Abstract
The three-dimensional structure of the fMet-tRNAfMet -binding domain of translation initiation factor IF2 from Bacillus stearothermophilus has been determined by heteronuclear NMR spectroscopy. Its structure consists of six antiparallel β-strands, connected via loops, and forms a closed β-barrel similar to domain II of elongation factors EF-Tu and EF-G, despite low sequence homology. Two structures of the ternary complexes of the EF-Tu⋅aminoacyl-tRNA⋅ GDP analogue have been reported and were used to propose and discuss the possible fMet-tRNAfMet-binding site of IF2.
Keywords: bacterial translation/NMR/protein/protein biosynthesis/ribosome
Introduction
Translation initiation in bacteria is mediated by initiation factors IF1, IF2 and IF3. The interaction of these proteins with the ribosomal subunits and with other components of the translational apparatus in several defined steps ultimately leads to the formation of a 70S initiation complex that carries the mRNA and the N-formylmethionine (fMet)-tRNAfMet in the correct position, thereby enabling the entry of the ribosome into the elongation phase of translation (for reviews, see Gualerzi and Pon, 1990; Gualerzi et al., 2000).
IF1 is involved in the association/dissociation processes of the ribosomal subunits (van der Hofstad et al., 1978 and references therein) and stimulates IF2 binding to the 30S subunit (Zucker and Hershey, 1986). The binding of IF1 to the ribosomal A-site (Moazed et al., 1995) and the similarity of the structure of IF1 (Sette et al., 1997) to that of domain IV of EF-G led to the proposal that in association with IF2 it may mimic the A-site-bound tRNA, thereby preventing premature binding of an aminoacyl-tRNA to the 30S A-site (Brock et al., 1998).
IF3 is involved in several key events during the formation of the 30S initiation complex and acts as a ‘fidelity factor’, destabilizing initiation complexes containing aminoacyl-tRNAs other than fMet-tRNA and non-canonical initiation triplets (Gualerzi et al., 1971; Hartz et al., 1989; La Teana et al., 1993).
IF2 is the largest of all bacterial translational factors and its main function is to stimulate interaction of the ribosomes with initiator fMet-tRNAfMet, which is accomplished through an acceleration of codon–anticodon base pairing at the ribosomal P-site (Gualerzi and Pon, 1990; Gualerzi et al., 2000). The activity of IF2 largely depends on a specific interaction that takes place on the ribosome between the factor and the initiator tRNA molecule. In turn, the affinity and specificity of this interaction are due to the recognition of the formylated α-amino group of fMet-tRNAfMet by the protein (RajBhandary and Chow, 1986; Gualerzi and Pon, 1990; Schmitt et al., 1996).
Following limited proteolysis in the presence of GTP, IF2 was found to consist of three domains (Gualerzi et al., 1991). The N-terminal part (1–154 in Bacillus stearothermophilus) is not conserved in length and sequence among prokaryotes. Its function is poorly understood (Gualerzi et al., 1991), but recently it was suggested that the N-terminal domain enhances the interaction of IF2 with the 30S and 50S ribosomal subunits (Moreno et al., 1999). The central part (155–519) (Gualerzi et al., 1991) contains the GDP/GTP-binding motifs (237–406) characteristic of many GTPases (Vachon et al., 1990). It also contains the binding site for the 50S subunit and displays some residual affinity for the 30S subunit (Gualerzi et al., 1991). The central localization of the GTP/GDP-binding motifs distinguishes IF2 from elongation factors EF-Tu and EF-G as well as termination factor RF-3, in which these binding motifs are localized at the N-terminal part of the molecule (Sprinzl, 1994). The C-terminal part of IF2 (520–741) contains the entire fMet-tRNAfMet-binding site of IF2 and is resistant to limited proteolysis (Gualerzi et al., 1991). However, it consists of two domains, IF2 C-1 and IF2 C-2, the latter of which has been shown to contain all molecular determinants necessary and sufficient for the recognition and binding of fMet-tRNAfMet (Misselwitz et al., 1997; Spurio et al., 2000).
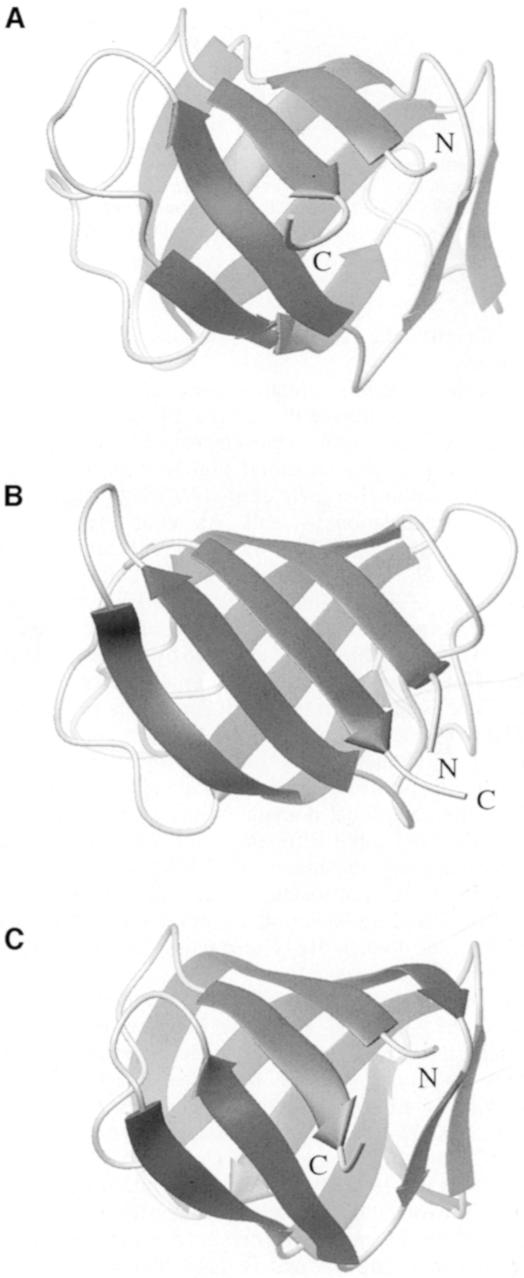
In this study, we present the solution structure of B.stearothermophilus IF2 C-2 and show that it adopts a β-barrel fold displaying a high degree of structural similarity with domain II of elongation factors EF-Tu (Berchtold et al., 1993; Kjeldgaard et al., 1993; Nissen et al., 1995, 1999) and EF-G (Ævarsson et al., 1994; Czworkowski et al., 1994; Al-Karadaghi et al., 1996), despite low sequence identity. The known ternary complexes of the EF-Tu⋅aminoacyl-tRNA⋅GDP analogue (Nissen et al., 1995, 1999) were used to propose and discuss the possible fMet-tRNAfMet recognition site of IF2.
