Abstract
This paper describes a method for building efficient representations of large sets of brain images. Our hypothesis is that the space spanned by a set of brain images can be captured, to a close approximation, by a low-dimensional, nonlinear manifold. This paper presents a method to learn such a low-dimensional manifold from a given data set. The manifold model is generative—brain images can be constructed from a relatively small set of parameters, and new brain images can be projected onto the manifold. This allows to quantify the geometric accuracy of the manifold approximation in terms of projection distance. The manifold coordinates induce a Euclidean coordinate system on the population data that can be used to perform statistical analysis of the population. We evaluate the proposed method on the OASIS and ADNI brain databases of head MR images in two ways. First, the geometric fit of the method is qualitatively and quantitatively evaluated. Second, the ability of the brain manifold model to explain clinical measures is analyzed by linear regression in the manifold coordinate space. The regression models show that the manifold model is a statistically significant descriptor of clinical parameters.
Keywords: Neurological Image Analysis, Population Analysis, Brain MRI, Manifold Learning, Computer Aided Clinical Diagnosis
1. Introduction
Many neuroimaging applications require a summary or representation of a population of brain images. The conventional approach is to build a single template, or atlas, that represents a population [1, 2, 3]. Recent work introduced clustering-based approaches, which compute multiple templates in a data driven fashion [4, 5]. Each template represents a part of the population. In a different direction, researchers proposed kernel-based regression of brain images with respect to an underlying parameter [6, 7, 8]. This yields a continuous curve in the space of brain images that estimates the conditional expectation of a brain image given the parameter value. A natural question that arises based on these investigations is whether the space spanned by a set of brain images can be approximated by a low-dimensional manifold. Our hypothesis is that a low-dimensional, nonlinear model can effectively represent variability in brain anatomy. This paper describes a method to learn a manifold model from a given population and evaluate its geometric fit and statistical properties.
Recent work on statistical analysis of brain images with clinical data shows that shape is a statistically significant predictor for various clinical parameters [9, 10, 11, 12, 13, 14, 15, 16]. As such, we are interested in finding a manifold representation that captures shape variability across large sets of images. Such a manifold model is interesting in several ways. The manifold parametrization gives rise to a coordinate system for the data that indicates its position on the manifold. If the manifold is constructed to reflect shape differences, then brain shape can be quantified in this new coordinate system. In this way, the manifold coordinates can be used as a proxy for statistical analysis of brain populations. The manifold coordinates of a particular image (e.g. patient in a clinical setting) could be used to compare against examples in a database with similar brain shape and known clinical history and to predict the likelihood of specific pathologies. Projections of images onto the manifold of brain images could also be used to construct priors or atlases that would aid in automatic processing or clinical studies.
The approach in this paper builds on existing manifold learning techniques to obtain a manifold model from a set of brain MR images. Manifold learning is a specific approach to nonlinear dimensionality reduction based on the assumption that data points are sampled from a low-dimensional manifold embedded in a high-dimensional ambient space. The aim is to uncover the low-dimensional manifold structure from the samples in the high-dimensional ambient space. Many methods for manifold learning have been proposed in the machine learning literature. Much of the recent work [17, 18, 19, 20] has focused around global or spectral methods. These methods have a closed form solution based on the spectral decomposition of a matrix that is compiled based on local properties of the input data. The bulk of manifold learning research has focused on low-dimensional manifolds embedded in high-dimensional spaces with Euclidean metrics. The space of brain images, however, does not fit directly into this setting. Research in shape analysis has shown that a Euclidean metric is not suitable for capturing shape differences in images [21]. In computational anatomy, researchers commonly resort to metrics based on coordinate transformations that align the images [4, 5]. Therefore, the low-dimensional manifold we aim to learn is embedded in the space of images with a metric induced by coordinate transformations on the image-domain, as illustrated Figure 1. Note that the structure induced by image metrics based on transformations is often described as a manifold — in this paper we reserve this term to refer to the manifold of brain images as described by the data.
Figure 1.
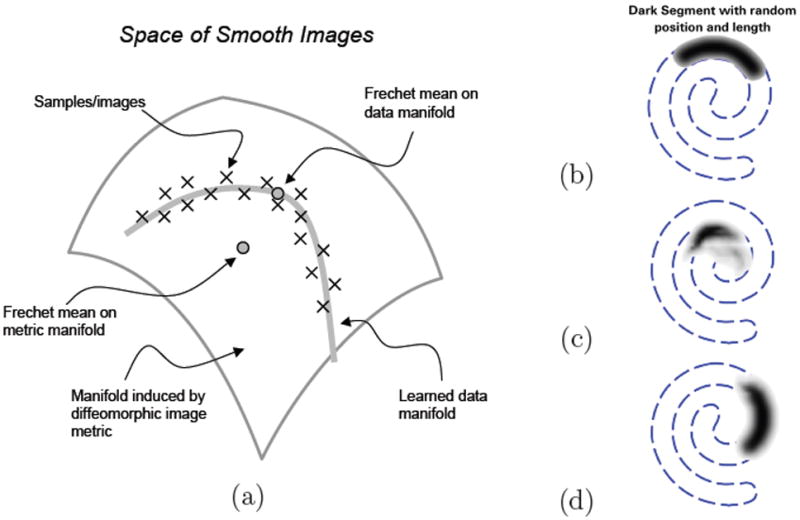
(a) Illustration of image data on a low-dimensional manifold embedded in a diffeomorphic space. (b) A set of images consists of random length/position segments from a spiral. (c) The Fréchet mean in the space induced by a metric based on coordinate transformations is not like any example from the set. (d) Fréchet mean on data-driven manifold reflects the image with average parameter values.
In many neuroimaging applications, it is necessary to map from ambient space to manifold coordinates or to construct brain images given manifold coordinates, which requires a generative model. These are the capabilities provided for linear models by principal component analysis (PCA), for instance, that allow one to compute loadings (manifold coordinates), to project data on the linear subspace, and to compute the residual or approximation error associated with these projections. Manifold learning methods thus far are mostly concerned with finding a low-dimensional parametrization of the data, but do not generally provide the tools to project or construct new data points. We propose to use the manifold coordinates obtained by one of the global manifold learning approaches to construct a generative manifold model that explicitly describes the manifold as a parametric surface in the ambient space. The parametric surface is formulated as a kernel regression over the manifold coordinates, that is, an estimate of the mean of the distribution of the manifold given the manifold coordinate. The reverse, a coordinate mapping from ambient space to manifold coordinates, is achieved in a similar manner. Weighted averaging of the manifold coordinates of the initial data set, again by kernel regression, yields a continuous mapping in the ambient space. The proposed methodology for the generative manifold model is motivated by the statistical representation of scattered data using principal surfaces [22]. In previous work [23], we describe a formulation by which generative manifold models via kernel regression give rise to principal surfaces, a nonlinear extension to PCA, as formally defined by Hastie [22]. Thus, the proposed framework for brain image analysis represents a statistically sound, higher-order representation of the space of brains.
In [24] we showed that it is possible to approximate the space of brain images by a low dimensional manifold. In this paper we present the method in greater depth. Additionally we show that the learned manifold can be used for statistical analysis on brain image populations. We apply the proposed approach to the OASIS and ADNI brain database, and show that the learned manifold provides a good fit in terms of projection distance and as a proxy for statistical analysis. Linear regression of the learned manifold coordinates with several clinical parameters provides strong evidence that the proposed manifold representation of brain image data sets captures important clinical trends.
