Abstract
Transport receptors of the importin β superfamily account for many of the nuclear import and export events in eukaryotic cells. They mediate translocation through nuclear pore complexes, shuttle between nucleus and cytoplasm and co-operate with the RanGTPase system to regulate their interactions with cargo molecules in a compartment-specific manner. We used affinity chromatography on immobilized RanGTP to isolate further candidate nuclear transport receptors and thereby identified exportin 4 as the most distant member of the importin β family so far. Exportin 4 appears to be conserved amongst higher eukaryotes, but lacks obvious orthologues in yeast. It mediates nuclear export of eIF-5A (eukaryotic translation initiation factor 5A) and possibly that of other cargoes. The export signal in eIF-5A appears to be complex and to involve the hypusine modification that is unique to eIF-5A. We discuss possible cellular roles for nuclear export of eIF-5A.
Keywords: eIF-5A/exportin/importin/nuclear export/nuclear pore complex
Introduction
Importin β-related nuclear transport receptors mediate many of the nucleocytoplasmic transport events (reviewed in Dahlberg and Lund, 1998; Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999). These receptors shuttle between nucleus and cytoplasm, interact with nuclear pore complexes (NPCs) and recognize and bind cargo molecules. According to the direction in which they carry a cargo, they can be classified as importins or exportins. The directionality of transport appears to be determined by a RanGTP gradient across the nuclear envelope (NE). Transport receptors are RanGTP-binding proteins that respond to this gradient by loading and unloading their cargo in the appropriate compartment; importins bind their import substrates at low RanGTP levels in the cytoplasm, release them upon RanGTP binding in the nucleus (Rexach and Blobel, 1995; Chi et al., 1996; Görlich et al., 1996b; Izaurralde et al., 1997; Schlenstedt et al., 1997; Siomi et al., 1997; Jäkel and Görlich, 1998) and return as cargo-free RanGTP–importin complexes to the cytoplasm (Izaurralde et al., 1997; Hieda et al., 1999). RanGTP–importin complexes are finally disassembled by the concerted action of cytoplasmic RanGAP and RanBP1 (or RanBP2), releasing the importin to bind and import another substrate molecule (Bischoff and Görlich, 1997; Floer et al., 1997; Lounsbury and Macara, 1997). Exportins are regulated in a precisely converse manner to importins. They bind their export substrates preferentially at high RanGTP concentrations in the nucleus and exit the nucleus as trimeric cargo–exportin–RanGTP complexes (Fornerod et al., 1997a; Kutay et al., 1997a, 1998; Arts et al., 1998a; Kaffman et al., 1998). The cytoplasmic disassembly of such complexes also requires RanGAP and RanBP1 (or RanBP2) (Bischoff and Görlich, 1997; Kutay et al., 1997a), and results in cargo release from the exportins and GTP hydrolysis, and allows the exportins to re-enter the nucleus and to accomplish further rounds of export.
The overall sequence similarity between the various transport receptors is low and, in many cases, restricted to the N-terminal RanGTP-binding motif (Fornerod et al., 1997b; Görlich et al., 1997). This can be explained at least in part by the fact that these receptors bind very different cargoes, such as the basic IBB domain in the case of importin β (Görlich et al., 1996a; Weis et al., 1996), tRNA in the case of exportin-t (Arts et al., 1998a; Kutay et al., 1998) or a leucine-rich nuclear export signal (NES) in the case of CRM1 (Fischer et al., 1995; Wen et al., 1995; Fornerod et al., 1997a; Stade et al., 1997). The RanGTP-binding motif can thus be considered as a diagnostic feature of importin β-related transport receptors, and in fact it allowed the identification of most of the 14 family members from the yeast Saccharomyces cerevisiae (Fornerod et al., 1997b; Görlich et al., 1997). Higher eukaryotes employ an even larger number of transport receptors; at least 22 in the case of mammals (see Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; this study and our unpublished data). It has been a major effort in the field of nuclear transport to allocate functions to each of these receptors and to identify the transport signals that they recognize.