Results and discussion
Assignment of IF2 C-2
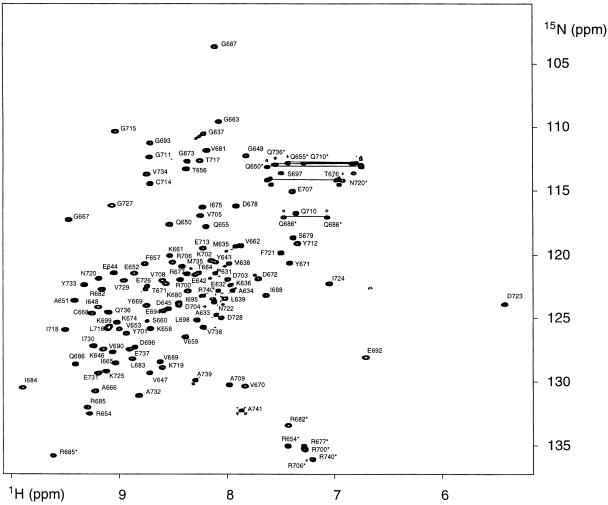
Sequential assignment of IF2 C-2 (E632–A741) was performed on a 15N-labelled sample, using two-dimensional NOE and TOCSY spectra, in combination with heteronuclear three-dimensional TOCSY-(1H,15N)-HSQC and 3D NOESY-(1H,15N)-HSQC spectra. The assignment was checked by use of 3D HNCA on a 15N (100%)/13C (10%)-labelled sample. A representative 2D (1H,15N)-HSQC spectrum is shown in Figure 1.
Fig. 1. 2D (15N,1H)-HSQC spectrum of IF2 C-2 in 20 mM KPi pH 5.2, 200 mM KCl, 312 K. The spectrum was recorded at a 1H frequency of 600 MHz. The peaks labelled with asterisks indicate side chain peaks, and the lines connect glutamine and asparagine side chain peaks.
A complete sequential assignment of IF2 C-2 has been obtained for all backbone atoms, while the side chain 1H and 15N nuclei were assigned for all residues in the structured part of the protein (Y643–A741), which shows good dispersion in the NMR spectra. The γ-methyl groups of V734, the δ-methyl groups of L183 and the δ-methyl groups of six out of 12 isoleucines (I648, I665, I684, I688, I695 and I724) could not be assigned due to overlap. The 1H, 15N, Cα and CO resonance chemical shifts of IF2 C-2 at pH 5.2, 312 K are available at the BioMagResBank (BMRB) as supplementary material, accession No. 4697.
Most medium- and long-range NOEs were obtained using a 3D NOESY-(1H,15N)-HSQC spectrum. A 2D NOE spectrum recorded in D2O was used to collect mainly NOEs involving the side chains protons.
Structure determination of IF2 C-2
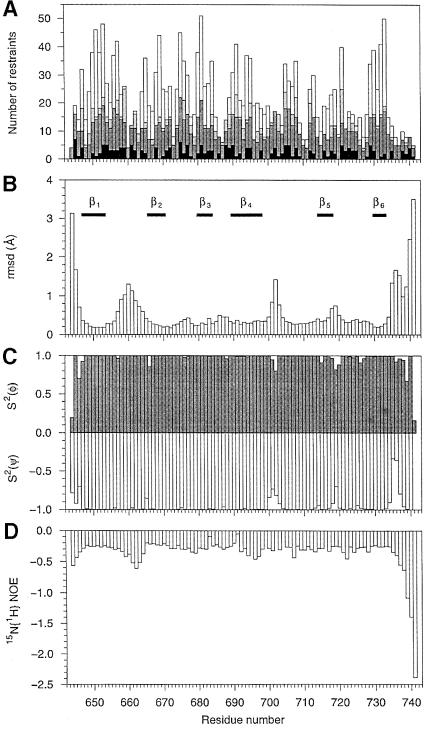
The structure was determined using medium- and long-range NOEs, restraints derived from amide proton exchange data and dihedral restraints. A total of 1228 NOEs were obtained from the analysis of the different NOESY spectra, and 71 ϕ and 35 χ1 dihedral restraints were derived from coupling constants. In addition, 27 hydrogen bonds were used in the calculations: the HN donor was obtained from amide proton exchange data, whilst the oxygen acceptor was determined on the basis of short NOE distances. The distribution of the distance restraints over the sequence of IF2 C-2 is displayed in Figure 2A.
Fig. 2. Structural statistics, distribution of NMR restraints and heteronuclear (1H,15N) NOE data of IF2 C-2. (A) Distribution of meaningful and non-redundant distance restraints, as defined by Aqua (Laskowski et al., 1996), as a function of the primary structure of IF2 C-2; the intra-residue, sequential, medium- and long-range distance restraints obtained from the NOE spectra are indicated by black, dark grey, light grey and white bars, respectively. All restraints, except for the intra-residue restraints, appear twice (i.e. at each residue). (B) R.m.s.d. between the conformers of the ensemble and the average structure as a function of the protein sequence. The superimposed position is calculated using the backbone atoms of V647–E657 and I665–Y733. (C) Angular order parameter for the angles ϕ (grey bars) and ψ versus the protein sequence. (D) Heteronuclear (1H,15N) NOEs versus the protein sequence, recorded at 309 K and a 1H frequency of 500 MHz.