2. Related Work
An important aspect of our work is the ability to measure image differences in a way that captures shape. It is known that the L2 metric does not adequately capture shape differences [21]. There are a variety of alternatives, most of which consider coordinate transformations instead of—or in addition to—intensity differences. A large body of work [25, 26, 27, 28] has examined distances between images using high-dimensional image warps that are constrained to be diffeomorphisms. Thus, distances between images are measured by computing a minimal diffeomorphic map, according to an appropriate metric on the transformation, between two images. This metric defines an infinite-dimensional manifold consisting of all shapes that are equivalent under a diffeomorphism. In a similar fashion, we define a metric based on elastic deformations. Our hypothesis, however, is that the space of brains is essentially (or approximately to within some level of noise) of significantly lower dimension. That is, not all images reachable by a diffeomorphic deformation are likely to be brains.
An alternative metric for image differences is proposed by [29, 30], specifically for the task of manifold learning for sets of time-series data. They use a set of local features (phase maps produced from Gabor filters), which are designed to capture the local offsets of edges and other features of interest. Although this is a computationally efficient way to capture local shape differences, this approach does not lend itself to kernel-based construction of images, which is essential to the work in this paper.
Several authors [6, 7, 8] have proposed kernel-based regression of brain images. The main distinction of the work described in this paper is that the underlying parametrization is learned from the image data. Our interest is to uncover potentially interesting structures from the image data and sets of parameters that can be compared against clinical variables. Kernel regression is used to explicitly represent the learned manifold in the ambient space.
The very limited work on learned manifolds for medical image analysis considers only manifolds that are generated from known variables with individual patients. For instance, Zhang et al. [30] use manifold learning specifically to improve segmentation in cardiac MR images. In that application the breathing and heartbeat form a 2D manifold. In this paper, we propose to discover an unknown manifold structure from a large database of brain images in a way that explicitly learns the structure of inter subject variability. In LEAP [31], the Laplacian eigenmaps algorithm [18] is employed to build an information diffusion based segmentation approach. The embedding quality is tested based on a clustering measure and leads to highly accurate segmentation results and indicates that the manifold assumption is valid. This work proposes a generative approach to examine the underlying manifold hypothesis.
3. Methodology
The approach proposed in this paper is best described as a two step process. First we learn a parametrization that captures the manifold structure in the data. Second we build a generative model, based on the learned parametrization, that explicitly represents the manifold as a parametric surface in the ambient space. The model provides the tools to construct data points on the manifold given parameters and to infer parameters given data points in the ambient space. We refer to the parameter space of the surface as the manifold coordinates C ⊂ ℝd where d is the intrinsic dimensionality of the manifold, the ambient space as A and the manifold as M ⊂ A.
For the first step, we use isomap [20], a manifold learning algorithm that finds a low-dimensional Euclidean configuration of points that approximately preserves geodesic distances. Isomap computes a piecewise linear approximation to the geodesic distance through the construction of a nearest neighbor graph using the metric in the ambient space. The assumption is that the geodesic distances between nearby samples can be accurately approximated by the distance in the ambient space. This corresponds exactly to the definition of a manifold—the metric is locally linear. Metric multidimensional scaling [32] on the pairwise approximate geodesic distances yields a distance preserving d-dimensional Euclidean configuration of points. Thus, applying isomap yields a discrete mapping zi = f̃ (yi) defined only on the original input samples, the set of images Sy = {y1, … yn} ∈ A.
The second step is the construction of the generative manifold model. The brain images Sy are samples from a random variable Y with an arbitrary density p(y) defined on A. Assume we are given a coordinate mapping f : A → C, representing a d-dimensional parametrization of A. Then the conditional expectation g(x) = E[Y ∣f (Y) = x] is a direct and explicative way to model a d-dimensional surface in A. Each point on the manifold represents the average over all brain images with the same parameter f(Y). For example, if f maps to age, g(x) is the average brain image at each age x. This is illustrated in [7] using manifold kernel regression as an unbiased estimate of the conditional expectation. However, the mapping f̃ from isomap is only defined on the input data points, furthermore it is only an approximation to the manifold coordinates. Hence we model the coordinate mapping f on the complete ambient space as a kernel regression on the discrete zi = f̃ (yi).
In summary the methodology consists of the following steps:
Compute Sz = {z1, … , zn} by isomap on Sy.
-
Build the generative manifold model with
the coordinate mapping f by kernel regression over Sz and
the explicit manifold representation g(x) = E[Y ∣f(Y) = x] estimated by manifold kernel regression.
An important observation is that for both steps we do not need an explicit representation of the data in the ambient space. Isomap as well as the kernel regressions only rely on distances. Thus an adaption to an image space only needs an appropriate metric on A that captures shape differences. Furthermore both isomap and kernel regression only require distance to be accurate between nearby samples, large distances are not directly used but approximated using the nearest neighbors graph. We introduce a distance measure that captures shape differences between images based on small deformations using an elastically regularized coordinate transformation and show how to build the generative manifold model in this setting. The proposed distance measure is an approximation to the diffeomorphic metric for small deformations, justifying the notion of learning a submanifold of the space of diffeomorphisms.
3.1. Diffeomorphic Space and Approximation with Elastic Deformations
For sets of brain images, we restrict our attention to the set of square-integrable functions on the domain Ω ⊂ ℝ3, i.e., the infinite-dimensional space L2(Ω). In shape analysis and computational anatomy tasks [1, 7, 2] diffeomorphic mappings are often used to assess shape differences. A diffeomorphic coordinate transformation between two images can be written as ϕ(r, 1), where
| (1) |
and υ(r, t) is a smooth, time varying vector field. The diffeomorphic framework defines a Riemannian metric ∥υ(r, τ)∥Q, based on a differential operator Q, on the space of diffeomorphic transformations. The Riemannian metric leads to the geodesics , here e is the identity transformation. This induces a metric d between images yi and yj
| (2) |
with the squared Euclidean distance of the image intensities. The metric (2) is only well defined for images within the orbit of the diffeomorphic transformations for a given template image. In this setting, the ambient space A is a manifold in L2(Ω) induced by the Riemannian manifold of diffeomorphic transformations acting on a template image.
Taking advantage of the fact that the proposed method relies only on distances between nearby samples, we propose to compute local approximations to the diffeomorphic metric. For two images that are very similar, the diffeomorphism is close to the identity and υ is small. Because the velocities of the geodesics are smooth in time [25], we can approximate the integrals for the coordinate transform and geodesic distance by a single vector field
| (3) |
Thus we can locally approximate the diffeomorphic distances based on an elastically-regularized deformation u
| (4) |
with ∥·∥Q as in the diffeomorphic setting and ε allows for noise in the images. Therefore the proposed approach effectively learns a submanifold, estimated from the data, within the ambient space A of diffeomorphisms.
For symmetry we define the distance as
| (5) |
For this work we choose . The free parameter α ∈ [0, 1], regulates the weight of the penalty between magnitude and smoothness of the warp. In all our experiments we set α = 0.9. For α = 1 the magnitude is disregarded and a simple translation would yield a zero distance. In our experiments we found that a penalty on magnitude adds important information, such as for example the magnitude of the change in ventricle size. Note that it is an open problem to show that (5) satisfies the triangle inequality and hence is a metric or a semimetric.