While characterizing a novel mammalian exportin (see below), we identified eIF-5A (eukaryotic translation initiation factor 5A) as a potential export substrate. eIF-5A was originally isolated as a candidate translation factor from a polyribosome-bound fraction (Kemper et al., 1976; Benne et al., 1978) and was suggested to be involved in the formation of the first peptide bond. Subsequent studies, however, indicated that translational initiation is not directly affected by a loss of eIF-5A function (Kang and Hershey, 1994; Zuk and Jacobson, 1998) and that eIF-5A is not required to assemble translation initiation complexes. Bona fide translation initiation factors are usually present substoichiometrically to ribosomes and associate with them only during initiation. In contrast, eIF-5A is present in excess over ribosomes (Hershey, 1994) and largely bound to cytoplasmic, puromycin-sensitive structures that might represent (rER-bound) polysomes (Shi et al., 1997). One should therefore assume that if eIF-5A functions in translation, then it is at some step subsequent to initiation.
eIF-5A carries a uniquely modified lysine that is referred to as hypusine [Nε-(4-amino-2-hydroxybutyl) l-lysine; Shiba et al., 1971; Park et al., 1982; Cooper et al., 1983]. eIF-5A is apparently the only hypusine-containing protein. It is present in eukaryotes (Gordon et al., 1987) and archaebacteria (Bartig et al., 1992) and is in fact one of the best conserved proteins between the two kingdoms. eIF-5A itself and its modification pathway are essential for viability in yeast (Schnier et al., 1991; Wöhl et al., 1993; Sasaki et al., 1996).
The X-ray structures of eIF-5A from two archaea species have been solved (Kim et al., 1998; Peat et al., 1998). They show eIF-5A to be composed of two compact domains that are linked by a flexible hinge. N-terminal domain I contains the hypusine modification site in an extended, protruding and highly conserved loop. The modification is unlikely to have a major effect on the eIF-5A structure. Instead, its absolute conservation indicates that the hypusine mediates essential interactions with other macromolecules. Hypusine is a 2-fold positively charged amino acid and resembles nucleic acid-binding polyamines such as spermine and spermidine. Domain II is similar to the RNA-binding motif found in the prokaryotic cold shock protein CspA, which has been suggested to function as an RNA chaperone (Jiang et al., 1997). Domain II and the hypusine-containing loop from domain I might thus constitute a bipartite RNA-binding site (Kim et al., 1998; Peat et al., 1998). An RNA-binding activity of eIF-5A has indeed been detected in vitro and has been found to depend on the hypusine modification (Liu et al., 1997). However, it is still unclear whether this RNA-binding activity reflects a genuine function of eIF-5A and if so what the physiological RNA ligand(s) of eIF-5A might be.
Eubacteria lack a hypusine-modified eIF-5A equivalent. However, the sequence similarity between eubacterial EF-P and archaebacterial/eukaryotic eIF-5A is significant enough to assume safely that the two represent homologous proteins (Kyrpides and Woese, 1998). EF-P is essential for viability in Escherichia coli (Aoki et al., 1997) and present in all eubacterial genomes examined so far. eIF-5A/EF-P can thus be considered a universally conserved and essential protein; it is apparently the only known protein that falls into this category whose function has remained elusive.
Loss of eIF-5A function is ultimately lethal (Schnier et al., 1991; Kang and Hershey, 1994; Sasaki et al., 1996; Zuk and Jacobson, 1998; Jansson et al., 2000). In their mortal phase the cells show diverse defects. Saccharomyces cerevisiae cells stop cell division, but continue to enlarge in size. One specific defect is an impaired degradation of mRNA, particularly of short-lived messages (Zuk and Jacobson, 1998). The defect apparently occurs between mRNA decapping and degradation by the Xrn1p exonuclease and could be consistent with the assumed RNA-binding activity of eIF-5A. However, it is unclear whether RNA turnover is the primary function that makes eIF-5A indispensable for viability in all organisms. Upon eIF-5A inactivation in yeast, the overall rate of translation is reduced by ∼30%, but is not immediately abolished (Kang and Hershey, 1994; Zuk and Jacobson, 1998), suggesting that translational initiation and elongation can proceed in the absence or at very low concentrations of eIF-5A. The intermediate effect on the overall translation rate and the absolute requirement of eIF-5A for viability could possibly be explained by a failure of eIF-5A-deficient cells to synthesize a subset of proteins or to synthesize them in a biologically active form.
Here we report the identification of a novel mammalian member of the importin β superfamily, which we refer to as exportin 4 (Exp4). We show that Exp4 functions as a nuclear export receptor for eIF-5A and perhaps for other substrates as well. As eIF-5A appears to be an RNA-binding protein, it might function as an export adapter in RNA export. Alternatively, Exp4-mediated eIF-5A export might be used to restrict eIF-5A activity to the cytoplasm.