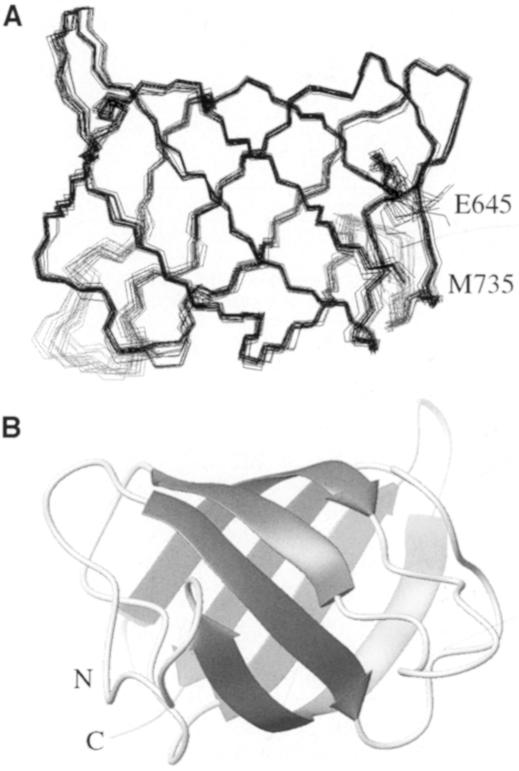
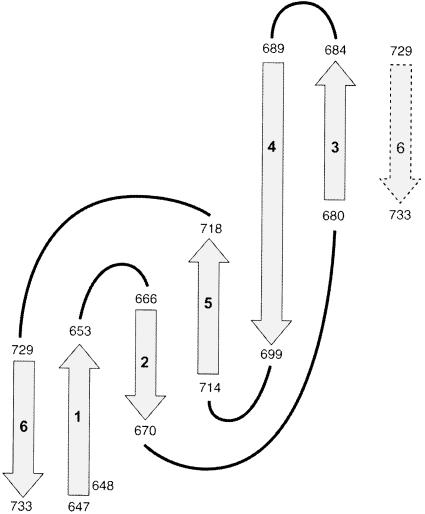
Using torsion angle dynamics, 50 structures were calculated, 20 of which were selected on the basis of low energy coinciding with few constraint violations. A summary of structural statistics is given in Table I. Figure 3A shows the overlay of the structure ensemble, while Figure 3B displays a ribbon diagram of a representative member rotated 180° about the z-axis. The solution structure of IF2 C-2 consists of six antiparallel β-strands forming a closed β-barrel. The six β-strands span the following residues: β1, V647–V653; β2, A666–V670; β3, K680–I684; β4, V689–K698; β5, C714–I718; and β6, V729–Y733. Residues 647 and 648 have similar NOEs to residues 733, indicating the presence of a β-bulge at the N-terminus of strand β1. The overall organization of the β-barrel consists of a greek-key motif, arrayed as 1 3 –1 –1 –3 (β6 β1 β2 β5 β4 β3), and is sketched in Figure 4. Strands β3 and β4 connected via type IV β-turns.
Table I. Restraints and structural statistics for IF2 C-2.
| Restraints | |
| distance restraints | 1228 |
| medium range | 101 |
| long range | 497 |
| dihedral angle restraints | 106 |
| φ | 71 |
| χ1 | 35 |
| hydrogen bonds | 27 |
| stereospecific assignments | 44% |
| β protons | 20/54 |
| γ methyl group of valine | 10/13 |
| δ methyl group of leucine | 0/2 |
| Structural statistics | |
| number of distance restraint violations >0.5 Å | 1 |
| maximum distance constraint violation (Å) | 0.54 |
| r.m.s. deviation from average structureb (Å) | |
| trace Cα | 0.32 |
| all heavy atoms | 0.95 |
| residues with φ/ψ dihedral angles inb | |
| most favoured regions | 59.2% |
| additional allowed regions | 31.0% |
| generously allowed regions | 8.6% |
| disallowed regions | 1.2% |
aRestraint and structural statistics are presented for the 20 best structures of the ensemble.
bFor residues 647–657 and 665–733.
Fig. 3. Solution structure of IF2 C-2. (A) Superposition of the Cα trace of the ensemble of 20 structures. The structures were overlaid using the residues as defined in Figure 2B. (B) Ribbon plot of the representative structure of the ensemble shown in (A), after 180° rotation about the z-axis. The β-strands are represented as arrows. The N- and C-termini are indicated in each structure.
Fig. 4. Overall organization of the antiparallel β-strands of IF2 C-2. The arrows indicate the β-strands that are numbered. The start and end of each strand is indicated. Strand β-1 starts with a β-bulge (residues 647 and 648).
The root mean square deviation (r.m.s.d.) versus the average structure for the protein core (residues V647–E657 and I665–Y733) is 0.32 Å for the backbone atoms and 0.95 Å for all heavy atoms (Figure 2B). The superposition of the ensemble of structures highlights the disordered parts: the N- and C-termini, and essentially two loops, i.e. the β1–β2 loop (residues 658–664) and the β4–β5 loop (residues 699–704). In fact the backbone angular order parameter (Figure 2C) indicates that the disorder in the β1–β2 loop is lower than suggested by the overlay chosen to calculate Figure 2B.The backbone r.m.s.d. calculated for this loop (residues 658–664) is 0.1 Å, indicating that the inner structure of this loop is well defined. The heteronuclear NOE data (Figure 2D) show that the disorder at the N- and C-terminal regions as well as the β1–β2 loop is due to flexibility with contributions on the nanosecond time scale. Apparently, the complete loop moves with respect to the IF2 C-2 molecule, the hinge being located at positions 657 and 665, as can be seen from Figure 2C. The observed disorder for the β4–β5 loop (r.m.s.d. = 0.4 Å) cannot be explained by the current 15N relaxation data and may be due merely to a lack of dihedral angle values for the residues 699, 702 and 704, which show considerable overlap in the (1H,15N)-HSQC spectra.
