using a confocal microscope (Ultraview, Perkin Elmer, UK).
Immunohistochemistry
Rats were anesthetized with pentobarbital (100 mg/kg, i.p.) and perfused with ice-cold phosphate-buffered saline followed by 4% paraformaldehyde. Brains were removed and post-fixed in 20% sucrose/4% paraformaldehyde solution for 20–48 h. Brains were then frozen, cut into 30-μm sections on a cryostat and mounted on gelatin-coated slides. Brain sections were rinsed with 1 × PBS for 10 min and permeabilized with pre-cold EtOH/CH3COOH (95:5%) for 10 min followed by 1 × PBS for 10 min for three times. The sections were preincubated in a blocking solution containing 3% normal goat serum, 3% BSA and 0.2% Triton X-100 in 1 × PBS for 2 h followed by 1 × PBS for 10 min for three times. For immunofluorescence detection of the nucleus, tissue sections were added with 20 μl of the VECTASHIELD mounting medium with DAPI (1.5 μg/ml; Vector Laboratories, Burlingame, CA). To confirm PIAS1 siRNA transfection, the Cy3-labelled PIAS1 siRNA (synthesized and conjugated by Ambion) was used for intra-hippocampal injection and brain sections were prepared 72 h after siRNA injection for visualization of Cy3 fluorescence under a confocal microscope. For immunofluorescence detection of Flag-STAT1Y701F transfection, tissue sessions were incubated with a mouse monoclonal anti-Flag antibody M2 (1:100, Sigma) in blocking buffer at 4°C overnight. Sessions were washed three times in 1 × PBS and incubated in goat anti-mouse FITC-conjugated IgG antibody (1:100, Sigma) in 1 × PBS for 1h. Digital photomicrographs were taken with an Olympus digital C-3030 camera mounted on a Zeiss microscope.
Statistics
Behavioural data were analysed by analysis of variance (ANOVA) with repeated measure followed by post-hoc Newman–Keuls multiple comparisons (represented by Q-value). Biochemical data were analysed with one-way ANOVA followed by Newman–Keuls comparisons or with Student's t-test.
Supplementary Material
Acknowledgments
We thank Dr KJ Tsai for his technical help. This study was supported by grants from the National Science Council of Taiwan (NSC98-2321-B-001-036 and NSC99-2321-B-001-018).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA (1996) A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Human Gene Ther 7: 1947–1954 [DOI] [PubMed] [Google Scholar]
- Arvin B, Neville LF, Barone FC, Feuerstein GZ (1996) The role of inflammation and cytokines in brain injury. Neurosci Biobehav Rev 20: 445–452 [DOI] [PubMed] [Google Scholar]
- Burger C, Lopez MC, Feller JA, Baker HV, Muzyczka N, Mandel RJ (2007) Changes in transcription within the CA1 field of the hippocampus are associated with age-related spatial learning impairments. Neurobiol Learn Mem 87: 21–41 [DOI] [PubMed] [Google Scholar]
- Castellucci VF, Kennedy TE, Kandel ER, Goelet P (1988) A quantitative analysis of 2-D gels identifies proteins in which labeling is increased following long-term sensitization in Aplysia. Neuron 1: 321–328 [DOI] [PubMed] [Google Scholar]
- Cavallaro S, D'Agata V, Manickam P, Dufour F, Alkon DL (2002) Memory-specific temporal profiles of gene expression in the hippocampus. Proc Natl Acad Sci USA 99: 16279–16284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CC, Ma YL, Lee EHY (2010) BDNF enhances Bcl-xL expression through protein kinase CK2-activated and NF-κB-mediated pathway in rat hippocampus. Brain Pathol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP, Taylor CT (2003) Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci USA 100: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis HP, Squire LR (1984) Protein synthesis and memory: a review. Psychol Bull 96: 518–559 [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA (2007) Neuronal competition and selection during memory formation. Science 316: 457–460 [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, Josselyn SA (2009) Selective erasure of a fear memory. Science 323: 1492–1495 [DOI] [PubMed] [Google Scholar]
- Huang AM, Wang HL, Tang YP, Lee EH (1998) Expression of integrin-associated protein gene associated with memory formation in rats. J Neurosci 18: 4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN (2001) The Stat family in cytokine signaling. Curr Opin Cell Biol 13: 211–217 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC, Girault JA (2003) PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J Biol Chem 278: 47434–47440 [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8: 713–718 [DOI] [PubMed] [Google Scholar]
- Kawai-Kowase K, Kumar MS, Hoofnagle MH, Yoshida T, Owens GK (2005) PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix–loop–helix proteins. Mol Cell Biol 25: 8009–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EHY, Hung HC, Lu KT, Chen WH, Chen HY (1992) Protein synthesis in the hippocampus associated with memory facilitation by corticotropin-releasing factor in rats. Peptides 13: 927–937 [DOI] [PubMed] [Google Scholar]