Results
Identification and molecular cloning of Exp4
Many of the nuclear transport pathways are mediated by RanGTP-binding proteins of the importin β superfamily. To identify further mammalian family members, we used affinity chromatography on immobilized RanGTP to enrich them from a HeLa cell extract (see Figure 1). The RanGTP-bound fractions were separated by SDS–PAGE and bands between 90 and 140 kDa were cut out. Peptide sequencing identified not only the already known transport receptors, but also a novel protein, which for reasons detailed below we refer to as Exp4.

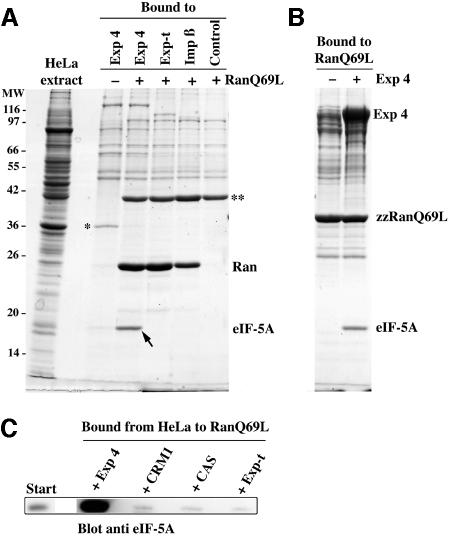
Fig. 1. Identification of exportin 4 (Exp4) amongst RanGTP-binding proteins from HeLa cells. A HeLa cell extract was subjected to binding to either immobilized RanGDP (wild-type protein) or RanGTP (Q69L mutant). Starting material and bound fractions were analysed by SDS–PAGE followed by Coomassie Blue staining. Protein bands specifically recovered on the RanGTP beads were analysed by microsequencing. Identified proteins are indicated. Exp4 is a novel protein.
We then used this partial sequence information to isolate a full-length Exp4 cDNA from a mouse cell line (see Materials and methods). It codes for a 129.9 kDa protein (Figure 2) with an isoelectric point of 4.9. Exp4 shares distant, but still significant similarity with other members of the importin β superfamily. The region of homology is restricted to the N-terminal RanGTP-binding motif and gives the best matches to exportin 1 (CRM1) and to the tRNA export receptor exportin-t. Other parts of the proteins do not show significant homologies to any other known protein. The multiple alignment of the known importin β-like factors (based on full-length sequences) indicates that Exp4 is the most distant member of this superfamily identified so far (not shown).
Fig. 2. Primary sequence of Exp4. The partial sequence information from human Exp4 was used to clone a full-length cDNA coding for mouse Exp4. The figure shows the deduced amino acid sequence of the open reading frame. Peptides obtained by protein sequencing are underlined.
Identification of an Exp4-specific export substrate
At this stage it was unclear whether Exp4 would mediate import, export or indeed nucleocytoplasmic transport at all. We therefore expressed Exp4 in E.coli and first established that the recombinant protein interacts with RanGTP in a manner that closely resembles other importin β-related factors (see below). As detailed in the Introduction, these transport receptors use RanGTP binding to regulate the interactions with their respective cargoes. To test whether Exp4 would behave in an analogous way, and to identify potential transport substrates, we tested which components from a HeLa extract it would bind to. The binding was performed with or without addition of the GTPase-deficient RanQ69L mutant (loaded with GTP). This mutant remains in the GTP-bound form even in the presence of cytoplasmic RanGAP (Bischoff et al., 1994; Klebe et al., 1995) and can thus be used to mimic a nuclear environment. As seen from Figure 3A, two proteins bound to the immobilized Exp4 in a Ran-regulated manner. These were identified by peptide sequencing as thymidylate synthase and eIF-5A. Thymidylate synthase appeared preferentially bound to the Ran-free form of Exp4 and could thus represent an import substrate for this transport receptor. However, subsequent tests could not detect any import activity of Exp4 towards recombinant thymidylate synthase (data not shown). We therefore did not characterize this interaction further.
Fig. 3. Identification of eIF-5A as a putative export substrate for Exp4. (A) zz-tagged fusions of Exp4, exportin-t and importin β were immobilized on IgG Sepharose and used for binding from a HeLa cell extract. Analysis was as in Figure 1. Load in the bound fractions corresponds to 35 times the starting material. Note, eIF-5A was specifically recovered with immobilized Exp4 in the presence of the RanQ69L mutant (3.2 µM GTP form). eIF-5A binding was not detectable when RanQ69L was omitted or Exp4 replaced by another nuclear transport receptor. ‘*’ indicates thymidylate synthase; ‘**’ indicates actin that also bound non-specifically in the presence of RanQ69L to the control beads that did not contain any zz fusion protein. (B) Binding from HeLa extract to immobilized zzRanQ69L (GTP). When a saturating concentration (1 µM) of exogenous Exp4 had been added, both Exp4 and a prominent eIF-5A band were recovered in the bound fraction, indicating the formation of a trimeric eIF-5A–Exp4–zzRanGTP complex. (C) Binding from HeLa extract to immobilized RanGTP after addition of 1 µM exogenous Exp4, CRM1, CAS or exportin-t. eIF-5A present in the bound fractions was detected by western blotting. Note, eIF-5A assembled into an export complex with Exp4 but not with CRM1, CAS or exportin-t.