Structural coordinates and NMR data have been deposited in the Protein Data Banks: 1D1N.
Structural homology to other translational factors
Scanning the PDB via the DALI server (Holm and Sanders, 1993) enabled us to show a structural similarity between IF2 C-2, domain II of EF-G (Ævarsson et al., 1994; Czworkowski et al., 1994; Al-Karadaghi et al., 1996) and domain II of EF-Tu (Berchtold et al., 1993; Kjeldgaard et al., 1993; Nissen et al., 1995, 1999) only. Sequence similarity displayed by the three translation factors in the relevant region of the molecule is very low: ∼13% identity and 17% homology. The 3D structures of the corresponding domains of EF-Tu, IF2 and EF-G are presented in Figure 5A, B and C, respectively. They have an identical topology and superimpose fairly well on one another. The best alignment between IF2 C-2 and either EF-G or EF-Tu, as given by the DALI Server, gave r.m.s.d. values for the Cα atoms of 1.20 and 1.36 Å, respectively. Figure 6 shows this sequence alignment from which one can identify the equivalent residues of these three domains.
Fig. 5. Structural homology between IF2 C-2 and the elongation factors EF-Tu and EF-G. Ribbon representations of the Cα traces of: (A) domain II of T.aquaticus EF-Tu (Nissen et al., 1995); (B) B.stearothermophilus IF2 C-2; and (C) domain II of T.thermophilus EF-G (Ævarsson et al., 1994). The N- and C-termini are indicated in each structure.
Fig. 6. Partial alignment of the primary sequences of T.aquaticus EF-Tu (top row), B.stearothermophilus IF2 C-2 (middle row) and T.thermophilus EF-G (bottom row), derived from the structure superposition shown in Figure 5. Conserved residues are highlighted in black, homologous residues in grey. The IF2 C-2 residues used for the alignment were 645–648, 650–695, 689–719, 721–724 and 726–734 for the alignment with residues 306–358 and 363–393 of T.thermophilus EF-G, and residues 646–689 and 691–732 for the alignment with residues 218–257, 259–294 and 297–306 of T.aquaticus EF-Tu.
It is clear that this similarity could all be coincidental, but it should be stressed that EF-Tu, EF-G and IF2 are all factors involved in protein biosynthesis. The three proteins display considerable structural similarity, in particular in domain structure, but there exist significant differences in domain organization as well. All three proteins have GTPase activity (La Cour et al., 1985; Jurnak, 1985; Vachon et al., 1990; Bourne et al., 1991; Gualerzi et al., 1991). The GTP/GDP-binding motifs of EF-Tu and EF-G are both located at the N-terminal part of these molecules and are followed directly in sequence by domain II. In IF2, the GTP/GDP-binding motif has a more central location and there is an insertion of ∼200 residues between the GTP/GDP-binding motif and the C-2 domain. In EF-Tu and EF-G, domain II is followed by one and three domains, respectively, while the corresponding motif of IF2 is the C-terminal domain. With respect to function, there are even more differences. EF-Tu functions as a carrier that brings the aminoacyl-tRNA to the A-site of the ribosome. EF-G translocates the peptidyl-tRNA from the A- to the P-site. However, it was never shown that EF-G, and thus its domain II, is interacting directly with the tRNA in this process.
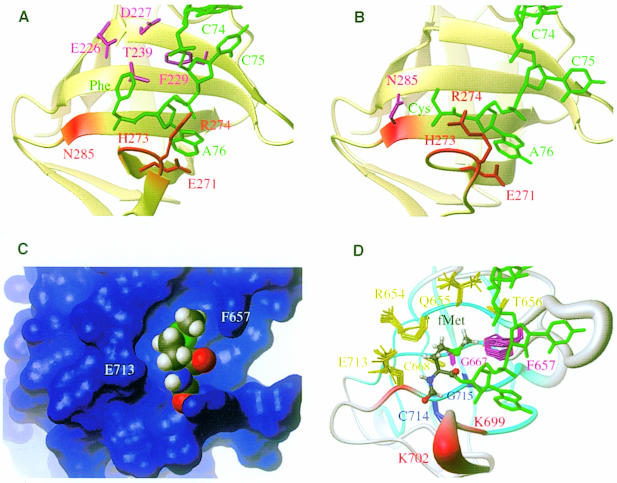
Since IF2, like EF-Tu, interacts with an aminoacyl-tRNA, the structural similarity between IF2 C-2 and domain II of EF-Tu enables us to propose the fMet-tRNAfMet-binding site of IF2. The three-dimensional structure of EF-Tu in a ternary complex with a GTP analogue and two different aminoacyl-tRNAs (yeast Phe-tRNAPhe and Escherichia coli Cys-tRNACys) is known (Nissen et al., 1995, 1999). The recognition site and the residues involved in the binding of Phe-tRNAPhe and Cys-tRNACys by domain II of Thermus aquaticus EF-Tu are depicted in Figure 7A and B, respectively. The overall three-dimensional structure of both ternary complexes is very similar, although the elbow angle of the tRNA is different, yielding a different orientation of the anticodon loop. However, the CCA acceptor stems and the attached amino acids bind similarly to domain II in both complexes.