To compute the elastic deformation distance for a pair of discrete images, we use a gradient descent scheme. For the L2 constraint , the squared image intensity differences we introduce a Lagrange multiplier λ
| (6) |
Taking the first variation of (6) with respect to u results in the partial differential equation:
| (7) |
and the derivative of L with respect to the Lagrange multiplier λ is
| (8) |
We alternate between solving the PDE in (7) with finite forward differences for a fixed λ and update λ based on (8) until the constraint is satisfied. We use a multiresolution, coarse to fine, optimization strategy to avoid local minima. Thus to register two images we have the steps
Initialize u with the identity transform u(x) = 0
Set λ = 10. This initial value depends on the range of the image intensities with a trade off between fast convergence for large λ and accuracy using a smaller λ.
-
For each scale from coarse to fine:
Solve the PDE in equation (7).
If the mean squared error between target and moving image is larger than ε increase λ and repeat (a).
Implementation details for the numerical solution of PDE’s resulting from the minimization of transformations based on regularized vector fields are described in detail in Modersitzki [33].
3.2. Generative Manifold Model
The generative manifold model is based on a coordinate mapping f : A → C and formulating the d-dimensional manifold M as the conditional expectation g(x) = E[Y ∣f(y) = x]. The composition of the two mappings g ο f provides a projection operator onto the manifold.
We compute the conditional expectation g(x) = E[Y∣f(y) = x] of a finite set of samples using a Nadaraya-Watson kernel regression, which converges to the expectation asymptotically. For the coordinate mapping f, we can use the point samples from Sz = {zi, z2, … , zn}, the output of isomap on the pairwise elastic distances, but they must be extended to A by some suitable scattered data interpolation. In practice, we extend the data through an approximation that smooths out the noisy manifold coordinates obtained by isomap on the sample data. Thus, we also formulate f using Nadaraya-Watson kernel regression.
The level sets of the function f define equivalence classes of brain images that have the same manifold coordinates. The conditional expectation g(x) = E[Y ∣f(y) = x] represents the distribution of a given class of brains (i.e. choice of manifold parameters) by its average. This formulation reflects the fact that brain images will generally not lie exactly on a low-dimensional manifold. The assumption is that a collection of brain images can be captured by a probability density that includes two components: the location on the manifold and a noise term which is transverse to the manifold. In this way the proposed manifold resembles a nonlinear generalization of PCA, such as the method of principal surfaces. Informally, principal surfaces, as defined by Hastie [22], pass through the middle of a distribution and minimize residual variance. In recent work [23], we show the formal relationships between the generative manifold model based on kernel regression mappings and principal surfaces. However, that work requires an optimization over the parameters Sz of f and is beyond the scope of this paper.
Note that the ambient space A is not Euclidean and thus the independent variables of f and the dependent variables of g are not in Euclidean space but in a space endowed with a metric based on coordinate transformations. For the coordinate mapping f this amounts to
| (9) |
Recall that d here is the elastic deformation metric as defined in section 3.1. For the mapping g we adopt the manifold kernel regression formulation in [7] based on computing a weighted Fréchet mean. The Fréchet mean is defined as
| (10) |
using the kernel regression weights based on the manifold coordinates. Thus, the kernel regression g with distances based on elastic deformations yields
| (11) |
The minimization in Equation 11 is computed by the iterative method described in [7]. This is a greedy algorithm that alternates between computing transformations υi that minimize d(y, yi) for a fixed target image y and updating the target image by . We use an isotropic Gaussian kernel for both mappings , i.e., and . The bandwidths for the kernels are selected based on the average k-th nearest neighbors distance with k = 15, i.e., and with nnk being the k-th nearest neighbor of xi and yi respectively.
The image in Figure 1(b) summarizes an image data set consisting of 100 images of dark spiral segments with varying length and location. Figure 2 shows images constructed by the generative manifold model learned from the 100 images. Thus, the images in Figure 2 depict samples on the manifold embedded in the ambient space. Figure 1 also shows the Fréchet mean of the data in the diffeomorphic space and the Fréchet mean on the learned manifold. The Fréchet mean of the learned manifold suggest that it can be beneficial and necessary to model image data sets by a low-dimensional manifold.
Figure 2.

Sampling along the first dimension of the manifold learned from images of spiral segments. The images shown are constructed, as discussed in section 3, from the learned manifold parametrization.
3.3. Discussion of the Manifold Model
The distance measure fundamentally defines which features are modeled. The metric used in this work is a trade-off between measuring length of the vector field and smoothness. Spatial normalization removes the effects of scale, orientation and translation prior to computing the distance measures. If this pre-processing step is omitted the manifold estimation will incorporate scale, orientation and translation into the resulting model and, depending on sample size, can result in masking out more subtle changes.
Note that the proposed approach is not restricted to work within the diffeomorphic framework. Other distance measures can readily be incorporated into the general manifold estimation framework. This provides opportunities for adaption to different image modalities or a more supervised approach. For example, the metric can be tailored to a specific region of interest or information theoretic measures such as Bregman divergences can be employed. For a supervised approach clinical information could be integrated into the metric, leading to a manifold estimation with a trade-off between geometric, image information based, fit and prediction of clinical information. However, this would require a careful cross-validation strategy to avoid overfitting.
The manifold model provides additional statistical and geometrical information. For example, a density estimation in the coordinate space, as shown in Figure 11, can be employed to detect subjects with low probabilities. For a geometric notion of an outlier, the projection distance can be used, as illustrated in Figure 5. The notion of projection distance and density on manifold could be combined to build a full density estimate in the ambient space, based on density on the manifold and variance orthogonal to the manifold.
Figure 11.
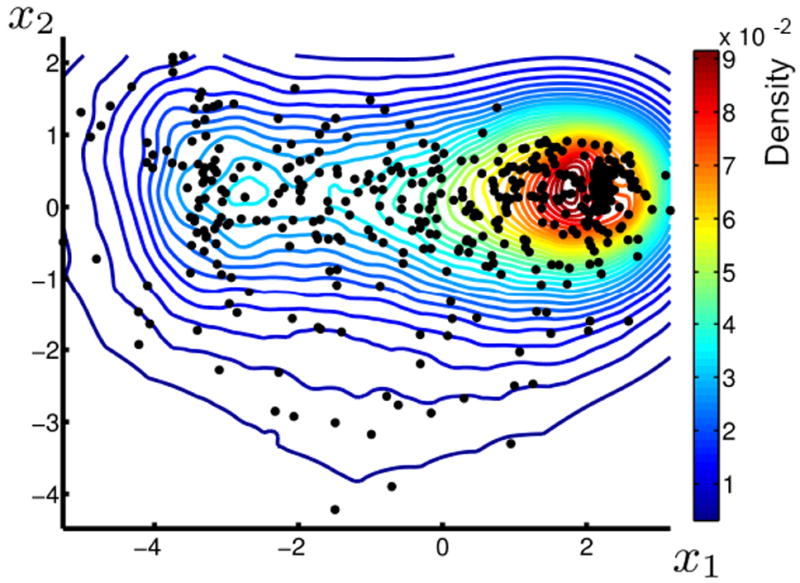
Kernel density estimate of subjects from the OASIS database projected onto a 2D manifold representation.
Figure 5.