- Liang KC, Hu SJ, Chang SC (1996) Formation and retrieval of inhibitory avoidance memory: differential roles of glutamate receptors in the amygdala and medial prefrontal cortex. Chin J Physiol 39: 155–166 [PubMed] [Google Scholar]
- Liao J, Fu Y, Shuai K (2000) Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT1) (PIAS1) in cytokine-induced PIAS1–Stat1 interaction. Proc Natl Acad Sci USA 97: 5267–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K (1998) Inhibition of stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA 95: 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, Wu H, Shuai K (2004) PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol 5: 891–898 [DOI] [PubMed] [Google Scholar]
- Liu B, Yang Y, Chernishof V, Ogorzalek Loo RR, Jang H, Tahk S, Yang R, Mink S, Shultz D, Bellone CJ, Loo JA, Shuai K (2007) Proinflammatory stimuli induce IKKα-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell 129: 903–914 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ma YL, Tsai MC, Hsu WL, Lee EH (2006) SGK protein kinase facilitates the expression of long-term potentiation in hippocampal neurons. Learn Mem 13: 114–118 [DOI] [PubMed] [Google Scholar]
- Matthies H (1989) Neurobiological aspects of learning and memory. Annu Rev Psychol 40: 381–404 [DOI] [PubMed] [Google Scholar]
- Meinecke I, Cinski A, Baier A, Peters MA, Dankbar B, Wille A, Drynda A, Mendoza H, Gay RE, Hay RT, Ink B, Gay S, Pap T (2007) Modification of nuclear PML protein by SUMO-1 regulates Fas-induced apoptosis in rheumatoid arthritis synovial fibroblasts. Proc Natl Acad Sci USA 104: 5073–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U (2003) DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev 17: 1992–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser E, Moser MB, Andersen P (1993) Spatial learning impairment parallels the magnitude of dorsal hippocampal lesions, but is hardly present following ventral lesions. J Neurosci 13: 3916–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Onodera T, Nishimura S, Thompson RF, Itohara S (2006) Molecular evidence for two-stage learning and partial laterality in eyeblink conditioning of mice. Proc Natl Acad Sci USA 103: 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK (2005) A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437: 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheschonka A, Tang Z, Betz H (2006) Sumoylation in neurons: nuclear and synaptic roles? Trends Neurosci 30: 85–91 [DOI] [PubMed] [Google Scholar]
- Sharrocks AD (2006) PIAS proteins and transcriptional regulation—more than just SUMO E3 ligases? Genes Dev 20: 754–758 [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B (2005) Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol 5: 593–605 [DOI] [PubMed] [Google Scholar]
- Song L, Bhattacharya S, Yunus AA, Lima CD, Schindler C (2006) Stat1 and SUMO modification. Blood 108: 3237–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai DJC, Su CC, Ma YL, Lee EHY (2009) SGK1 phosphorylation of IκB kinase α and p300 upregulates NF-κB activity and increases N-methyl-D-aspartate receptor NR2A and NR2B expression. J Biol Chem 284: 4073–4089 [DOI] [PubMed] [Google Scholar]
- Tang Z, El Far O, Betz H, Scheschonka A (2005) PIAS1 interaction and sumoylation of metabotropic glutamate receptor 8. J Biol Chem 280: 38153–38159 [DOI] [PubMed] [Google Scholar]
- Tsai KJ, Chen SK, Ma YL, Hsu WL, Lee EHY (2002) sgk, a primary glucocorticoid-induced gene, facilitates memory consolidation of spatial learning in rats. Proc Natl Acad Sci USA 99: 3990–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T (1996) Discovery of genes involved with learning and memory: an experimental synthesis of Hirschian and Benzerian perspectives. Proc Natl Acad Sci USA 93: 13460–13467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungureanu D, Vanhatupa S, Gronholm J, Palvimo J, Silvennoinen O (2005) SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood 106: 224–226 [DOI] [PubMed] [Google Scholar]
- Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, Janne OA, Palvimo JJ, Silvennoinen O (2003) PIAS proteins promote SUMO-1 conjugation to STAT1. Blood 102: 3311–3313 [DOI] [PubMed] [Google Scholar]
- Weber S, Maass F, Schuemann M, Krause E, Suske G, Bauer UM (2009) PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes Dev 23: 118–132 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang YC, Lin CH, Lee EHY (2006) Serum- and glucocorticoid-inducible kinase 1 (SGK1) increases neurite formation through microtubule depolymerization by SGK1 and by SGK1 phosphorylation of tau. Mol Cell Biol 26: 8357–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Warner DR, Roberts EA, Pisano MM, Greene RM (2005) Novel interaction between nuclear coactivator CBP and the protein inhibitor of activated Stat1 (PIAS1). J Interferon Cytokine Res 25: 321–327 [DOI] [PubMed] [Google Scholar]
- Zimnik S, Gaestel M, Niedenthal R (2009) Mutually exclusive STAT1 modifications identified by Ubc9/substrate dimerization-dependent SUMOylation. Nucleic Acid Res 37: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.