Fig. 7. Ribbon plot of the recognition site of T.aquaticus EF-Tu in complex with yeast Phe-tRNAPhe (Nissen et al., 1995) (A) or with E.coli Cys-tRNACys (Nissen et al., 1999) (B), and B.stearothermophilus IF2 C-2 surface (C) or ribbon sausage (D) modelled formylmethionine and CCA acceptor arm. The β-strands are represented as arrows. Residues involved in the binding of the aminoacyl moiety main chain or of the last ribose are indicated in red, while residues involved in the binding of the aminoacyl side chain are in magenta. In (D), the CCA acceptor arm adopts the same conformation as in the EF-Tu⋅Cys-tRNACys complex. The β-strands are coloured in cyan and the coils and turns in grey. The size of the ribbon is proportional to the r.m.s.d. of the ensemble of 20 structures when superimposed on the mean structure. The side chain of the residues that might be involved in the recognition are displayed for the 20 structures. The conserved residues C714 and G715 are indicated in blue. The residues surrounding the pocket are indicated in yellow.
For IF2, several differences exist in the way in which it interacts with the fMet-tRNAfMet as compared with EF-Tu. In the GTP-bound form of EF-Tu, a pocket is present at the interface of the three domains, where the aminoacyl-tRNA is bound (Abel et al., 1996). Binding of fMet-tRNAfMet to IF2 requires only the last 100 C-terminal amino acids (Spurio et al., 2000). In addition, fMet-tRNAfMet binding by IF2 is not influenced by GTP/GDP (Petersen et al., 1979; Pon et al., 1985; Polekhina et al., 1996). It seems that there are few interactions between fMet-tRNAfMet and the other domains of IF2, or at least they do not contribute to the affinity.
IF2 recognition site for fMet-tRNAfMet
The analysis of the surface of this area in IF2 highlights the existence of a pocket localized at a position corresponding to the phenylalanine or cysteine side chain moiety in the ternary complexes of EF-Tu. A methionyl side chain (Figure 7C) can fill this pocket, which is connected to a channel that could represent the binding site for the CCA end of fMet-tRNAfMet, similar to the CCA-binding site of EF-Tu. Although EF-Tu uses other groove domains for tRNA binding, while IF2 probably does not, we use the assumption that the orientation of fMet-tRNAfMet in complex with IF2 is similar to that of the aminoacyl-tRNAs on EF-Tu in order to build a model of the fMet-tRNAfMet complex (Figure 7D). This model is discussed below.
In the ternary complexes of EF-Tu, the αNH2 groups of both Cys-tRNACys and Phe-tRNAPhe are hydrogen bonded to the amide proton of H273 and to the main chain carbonyl group of N285. In B.stearothermophilus IF2, these residues correspond to the non-conserved Y701 and to the partially conserved E713, respectively. The main chain carbonyl of E713 could participate in hydrogen bonding with the amide proton of formylmethionine as a donor. The α-NH of formylmethionine cannot be an acceptor of a hydrogen bond and, therefore, Y701 cannot be involved in a direct interaction with the initiator tRNA. In addition, EF-Tu interacts with the CCA ends of the aminoacyl-tRNAs via hydrogen bonds involving both the main and side chains of R274 (sometimes replaced by glutamine or lysine). These interactions are different in the two ternary complexes available. R274 corresponds to a conserved lysine in IF2 (K702 in B.stearothermophilus). Since K702 is located in the above-mentioned mobile β4–β5 loop, its interaction with fMet-tRNAfMet could stabilize this loop in a conformation similar to that observed for the corresponding EF-Tu loop in one of the ternary complexes. Finally, the side chain of the conserved E271 of EF-Tu, which corresponds to a conserved lysine of IF2 (K699 in B.stearothermophilus), is stacked over the adenine at position 76. The side chain carbonyl group is hydrogen bonded to the 2′ OH of the ribose. It is unlikely that the conserved K699 of IF2 could interact in a similar way, but it could possibly form a hydrogen bond with the phosphate of A76, similarly to the side chain of R274 in the Phe-tRNA complex.
The amino acid side chains of Phe-tRNAPhe and Cys-tRNACys are located similarly at the surface of EF-Tu, although they interact with different residues. Phenyl alanine is docked into a pocket lined with the side chains of E226, D227, F229 and T239, while the sulfur atom of cysteine is in van der Waals contact with the side chain of N285. In B.stearothermophilus IF2, these residues correspond to R654 (not conserved among IF2), Q655 (not conserved), F657 (conserved as a hydrophobic residue), G667 (conserved) and E713 (partially conserved), respectively. F657 could interact with the ε-methyl group of formylmethionine, whilst the absence of the side chain of G667 could create space for the methionyl side chain of formylmethionine. Together with two other conserved residues, G715 and C714, it forms the bottom of the pocket. Figure 7D shows the residues of IF2 that might be involved in the binding of formylmethionine and the last ribose of the initiator tRNA.
Conclusion
All the molecular determinants governing the thermodynamics and specificity of the IF2–fMet-tRNAfMet interaction (Spurio et al., 2000) are located in the last 100 amino acids of the C-terminal domain of translation initiation factor IF2. The 3D structure of IF2 C-2, elucidated here by heteronuclear multidimensional NMR spectroscopy, was found to be largely homologous to that of domain II of EF-Tu and domain II of EF-G, with which it can be superimposed with an r.m.s.d. of 1.20 and 1.36 Å, respectively. This homology could be coincidental but is more likely to be a result of the use of an ancestral structural module (in this case a six-stranded closed β-barrel) to fulfil both superficially similar (EF-Tu and IF2) and unrelated (EF-G and the other two factors) functions. Domain II of EF-Tu is involved in the binding of both the amino acid and CCA acceptor end of aminoacyl-tRNA, like IF2 C-2. In contrast to IF2, the full interaction of EF-Tu with aminoacyl-tRNA takes place in a pocket formed by the three domains of the EF-Tu molecule, and residues from all three domains are involved.