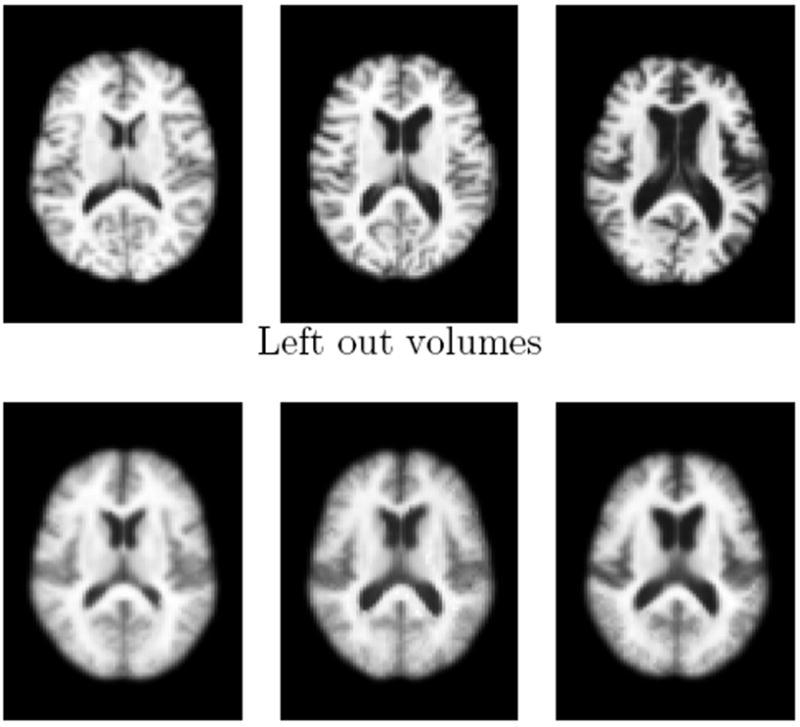
Reconstructions (bottom row) of three left out images (top row) with small, medium and large ventricle sizes. The corresponding projection distances are 1.07, 0.81 and 1.23 respectively. Note that the manifold model does not have enough data to represent the subject on the right, which shows a, for this data set atypical, separation between the anterior and posterior horn.
The use of Isomap for building the coordinate mapping f ensures that increasing the dimensionality of the manifold model will not change the lower dimensional coordinates. However, the kernel regression representation of the manifold does not result in a perfect reconstruction with increasing dimension for finite sample sizes.
Kernel regression converges asymptotically to the true conditional expectation with increasing sample size and decreasing kernel bandwidth. The bandwidth in the kernel regression plays an important role, a too small bandwidth results in an overfit of the data while a too large bandwidth results in an overly smooth estimate of the conditional expectation. For example, the limit case of a zero bandwidth results in direct reconstruction of the original data points. On the other hand, for infinite bandwidth the regression results in a point estimate, the sample mean. The selection of an optimal kernel bandwidth is only in theory possible, based on knowledge of the underlying density. In this work we select the kernel bandwidth based on the average distance of the k = 15 nearest neighbor. This is a heuristic that yields an estimate such that, on average, 15 points are within the 60th percentile of the Gaussian kernel and thus have a relatively large influence on the estimate. Changing k to 10 or 20 does not affect the outcomes significantly. Note that both dimensionality and kernel bandwidth affect the quality of the manifold fit. Thus it is possible to have a decreasing fit with increasing manifold dimension depending on the kernel bandwidth.
Additionally the kernel regression forces the estimated manifold to lie within the convex hull of the given data set. Thus, the projection of a brain image, with a pathology not present in the original data set, will result in a brain image that resembles an image from the original data set, as illustrated in Figure 5.
4. Results
The performance of the manifold model is evaluated in two ways. First, the geometric manifold fit is examined qualitatively and quantitatively in terms of projection distances. Second, the statistical explanatory power of the learned manifold parametrization with respect to clinical data is explored. The statistical significance is analyzed with linear regression of the manifold coordinates with the clinical parameters.
The evaluations were tested on 156 images of the ADNI Database and the full OASIS cross-sectional database.
4.1. OASIS Data Set
The OASIS brain database 1 consists of T1 weighted MRI of 416 subjects aged 18 to 96. 100 of the subjects over the age of 60 are diagnosed with mild to moderate dementia. The images are provided skull stripped, gain field corrected and registered to the atlas space of Talaraich and Tournoux [34] with a 12-parameter affine transform. Associated with the data are several clinical parameters. For the statistical analysis we restrict our attention to age, mini mental state examination (MMSE) and clinical dementia rating (CDR). Figure 3 shows the population characteristics in terms of age, MMSE and CDR.
Figure 3.
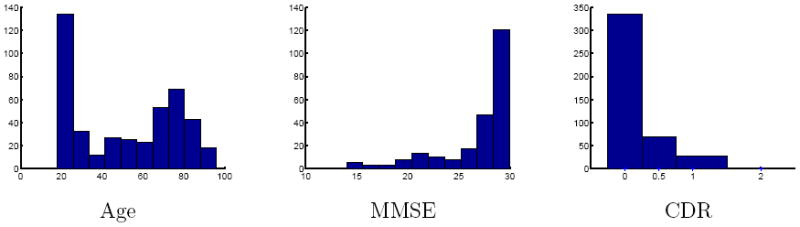
Histograms of age, MMSE and CDR for the OASIS data set.
4.2. ADNI Data Set
We used a subset of T1 weighted MRI of 156 subjects aged 57 to 88 of the ADNI database 2. The images are provided geometry distortion corrected, intensity nonuniformity corrected and histogram peak sharpend. We additionally processed the images with a histogram equalization by a piecewise linear fit to the three histogram peaks of CSF, gray matter and white matter and an affine registration to an unbiased atlas. The population contains subjects diagnosed normal (38 subjects), with mild cognitive impairment (MCI, 84 subjects) and early Alzheimer’s disease (AD, 34 subjects). For the statistical analysis we used age, MMSE and the diagnosis. Figure 4 shows the population characteristics in terms of age and MMSE. The 156 subjects were selected to represent a large portion of subjects diagnosed with MCI, with the aim to span the development from healthy to early AD.
Figure 4.
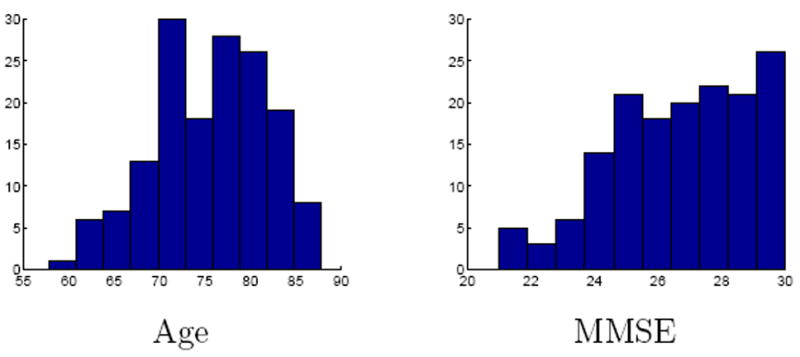
Histograms of age, MMSE and diagnosis for the ADNI data set.
4.3. Manifold Fit Evaluation
We compare the manifold fit against principal component analysis in Euclidean space by projection onto the d dominant (largest eigenvalue) eigenvectors. A nonlinear manifold representation in Euclidean space did not improve over PCA.
For PCA, we left one image out during the computation of the principal components and projected this image onto the principal components. This procedure was repeated to leave each image out once. For the generative manifold model, this involved computing isomap without the left out image yi, i.e. removing the row and column of the corresponding image from the pairwise distance matrix, and then compute the projection distance g(f(yi)) with g and f based on the reduced data set. Figure 5 shows three images from the ADNI data set and reconstructions based on a 2D manifold model.
OASIS Data
Figure 6 shows axial slice 80 on 2D manifold coordinates obtained by the proposed method. A visual inspection reveals that the learned manifold detects the change in ventricle size as the most dominant parameter (x1). It is unclear if the second dimension (x2) captures a global trend.