eIF-5A was selectively recovered with the RanGTP-bound form of Exp4, but not with any other transport receptor when tested under identical conditions (Figure 3A). This suggests that eIF-5A can specifically assemble into a trimeric eIF-5A–Exp4–RanGTP export complex and might thus represent an export substrate of Exp4.
To confirm these results, we performed the binding from the HeLa extract the other way around and immobilized RanQ69LGTP instead of Exp4 (Figure 3B and C). Without further addition, very little eIF-5A was recovered in the bound fraction, probably because concentration of endogenous Exp4 in the extract is low. However, when 1 µM Exp4 was added to the incubation mixture, a prominent, Coomassie-stainable eIF-5A band, along with Exp4 itself, was recovered in the RanGTP-bound fraction, indicating the formation of a trimeric eIF-5A–Exp4–Ran complex (Figure 3B). The identity of the eIF-5A band was verified by immunoblotting (Figure 3C). The same panel also shows that eIF-5A specifically assembles into complexes only with Exp4, but not with other exportins such as CAS, exportin-t or CRM1.
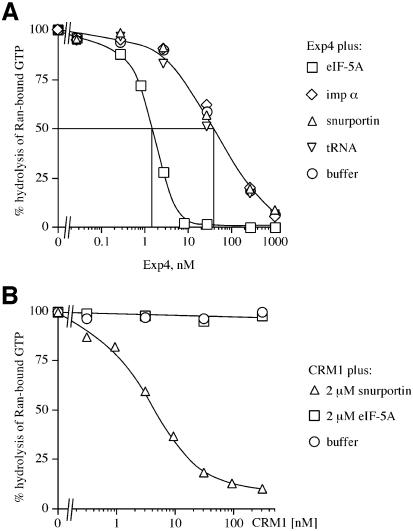
Figure 3A demonstrated that the eIF-5A–Exp4 interaction is greatly enhanced by the presence of RanGTP. To test whether this represents a truly co-operative binding between these three components, we employed a more quantitative assay. This assay is based on the observation that binding of an importin β-like factor to RanGTP prevents GTPase activation by RanGAP (Floer and Blobel, 1996; Görlich et al., 1996b). GTP hydrolysis can easily be quantified and used to calculate the proportion of Ran that is associated with the respective factor. From the dose dependence of the effects one can estimate dissociation constants of the complexes.
We first varied the concentration of Exp4 in the absence of eIF-5A (Figure 4A) and observed a half-maximum RanGTP binding at an Exp4 concentration of ∼40 nM, which corresponds to the apparent KD (equilibrium dissociation constant) for the RanGTP–Exp4 interaction. The KD was lowered to ∼1.5 nM in the presence of a saturating concentration of native, fully modified eIF-5A, indicating co-operative binding. This co-operative effect was highly specific for the eIF-5A–Exp4 interaction and was not observed for combinations of Exp4 with other export substrates, such as importin α, tRNA or the CRM1 substrate snurportin 1. Our data also indicate that eIF-5A is bound by Exp4 at least 1000 times better than by human CRM1 (compare Figure 4A with B), exportin-t or CAS (Figure 3C and data not shown).
Fig. 4. Quantitative characterization of the eIF-5A–Exp4–RanGTP interaction. (A) The assay exploits the observation that binding of RanGTP to an importin β-like factor prevents GTPase activation by RanGAP. Ran-[γ-32P]GTP (50 pM) was pre-incubated at 15°C with the Exp4 concentrations indicated in the absence or presence of either eIF-5A (fully hypusinated), snurportin 1, importin α (imp α) or tRNA (2 µM each). After 30 min, a 30 s GTPase reaction was started by addition of 40 nM S.pombe RanGAP. Hydrolysis of Ran-bound GTP was determined as released [32P]phosphate. Note that the presence of eIF-5A increased the affinity of Exp4 for RanGTP ∼30-fold, while snurportin 1, importin α and tRNA had no effect. (B) Measurements were performed exactly as in (A) except that CRM1 was added instead of Exp4. Note that snurportin 1 bound selectively to CRM1, while eIF-5A showed no binding.