The large conformational change associated with the transition from the GTP to the GDP form of EF-Tu (Abel et al., 1996; Polekhina et al., 1996) and the simultaneous participation of the three domains of EF-Tu in aminoacyl-tRNA binding are obviously interconnected and explain the profound influence that the guanosine nucleotides have on this interaction. For the opposite reason, the confinement of the entire fMet-tRNAfMet-binding site of IF2 within a small, structurally and functionally independent domain accounts for the insensitivity of IF2–fMet-tRNAfMet interaction to the presence of guanosine nucleotide and underlines the different role that GTP binding and hydrolysis might have in the functioning of the two translation factors.
Despite these differences, comparison of the IF2 C-2 structure with that of domain II of EF-Tu (within two ternary complexes with aminoacyl-tRNA and GDPNP) allowed us to propose a reasonable pattern of potential interactions, which could be responsible for the interaction of IF2 with fMet-tRNAfMet. The function of domain II of EF-G, on the other hand, does not involve an interaction with aminoacyl-tRNA but probably with the ribosome (Ævarsson et al., 1994). Thus, the remarkable homology of the 3D structures, particularly striking considering the almost complete absence of sequence homology, should not be over-interpreted at the cost of minimizing the specific and peculiar roles that each one of the three factors plays in translation. Despite these cautions, it is remarkable that the two domains of EF-Tu and IF2 involved in transferring the aminoacyl-tRNA to the ribosome are structurally so closely related.
Materials and methods
Sample preparation
Domain IF2 C-2 spanning from E632 to A741 of B.stearothermophilus IF2 was produced and purified as described (Spurio et al., 2000). Uniformly 15N-labelled samples were prepared using E.coli JM109 harbouring pcI and pXP401C-2 grown in M9 minimal medium supplemented with 15NH4Cl (0.9 g/l) and glucose (3 g/l). For 13C/15N labelling, 10% of the supplemented glucose was [13C]glucose.
NMR spectroscopy
NMR spectroscopy on 15N-labelled IF2 C-2 and 13C/15N-labelled (100% 15N/10% 13C) IF2 C-2 samples was carried out at 1.5 mM protein concentration in 20 mM KPi pH 5.2, 200 mM KCl and 95% H2O/5% D2O. Two-dimensional 1H and three-dimensional (1H,15N)-NMR spectra were recorded at 312 K on a Varian INOVA 750 MHz spectrometer and the triple-resonance spectra were recorded on either Bruker AMXT 600 MHz or Varian INOVA 500 MHz spectrometers. Spectral widths of 10 p.p.m. for the 1H and 33 p.p.m. for the 15N (thus folding the Nε signals of arginines) experiments were used. Proton chemical shifts are referred to the water resonance at 4.61 p.p.m.; nitrogen and carbon chemical shifts were calculated by indirect referencing (Wishart et al., 1995).
A 2D DQF-COSY spectrum (Piantini et al., 1982) was recorded with 800 t1 increments. Clean-TOCSY spectra (Griesinger et al., 1988), with mixing times of 50 and 75 ms, and NOESY spectra (Jeener et al., 1982), with mixing times of 50 and 100 ms, were recorded with 512 t1 increments of eight scans. These experiments were processed using squared sine-bell window functions, shifted by 60°, in both domains. Zero filling in t1 was applied to obtain a final matrix 2048 × 1024 data points. Two 3D TOCSY-(1H,15N)-HSQC spectra were recorded with mixing times of 25 and 50 ms, a 3D NOESY-(1H,15N)-HSQC spectrum with a mixing time of 100 ms and a 3D ROESY-(1H,15N)-HSQC (Bothner-By et al., 1984; Marion et al., 1989) spectrum with a mixing time of 50 ms. These 3D spectra were recorded with 256 increments in the indirect 1H dimension and 152 increments in the 15N dimension. After processing, the final matrices consisted of 1024 × 256 × 512 points. To determine the 3JHNHα and 3JHNHβ coupling constants, 3D HNHA (Vuister and Bax, 1993) and 3D HNHB (Archer et al., 1991) spectra were obtained. The 10% 13C-labelled sample was used to perform the stereospecific assignment of the valine methyl group by use of a 2D HCCH (Kay et al., 1993). The sequential backbone assignment was checked by use of 3D HNCA (Grzesiek and Bax, 1992) and 3D HNCO (Grzesiek and Bax, 1992) spectra. With the exception of the 3D HNHA and 3D HNHB spectra, all other spectra were obtained with sensitivity enhancement schemes in conjunction with gradient coherence selection (Kay et al, 1993).
The heteronuclear (1H,15N) cross-relaxation rate constants were the average results of two series of spectra recorded with and without 2.5 s of proton saturation of the amide protons. The relaxation data were analysed using in-house developed software.
Processing was carried out on Silicon Graphics O2 work stations using the software NMRPIPE (Delaglio et al., 1995) and analysed with the NMR analysis program REGINE on Silicon Graphics stations.