Figure 6.
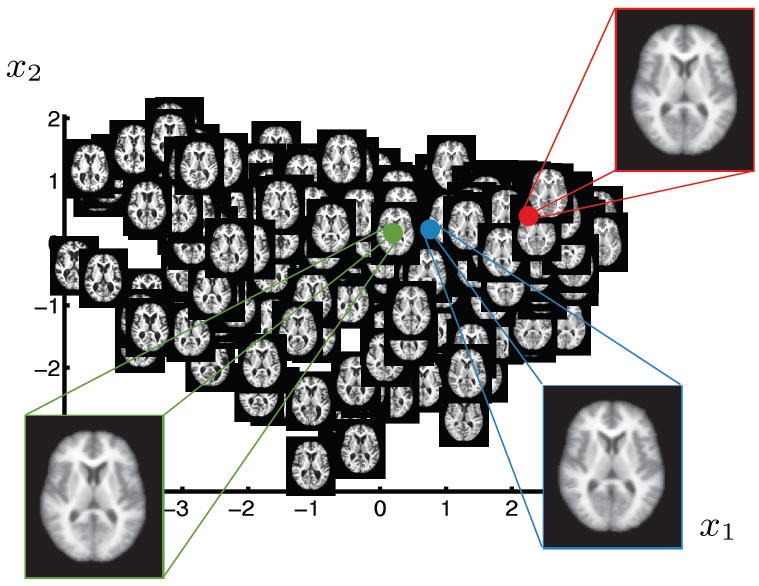
2D parametrization of OASIS brain MRI database obtained by the proposed approach. For each brain image axial slice number 80 is visualized at the corresponding location on the 2D manifold. The insets show the mean (green), median (blue) and mode (red) of the learned manifold and the corresponding reconstructed images.
Figure 7 shows projection distances for the generative manifold model (a) and PCA (b) as a function of the dimensionality. The projection distance is measured as the mean of the distances between the original brain images and their projection on to the learned manifold and scaled by the average nearest neighbor distance:
| (12) |
with nn(yi) the nearest neighbor of yi. A reconstruction error smaller than one indicates that, on average, the projection onto the manifold is closer than the nearest neighbor. The projection distances for PCA are scaled by average nearest neighbor in Euclidean space. Note that the measurements are in different metric spaces, capturing different properties, and as such only provide a qualitative comparison in terms of behavior with increasing dimension. PCA shows very little reduction in projection distance with increased dimension, barely one percent in the first three dimension. While the improvements for the manifold model are not drastic either, roughly ten percent in the first three dimensions, they are an order of magnitude larger than for PCA. The PCA projection distances do not indicate a linear low-dimensional subspace, rather the almost linear decrease suggests a isotropic normal distribution of the data. The change in gradient at dimension three for the manifold model suggests that the available data can be approximately described with a three dimensional manifold. Note that the manifold based approach, due to the kernel regression, will not lead to a 0 projection distance with increasing dimension. Beyond dimension 3 the reduction in projection distances is marginal for the proposed approach. This indicates that a 3 dimensional manifold is close to achieving the minimal projection distance possible for the given amount of data and the chosen kernel bandwidth. The projection distances for the manifold model are smaller than the average nearest neighbor distance, an indication that the learned manifold accurately captures the data. We do not postulate that the space of brains is captured by a 3D manifold. The approach learns a manifold from the available data and thus it is likely that given more samples we can learn a higher-dimensional manifold for the space of brains.
Figure 7.
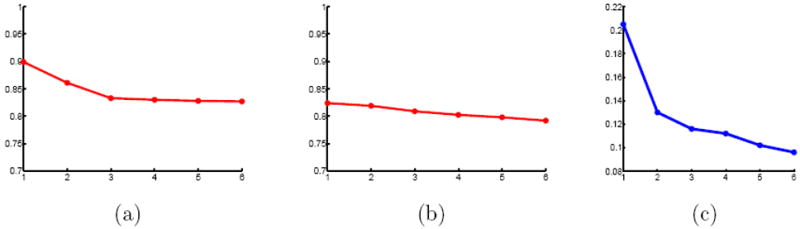
Projection distances, scaled by average nearest neighbor distance (12), for the OASIS data set against dimensionality for (a) the proposed method and (b) PCA. The isomap residual (c) shows the distortion required to embed the manifold in Euclidean space.
Figure 7 (c) shows the isomap residual as a function of the number of dimensions. The isomap residual measures the distortion required to embed the pairwise elastic deformation distances D into a d-dimensional Euclidean space by the 1 − R2 value between D and De, the pairwise distance of the data points in the d-dimensional Euclidean space. That is, how well do the distorted distances De explain the original distances D. The isomap residual roughly agree with the manifold projection distances and suggest also a two to three dimensional manifold.
Figure 8 shows axial slices of brain images generated with the proposed method by applying the mapping g to regularly sampled grid locations on the 2D manifold coordinates shown in Figure 6, i.e. a sampling of the learned brain manifold. The first dimension (x1) shows the change in ventricle size. The second dimension (x2) is less obvious. A slight general trend observable from the axial slices is less gray and white matter as well as a change in frontal horn ventricle shape from elongated to more circular.
Figure 8.
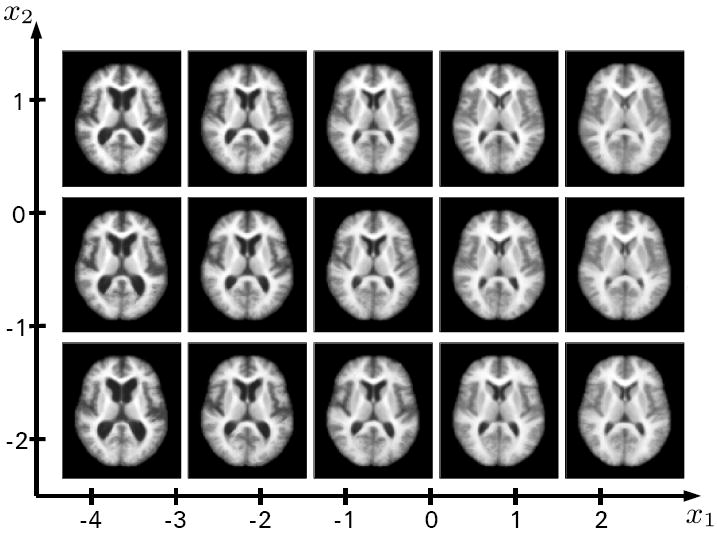
Reconstructions using the proposed method on equally spaced grid locations on the 2D representation shown in Figure 6.
ADNI Data
The results from the ADNI data show similar behavior as the results from the OASIS data set, although less pronounced. Figure 9 shows the projection distances as a function of the number of dimensions for the manifold model and PCA in Euclidean space as well as the isomap residuals. Again, PCA does not show any indication of a low-dimensional linear subspace. The projection distance for the manifold model are less telling than for OASIS data set. This might be explained by the differences in the two populations. Compared to the OASIS data set the ADNI data set spans a much smaller age range with stronger pathologies. It might be expected that the ADNI data set has larger variations in different directions due to the differences in anatomy caused by disease progressions. This is in contrast to the OASIS data set where a large portion of the variation is likely based on age. This explanation is supported by the statistical findings for the OASIS and ADNI data set in Section 4.4, where a description of MMSE or diagnosis in the ADNI data set requires additional dimensions beyond those that best describe age. In contrast, MMSE and CDR are contained within the dimensions that best describe age for the OASIS database.