Structure calculations
Distances were derived from the intensities using a NOESY with 50 ms mixing time and a 3D NOESY-(1H,15N)-HSQC spectrum with 100 ms mixing time, and classified as strong, medium and weak. The upper bound distance constraints were set to 2.8, 3.5 and 5.0 Å (6.5 Å for the 3D NOESY-HSQC, to take spin diffusion into account), respectively. Lower bounds were set to the sum of the van der Waals radii of the atoms involved. Pseudo-atom corrections were applied for the methylene, geminal and aromatic ring protons (Wüthrich et al., 1983). Constraints involving methyl groups were corrected for the three-proton intensity and a pseudo-atom correction of 0.3 Å was applied (Koning et al., 1990). In total, 1228 unique constraints were obtained, of which there were 417 sequential, 101 medium-range, 497 long-range NOEs and 54 ambiguous constraints. Twenty-seven hydrogen bonds were derived from the analysis of the amide proton exchange experiments, the oxygen acceptor being determined on the basis of the ternary structure and after analysis of the calculated structures. The hydrogen bonds were turned into two types of distance constraints: the HN–O distance was set between lower and upper bond distances of 1.5 and 2.4 Å, and the N–O distances to lower and upper bonds of 2.5 and 3.4 Å. From the ratio of the cross-peak to diagonal peak in the 3D HNHA experiment, 71 3JHNHα couplings were identified. The ϕ angles derived from 3JHNHαwere translated to dihedral constraints of –140 ± 20° for the β-sheet region and –60 ± 20° for residue 722. Also, 35 3JHNHβ couplings were identified by analysis of the 3D HNHB experiment. The combined analysis of the 3D TOCSY-HSQC (mixing time 25 ms), the 3D ROESY-HSQC and the 3D HNHB experiments resulted in the determination of the χ1 rotameric states, together with the stereospecific assignment of the β protons. Structures were calculated using the program X-PLOR (Brünger, 1992), version 3.9. An initial structure with proper geometry was generated and then derived into 50 structures, which were refined using torsion angle dynamics (Rice et al., 1994; Stein et al., 1997). First, 3000 steps of 15 fs of torsion angle dynamics were performed at high temperature (50 000 K). Then the structures were cooled down to 1000 K, in temperature steps of 100 K, using torsion angle dynamics. Finally, Cartesian molecular dynamics were used to cool down the structures from 1000 to 300 K, in temperature steps of 50 K. The structure refinement was done as an iterative procedure. An initial set of structures was calculated using ∼500 restraints. Inspections of the initial structures helped in reducing several ambiguities in the assignment of additional NOESY cross-peaks. With the additional restraints, a new set of structures was calculated. This was repeated 25 times, until the total set of NOEs was used. From the final 50 structures, 20 were selected on the basis of low restraint energy and few constraint violations.
The determination of the restraint violations was performed using the software Aqua (Laskowski et al., 1996). The analysis of the structures was done using Procheck-NMR (Laskowski et al., 1996), Promotif (Hutchinson et al., 1996) and MOLMOL (Koradi et al., 1996).
Acknowledgments
Acknowledgements
We thank Dr Gert Folkers for advice and helpful discussions, and Albert George for technical assistance. S.M. was the recipient of an EMBO long-term fellowship (1996-ALT456). This work was supported by grants from the EC ‘Biotechnology’ programme and from the EC programme ‘Access to Large Scale Facilities’ to R.B. and C.O.G. Additional financial support from the Netherlands Foundation for Chemical Research (NOW/CW) to R.B. and from the Italian CNR (Progetto Strategico), MURST-CNR Biotechnology Program L.95/95 and MURST (PRIN ‘Nucleic acid–protein interaction’) to C.O.G. are gratefully acknowledged.
References
- Abel K., Yoder,M.D., Hilgenfeld,R. and Jurnak,F. (1996) An α to β conformational switch in EF-Tu. Structure, 4, 1153–1159. [DOI] [PubMed] [Google Scholar]
- Ævarsson A., Brazhnikov,E., Garber,M., Zheltonosova,J., Chirgadze,Y., Al-Karadaghi,S., Svensson,L.A. and Liljas,A. (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J., 13, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karadaghi S., Ævarsson,A., Garber,M., Zheltonosova,J. and Liljas,A. (1996) The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure, 4, 555–565. [DOI] [PubMed] [Google Scholar]
- Archer S., Ikura,M., Torchia,D.A. and Bax,A. (1991) An alternative 3D NMR technique for correlating backbone 15N with side-chain Hβ resonances in larger proteins. J. Magn. Reson., 95, 636–641. [Google Scholar]
- Berchtold H., Reshetnikova,L., Reiser,C.O.A., Schirmer,N.K., Sprinzi,M. and Hilgenfeld,R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. [DOI] [PubMed] [Google Scholar]
- Bothner-By A.A., Stephens,R.L. and Lee,Y.M. (1984) Structure determination of tetrasaccharide: transient nuclear Overhauser effect in the rotating frame. J. Am. Chem. Soc., 106, 811–813. [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCornick,F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature, 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Brock S., Szkaradkiewicz,K. and Sprinzl,M. (1998) Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol., 29, 409–417. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR v3.1 Manual. Yale University Press, New Haven, CT. [Google Scholar]
- Czworkowski J., Wang,J., Steitz,T.A. and Moore,P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPIPE: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Griesinger C., Otting,O., Wüthrich,K. and Ernst,R.R. (1988) Clean-TOCSY for 1H-proton spin systems identification in macromolecules. J. Am. Chem. Soc., 110, 7870–7872. [Google Scholar]
- Grzesiek S. and Bax,A. (1992) Improved 3D triple resonance NMR techniques applied to a 31 kDa protein. J. Magn. Reson., 96, 432–440. [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Pon,C.L. and Kaji,A. (1971) Initiation factor dependent release of aminoacyl-tRNAs from complexes of 30S ribosomal subunits, synthetic polynucleotide and aminoacyl tRNA. Biochem. Biophys. Res. Commun., 45, 1312–1319. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O., Severini,M., Spurio,R., La Teana,A. and Pon,C.L. (1991) Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J. Biol. Chem., 266, 16356–16362. [PubMed] [Google Scholar]
- Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A., Spurio,R., Tomsic,J. and Pon,C.L. (2000) In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 477–494. [Google Scholar]
- Hartz D., McPheeters,D.S. and Gold,L. (1989) Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev., 3, 1899–1912. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sanders,C. (1993) Protein structures comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hutchinson E.G. and Thornton,J.M. (1996) Promotif—a program to identify and analyze structural motif in proteins. Protein Sci., 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeener J., Meier,B.H., Bachman,P. and Ernst,R.R. (1982) Investigations of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys., 71, 4546–4553. [Google Scholar]
- Jurnak F. (1985) Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science, 230, 32–36. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Xu,G.-Y., Singer,A.U., Muhandiram,D.R. and Forman-Kay,J.D. (1993) A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlation in H2O samples of proteins. J. Magn. Reson. B, 101, 333–337. [Google Scholar]
- Kjeldgaard M., Nissen,P., Thirup,S. and Nyborg,J. (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure, 1, 35–50. [DOI] [PubMed] [Google Scholar]
- Koning M.M.G., Boelens,R. and Kaptein,R. (1990) Calculation of the nuclear Overhauser effect and the determination of proton–proton distances in the presence of internal motions. J. Magn. Reson., 90, 111–123. [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- La Cour T.F., Nyborg,J., Thirup,S. and Clark,B.F. (1985) Structural details of the binding of guanosine diphosphate to elongation factor Tu from E.coli as studied by X-ray crystallography. EMBO J., 4, 2385–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann,J.A.C., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) Aqua and Procheck-NMR: programs for checking the quality of the programs solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1993) Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl Acad. Sci. USA, 90, 4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion D., Driscoll,P.C., Kay,L.E., Wingfield,P.T., Bax,A., Gronenborn,A.M. and Clore,G.M. (1989) Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three dimensional heteronuclear 1H–15N Hartman–Hahn multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1β. Biochemistry, 28, 6150–6156. [DOI] [PubMed] [Google Scholar]
- Misselwitz R., Welfle,K., Krafft,C., Gualerzi,C.O. and Welfle,H. (1997) Translational initiation factor IF2 from Bacillus stearothermophilus: a spectroscopic and microcalorimetric study of the C-domain. Biochemistry, 36, 3170–3178. [DOI] [PubMed] [Google Scholar]
- Moazed D., Samaha,R.R., Gualerzi,C. and Noller,H.F. (1995) Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- Moreno J.M.P., Dyrskjøtersen,L., Kristensen,J.E., Mortensen,K.K. and Sperling-Petersen,H.U. (1999) Characterization of the domains of E.coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Lett., 455, 130–134. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F.C. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen P., Thirup,S., Kjeldgaard,M. and Nyborg,J. (1999) The crystal structure of Cys-tRNACys–EF-Tu–GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure Fold. Des., 7, 143–156. [DOI] [PubMed] [Google Scholar]
- Petersen H.U., Roll,T., Grunberg-Manago,M. and Clark,B.F.C. (1979) Specific interaction of initiation factor IF2 of E.coli with formylmethionyl-tRNAfMet. Biochem. Biophys. Res. Commun., 91, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Piantini U., Sørensen,O.W. and Ernst,R.R. (1982) Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc., 104, 6800–6801. [Google Scholar]
- Polekhina G., Thirup,S., Kjeldgaard,M., Nissen,P., Lippmann,C. and Nyborg,J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure, 4, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Pon C.L., Paci,M., Pawlik,R.T. and Gualerzi,C.O. (1985) Structure–function relationship in Escherichia coli initiation factors. Biochemical and biophysical characterization of the interaction between IF-2 and guanosine nucleotides. J. Biol. Chem., 260, 8918–8924. [PubMed] [Google Scholar]
- RajBhandary U.L. and Chow,C.M. (1986) Initiator tRNAs and initiation of protein synthesis. In Söll,D. and RajBhandary,U.T. (eds), tRNA. Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 511–528. [Google Scholar]
- Rice L.M. and Brünger,A.T. (1994) Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins, 19, 277–290. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Guillon,J.M., Meinnel,T., Mechulam,Y., Dardel,F. and Blanquet,S. (1996) Molecular recognition governing the initiation of translation in Escherichia coli. Biochimie, 78, 543–554. [DOI] [PubMed] [Google Scholar]
- Sette M., van Tilborg,P., Spurio,R., Kaptein,R., Paci,M., Gualerzi,C.O. and Boelens,R. (1997) The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J., 16, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M. (1994) Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem. Sci., 19, 245–250. [DOI] [PubMed] [Google Scholar]
- Spurio R. et al. (2000) The C-terminal sub-domain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J. Biol. Chem., 275, 2447–2454. [DOI] [PubMed] [Google Scholar]
- Stein E.G., Rice,L.M. and Brünger,A.T. (1997) Torsion angle molecular dynamics: a new efficient tool for NMR structure calculation. J. Magn. Reson., 124, 154–164. [DOI] [PubMed] [Google Scholar]
- Vachon G., Laalami,S., Grunberg-Manago,M., Julien,R. and Cenatiempo,Y. (1990) Purified internal G-domain of translational initiation factor. Biochemistry, 29, 9728–9733. [DOI] [PubMed] [Google Scholar]
- van der Hofstad G.A.J.M., Buitenhek,A., van den Elsen,P.J., Voorma,H.O. and Bosch,L. (1978) Binding of labeled initiation factor IF-1 to ribosomal particles and the relationship to the mode of IF-1 action in ribosome dissociation. Eur. J. Biochem., 89, 221–228. [DOI] [PubMed] [Google Scholar]
- Vuister G. and Bax,A. (1993) Quantitative correlation: a new approach for measuring homonuclear three-bond J (HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc., 115, 7772–7777. [Google Scholar]
- Wishart D.S., Bigam,C.G., Yao,J., Abildgaard,F., Dyson,H.J., Oldfield,E., Markley,J.L. and Sykes,B.D. (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR., 6, 135–140. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Billeter,M. and Braun,W. (1983) Pseudo-structures for the 20 common amino acids for use in studies of protein conformations with nuclear magnetic resonance. J. Mol. Biol., 169, 949–961. [DOI] [PubMed] [Google Scholar]
- Zucker F.H. and Hershey,J.W.B. (1986) Binding of Escherichia coli protein synthesis initiation factor IF1 to 30S ribosomal subunits measured by fluorescence polarization. Biochemistry, 25, 3682–3690. [DOI] [PubMed] [Google Scholar]