Figure 9.
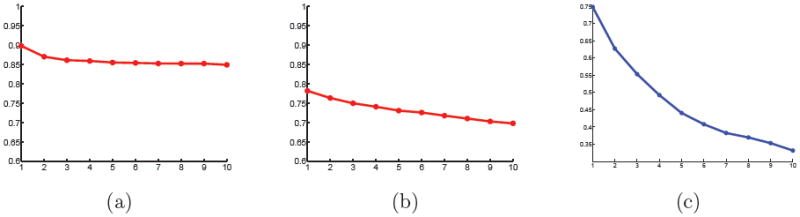
Projection distances on ADNI data set, scaled by average nearest neighbor distance, against dimensionality for (a) the proposed method and (b) PCA. (c) Isomap embedding distortion plotted as a function of dimensionality.
Figure 10 shows the parametrization along the first and second dimension and along the first and sixth dimension. The first and sixth dimension are the dimensions that best explain diagnosis. Again ventricle size is the most significant change observable in both cases along dimension one. In Figure 13 slices of reconstructed images along dimension one and 6 are shown, indicating some additional differences in ventricle shape.
Figure 10.
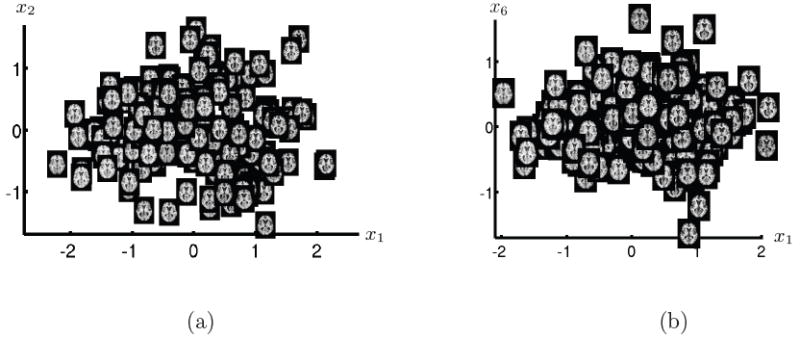
Parametrization along dimensions (a) 1, 2 and (b) 1, 6 of the manifold coordinates of the ADNI data set. For each brain image axial slice number 80 is visualized at the corresponding location on the 2D manifold. The parametrization in (b) shows the two statistical significant manifold coordinate dimensions to explain diagnosis.
Figure 13.
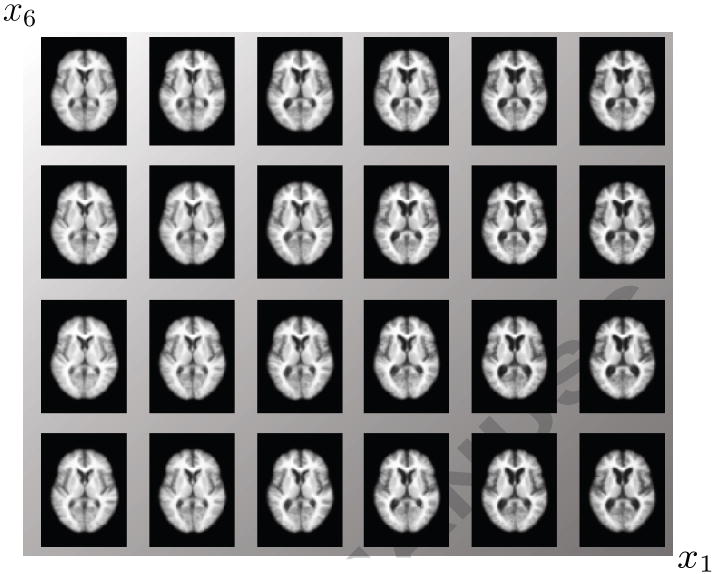
Slice number 80 of the reconstructed images along the statistically significant manifold coordinates (first and sixth dimension) for explaining the diagnosis. The shading indicates the gradient of the corresponding linear model on diagnosis (row 8 in Table 3).
4.4. Statistical Analysis
The coordinate space of the estimated manifold can be analyzed with traditional statistical approaches. Figure 11 shows a kernel density estimate of the distribution of brain images in the coordinate space of the 2D manifold estimated from the OASIS database.
To evaluate the statistical predictive power of the manifold coordinates, we fit multiple linear regression models of clinical data versus manifold coordinates. This includes regression of age, MMSE and CDR for the OASIS data set and age, MMSE and diagnosis for the ADNI data set. We compare the regression models on manifold coordinates to regression models on PCA coordinates. As for the geometric fit, regression models on a nonlinear manifold model (i.e., isomap) in Euclidean space did not improve over PCA. For further comparison we also regressed CDR (OASIS data set), diagnosis (ADNI data set), and MMSE with age.
The regression of PCA and manifold coordinates involves a model selection process to select the independent variables that best describe the clinical parameters. We use the Schwarz’s Bayesian information criterion (BIC) [35] with an exhaustive search to select the optimal model in each case. The BIC is −2 ln L(M)+k ln(n) with n the number of samples, k the number of free parameters of model M and L(M) the maximum likelihood of model M. The BIC is derived based on the asymptotics of a Bayes solution to the model selection problem. The BIC penalizes models with a large number of parameters k, an application of the principal of Occam’s razor. More precisely, the maximum likelihood of a model with a parameters needs to improve over a model with b parameters, with a ≥ b, n(a−b) times to achieve a smaller or equal BIC. In addition to the coordinates, we included age as an independent variable in the models involving MMSE and CDR/diagnosis in the model selection process. This essentially controls for age; independent variables (PCA or manifold coordinates) that do not improve the models over using age as an independent variable are ignored. In all cases ages did not show up in the best fitting models for PCA and the manifold coordinates. Thus, adding age information does not lead to an improved fit under the BIC criterion.
We denote the manifold coordinates with x = (x1, … , xm) and the PCA coordinates with l = (l1, … , lm). Thus the notation
| (13) |
describes a multiple linear regression model of age on the first three manifold coordinates.
OASIS data
Table 1 shows the optimal linear regression results for the OASIS data set. In this data set only 235 out of the 416 subjects have MMSE scores. The regression for MMSE is thus only performed on this subset of the population. For MMSE and CDR the manifold model is the statistically most significant (largest F), and it has the largest R2 statistic with the smallest residual but the differences between the PCA and manifold model are very small. For age, the best model is achieved with a 5D PCA, while the manifold representation achieves the best fit with the first three coordinates. This nicely supports the 3D manifold suggested by the reconstruction errors in Figure 7. Note that the manifold model achieves similar power with three parameters as does the PCA model with five parameters.
Table 1.
Optimal linear regression models from the OASIS data set. The PCA coordinates are denoted with li and the manifold coordinates with xi. A < ε entry denotes smaller than machine precision.
| Model | t | Residual | R2 | F | p-value | |
|---|---|---|---|---|---|---|
|
|
41.5 / -12.5 / 4.2 / -7.1 / -8.7 | 10.5 | 0.82 | 404.9 | < ε | |
|
|
-39.3 / 10.0 / -13.0 | 10.87 | 0.82 | 639.5 | < ε | |
| MMSE = a0 + a1age | -4.0 | 3.59 | 0.06 | 15.82 | 9.3e-05 | |
| MMSE = a0 + a1l1 | -6.6 | 3.40 | 0.16 | 43.13 | 3.3e-10 | |
| MMSE = a0 + a1x1 | 6.8 | 3.36 | 0.18 | 50.3 | 1.6e-11 | |
| CDR = a0 + a1age | 12.0 | 0.27 | 0.25 | 144.5 | < ε | |
| CDR = a0 + a1l1 | 14.9 | 0.26 | 0.34 | 223.9 | < ε | |
| CDR = a0 + a1x1 | -15.8 | 0.25 | 0.36 | 248.5 | < ε | |
The residual for MMSE and CDR is fairly large and it is not clear if the statistical significance is due to the relatively large sample size and large amount of healthy young subjects. Table 2 shows optimal linear models, as before, selected based on BIC and exhaustive search, for subjects aged 60 to 80. The results from this subset are comparable to the results on the full data. This suggests that the manifold is not only capturing the variation induced by the large differences in age among the subjects but also important clinical trends.
Table 2.
Optimal Linear regression models of the OASIS data set restricted to subjects aged 60 to 80.
| Model | t | Residual | R2 | F | p-value | |
|---|---|---|---|---|---|---|
|
|
5.1 / -2.0 | 4.3 | 0.18 | 13.3 | 6.1e-6 | |
|
|
-4.4 / 2.0 | 4.4 | 0.14 | 9.9 | 1.0e-4 | |
| MMSE = a0 + a1age | -1.0 | 4.09 | 0.008 | 1.0 | 0.3 | |
| MMSE = a0 + a1l1 | -4.0 | 3.86 | 0.12 | 15.9 | 1.2e-4 | |
| MMSE = a0 + a1x1 | 4.6 | 3.8 | 0.15 | 20.9 | 1.2e-5 | |
| CDR = a0 + a1age | 3.5 | 0.37 | 0.09 | 11.2 | 1.1e-3 | |
| CDR = a0 + a1l1 | 4.9 | 0.36 | 0.16 | 24.1 | 3.0e-6 | |
| CDR = a0 + a1x1 | -5.5 | 0.35 | 0.20 | 30.0 | 2.4e-7 | |
ADNI Data
Table 3 shows the optimal linear regression results for the ADNI data set. For MMSE and diagnosis the manifold model is the statistically most significant with the smallest residual. For age the best model is achieved with three dimensions of the PCA coordinates, while the manifold representation achieves the best fit with the first two coordinates. Note that the three PCA coordinates that best explain age are not the first three (dominant) principal components which capture the most variation.
Table 3.
Optimal linear regression models from the ADNI data set.
| Model | t | Residual | R2 | F | p-value | |
|---|---|---|---|---|---|---|
|
|
5.41 / -2.24 / -2.50 / | 5.5 | 0.21 | 13.5 | 7.3e-8 | |
|
|
4.83 / 2.34 | 5.7 | 0.16 | 14.39 | 1.8e-6 | |
| MMSE = a0 + a1age | 0.65 | 2.43 | 0.003 | 0.43 | 0.52 | |
| MMSE = a0 + a1l1 | -2.53 | 2.39 | 0.04 | 6.38 | 0.01 | |
| MMSE = a0 + a1x1 | -2.83 | 2.37 | 0.05 | 8.03 | 0.005 | |
| diagnosis = a0 + a1age | -0.74 | 0.68 | 0.003 | 0.54 | 0.46 | |
| diagnosis = a0 + a1l1 | 2.54 | 0.67 | 0.04 | 6.43 | 0.012 | |
| diagnosis = a0 + a1x1 + a6x6 | 3.55 / -3.30 | 0.64 | 0.13 | 11.74 | 1.8e-5 | |
For explanation of diagnosis note that the manifold model has a significantly lower p-value than either age or the PCA coordinates. Of further interest is the inclusion of the sixth dimension of the manifold model for explanation of diagnosis, which supports the claims made in Section 4.3.
Figure 12 shows residuals from the linear models based on manifold coordinates plotted against the residuals from the PCA regression. The regression residuals show roughly the same behavior, i.e large residuals in PCA correspond to large residuals in the manifold model of the same sign. However for the regression on diagnosis the residuals for the manifold model show a larger variation for subjects where PCA predicts the diagnosis fairly accurately. The manifold regression model yields a model with a more significant slope and thus some of the subject diagnosed with MCI tend more towards normal or AD, while the PCA model is closer to a constant model and is more likely to predict MCI.
Figure 12.
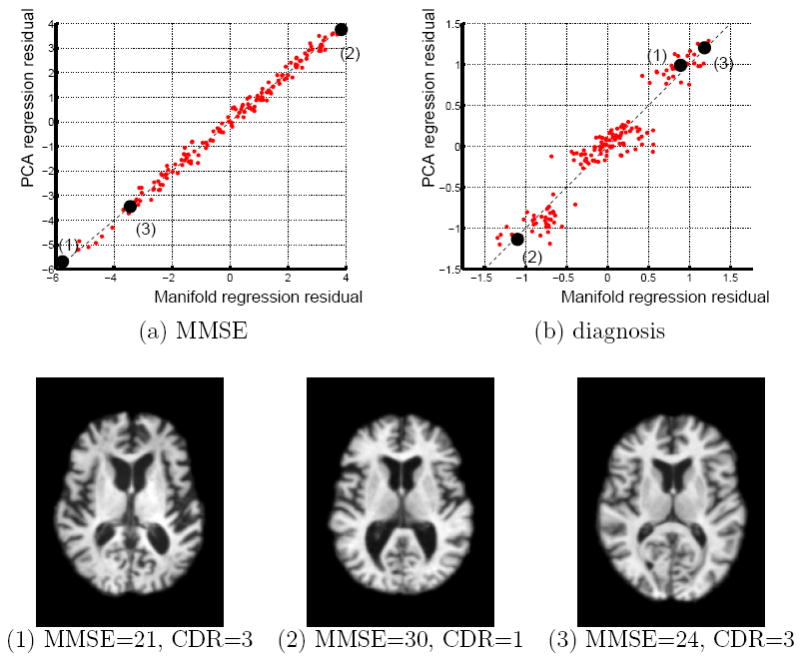
Residuals from the manifold regression model against the PCA regression model for (a) MMSE and (b) diagnosis. The bottom row shows three selected images with large residuals. The first image has a very low MMSE score that is not well represented in the image set. The second image shows a subject with large ventricle scores but a perfect MMSE scores. Both approaches relate ventricle size to lower MMSE scores and thus predict a lower MMSE score. The third subject shows the inverse, a subject with small ventricles but a relatively low MMSE score.
Figure 13 shows axial slices of constructed images on a grid sampled along dimension one and six of the manifold coordinates to illustrate the differences occurring in these dimensions. The shading represent the slope of the linear model of row 8 in Table 3. Dimension one captures the differences in ventricle size, while dimension six indicates an increase in the left lateral ventricle from top to bottom.
4.5. Computational Considerations
The distance computation between two images requires an elastic registration which takes with our multiresolution implementation about 1 minute on a 128 × 128 × 80 volume. The cost for learning the manifold from the 416 images of the OASIS database is high and requires about a week using a cluster of 50 2 GHz processors.
The implementation of the elastic registration, as well as an Isomap implementation, is available at http://www.cs.utah.edu/~sgerber/software.
5. Conclusions
This paper presents a novel approach to represent the space of brain images by a manifold model. The linear regression results show that the generative manifold model is able to uncover, using the raw image data only, statistically significant important relations to clinical data.
An open question is whether the manifolds shown here represent the inherent amount of information about shape variability in the data or whether they reflect particular choices in the proposed approach. In particular, implementation specific enhancements on image metric, reconstruction, and manifold kernel regression could lead to refined results.
There are various directions for improvements on the proposed method. The diffeomorphic metric is restricted to images that are within the orbit of a given template under the transformation. Recent work on metamorphosis [36] provides a elegant framework that yields a metric over the complete image space. The generative manifold model is directly applicable to an image space endowed with a metric based on metamorphosis. Another area of future work is to extend the approach to other image modalities. A metric that captures different properties or works with different image modalities can easily be integrated into the system.
An interesting question is whether the learned submanifold is truly nonlinear or if an approach such as principal geodesic analysis (PGA) [37], a generalization of PCA to Riemannian manifolds, leads to similar results. Furthermore, PGA could be used to do analysis on the learned manifold.
Acknowledgments
This work was supported by the National Institute of Health/NCBC grant U54-EB005149, the NSF grant CCF-073222 and the NIBIB grant 5RO1EB007688-02.
Data used in the evaluation of this work was funded by the Alzheimers Disease Neuroimaging Initiative (ADNI; Principal Investigator: MichaelWeiner; NIH grant U01 AG024904) as well as the Open Access Structural Imaging Series (OASIS; grants P50 AG05681, P01 AG03991, R01 AG021910, P20 MH071616, U24 RR021382).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lorenzen P, Davis BC, Joshi S. Unbiased atlas formation via large deformations metric mapping. MICCAI. 2005:411–418. doi: 10.1007/11566489_51. [DOI] [PubMed] [Google Scholar]
- 2.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23(Supplement 1):S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. mathematics in Brain Imaging. [DOI] [PubMed] [Google Scholar]
- 3.Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage. 2004;23(Supplement 1):S139–S150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Blezek DJ, Miller JV. Atlas stratification. Medical Image Analysis. 2007;11(5):443–457. doi: 10.1016/j.media.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabuncu MR, Balci SK, Golland P. Discovering modes of an image population through mixture modeling. MICCAI (2) 2008:381–389. doi: 10.1007/978-3-540-85990-1_46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill DLG, Hajnal JV, Rueckert D, Smith SM, Hartkens T, McLeish K. A dynamic brain atlas. MICCAI. 2002:532–539. [Google Scholar]
- 7.Davis B, Fletcher P, Bullitt E, Joshi S. Proceedings of the 2007 International Conference on Computer Vison. International Conference on Computer Vision; 2007. Population shape regression from random design data; pp. 1–7. [Google Scholar]
- 8.Ericsson A, Aljabar P, Rueckert D. Construction of a patient-specific atlas of the brain: Application to normal aging. ISBI. 2008:480–483. [Google Scholar]
- 9.Chou Y-Y, Lepor N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in alzheimer’s disease. NeuroImage. 2008;40(2):615–630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiology of Aging. 2008;29(4):514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apostolova LG, Thompson PM. Brain mapping as a tool to study neurodegeneration. Neurotherapeutics. 2007;4:387–400. doi: 10.1016/j.nurt.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron JC, Chtelat G, Desgranges B, Perchey G, Landeau B, de la Sayette V, Eustache F. In vivo mapping of gray matter loss with voxel-based morphometry in mild alzheimer’s disease. NeuroImage. 2001;14(2):298–309. doi: 10.1006/nimg.2001.0848. [DOI] [PubMed] [Google Scholar]
- 13.Busatto GF, Garrido GEJ, Almeida OP, Castro CC, Camargo CHP, Cid CG, Buchpiguel CA, Furuie S, Bottino CM. A voxel-based morphometry study of temporal lobe gray matter reductions in alzheimer’s disease. Neurobiology of Aging. 2003;24(2):221–231. doi: 10.1016/s0197-4580(02)00084-2. [DOI] [PubMed] [Google Scholar]
- 14.Callen D, Black S, Gao F, Caldwell C, Szalai J. Beyond the hippocampus: Mri volumetry confirms widespread limbic atrophy in ad. Neurology. 2001;57:1669–1674. doi: 10.1212/wnl.57.9.1669. [DOI] [PubMed] [Google Scholar]
- 15.Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. NeuroImage. 2001;14(3):595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- 16.Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the ravens maps: Methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;14(6):1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- 17.Roweis S, Saul L. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(550):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 18.Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 2003;15(6):1373–1396. [Google Scholar]
- 19.Schölkopf B, Smola A, Müller K-R. Nonlinear component analysis as a kernel eigen-value problem. Neural Computation. 1998;10:1299–1319. [Google Scholar]
- 20.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(550):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 21.Twining CJ, Marsland S. Constructing an atlas for the diffeomorphism group of a compact manifold with boundary, with application to the analysis of image registrations. J Comput Appl Math. 2008;222(2):411–428. [Google Scholar]
- 22.Hastie T, Stuetzle W. Principal curves. Journal of the American Statistical Association. 1989;84(406):502–516. [Google Scholar]
- 23.Gerber S, Tasdizen T, Whitaker R. Dimensionality reduction and principal surfaces via kernel map manifolds. Proceedings of the 2009 International Conference on Computer Vison. 2009 to appear. [Google Scholar]
- 24.Gerber S, Tasdizen T, Joshi S, Whitaker R. On the Manifold Structure of the Space of Brain Images. In: Yang GZ, Hawkes DJ, Rueckert D, editors. Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2009. pp. 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Younes L, Arrate F, Miller MI. Evolutions equations in computational anatomy. NeuroImage. 2009;45(1, Supplement 1):S40–S50. doi: 10.1016/j.neuroimage.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen GE, Rabbitt RD, Miller MI. Deformable Templates Using Large Deformation Kinematics. IEEE Transactions on Medical Imaging. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 27.Dupuis P, Grenander U. Variational problems on flows of diffeomorphisms for image matching. Q Appl Math. 1998;LVI(3):587–600. [Google Scholar]
- 28.Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vision. 2005;61(2):139–157. [Google Scholar]
- 29.Pless R. Image spaces and video trajectories: Using isomap to explore video sequences. ICCV; Washington, DC, USA: IEEE Computer Society; 2003. p. 1433. [Google Scholar]
- 30.Zhang Q, Souvenir R, Pless R. CVPR 2006. IEEE; 2006. On manifold structure of cardiac MRI data: Application to segmentation; pp. 1092–1098. [Google Scholar]
- 31.Wolz R, Aljabar P, Hajnal JV, Hammers A, Rueckert D. the Alzheimer’s Disease Neuroimaging Initiative, Leap: learning embeddings for atlas propagation. NeuroImage. 2010;49(2):1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox TF, Cox MAA. Monographs on Statistics and Applied Probability. Vol. 59. Chapman and Hall; London: 1994. Multidimensional Scaling. [Google Scholar]
- 33.Modersiztki J. Numerical Methods for Image Registration. Oxford University Press; Oxford: 2004. [Google Scholar]
- 34.Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. 3- Dimensional proportional system: an approach to cerebral imaging. Thieme; New York: 1988. [Google Scholar]
- 35.Schwarz G. Estimating the dimension of a model. Ann Statist. 1978;6(2):461–464. [Google Scholar]
- 36.Trouvé A, Younes L. Metamorphoses through lie group action. Foundations of Computational Mathematics. 2005;5:173–198. [Google Scholar]
- 37.Fletcher PT, Lu C, Pizer SM, Joshi S. Principal geodesic analysis for the study of nonlinear statistics of shape. IEEE Transactions on Medical Imaging. 2004;23(8):995–1005. doi: 10.1109/TMI.2004.831793. [DOI] [PubMed] [Google Scholar]