The hypusine modification in eIF-5A apparently contributes to Exp4 binding
The maturation of eIF-5A to the fully hypusinated form occurs in two steps. First, deoxyhypusine synthase transfers a 4-amino-butyl group from spermidine to the ε-amino group of lysine 50 (in the human sequence) to form deoxyhypusine (Park et al., 1982; Dou and Chen, 1990; Wolff et al., 1990). This reaction is evident in all eukaryotes and archaea species examined (Gordon et al., 1987; Bartig et al., 1990). The deoxyhypusine is then hydroxylated by an as yet unidentified enzyme to yield the mature hypusine. This second step occurs in all eukaryotes and crenarchaea species tested, but not in euryarchaea (Bartig et al., 1990), suggesting that deoxyhypusine can substitute for hypusine at least in some organisms.
The hypusine/deoxyhypusine modification of eIF-5A is essential for viability (Sasaki et al., 1996) and thus for eIF-5A function. We therefore wanted to test whether the hypusine residue is also involved in the interaction with Exp4. To this end we had to generate eIF-5A at various stages of modification. The fully modified eIF-5A was enriched by a multi-step procedure from HeLa cells to a purity of >95% (see Materials and methods and Supplementary material, available at The EMBO Journal Online). Unmodified eIF-5A was obtained by recombinant expression of the human protein in E.coli (note that eubacteria lack hypusination). For best comparability, eIF-5A was expressed with its authentic N- and C-termini (i.e. untagged) and purified by conventional chromatography. The recombinant protein was properly folded as judged by the following criteria. First, it was soluble when expressed in E.coli. Secondly, it showed similar chromatographic properties to the native protein and eluted in sharp peaks from the ion exchange and gel filtration columns (note, these chromatographic procedures probe protein shape and charge distribution). Thirdly, it was an efficient substrate for deoxyhypusination. To obtain deoxyhypusinated eIF-5A, we expressed human deoxyhypusine synthase in E.coli and used the purified enzyme to modify recombinant eIF-5A in the presence of NAD and spermidine (see Materials and methods).
We next wanted to determine apparent KDs for the interactions of the Exp4–RanGTP complex with the various forms of eIF-5A. For this purpose, we used the assay described for Figure 4A, but kept the concentration of Exp4 constant at 25 nM and instead varied that of eIF-5A in the assay. Increasing concentrations of eIF-5A promoted the RanGTP binding to Exp4. The half-maximum effect for the E.coli-expressed, unmodified eIF-5A was observed at a concentration of ∼75 nM, which can be taken as a rough estimate for the KD for dissociation of eIF-5A from the trimeric complex (see Table I). Deoxyhypusination of recombinant eIF-5A improved Exp4 binding ∼3-fold. The native, hypusine-modified eIF-5A bound the Exp4–RanGTP complex with a KD of ∼2 nM, i.e. ∼35 times more strongly than the unmodified protein. The hypusination site is located within eIF-5A at the middle of a flexible, highly exposed loop that appears not to be involved in secondary structures or other interactions with the rest of the eIF-5A molecule. This strongly suggests that the hypusine improves Exp4 binding not by changing eIF-5A conformation but instead by making a direct contact to Exp4. Our data also indicate that deoxyhypusine can only partially substitute for hypusine in the interaction with Exp4.
Table I. Apparent affinities of eIF-5A derivatives for Exp4–RanGTP complex.
| eIF-5A species | Apparent KD for dissociation from eIF-5A–Exp4–RanGTP complex | Relative affinity for Exp4 (native eIF-5A = 100) |
|---|---|---|
| Purified from HeLa cells, fully hypusinated | 2 nM | 100 |
| Recombinant, non-modified | 75 nM | 2.5 |
| Recombinant, deoxyhypusinated | 25 nM | 8 |
| Recombinant domain I (residues 1–83), deoxyhypusinated | ∼1–2 µM | 0.1 |
| Recombinant domain II | >10 µM | <0.01 |
As detailed in the Introduction, eIF-5A is composed of two domains. The N-terminal domain I (aa 1–83) harbours the hypusination site and was indeed an efficient substrate for deoxyhypusine synthase. Surprisingly, the isolated, deoxyhypusinated domain I bound Exp4 with only 1% affinity as compared with the identically modified full-length protein, while the complementary domain II (residues 83–154) alone showed <0.1% binding (see Table I). This suggests that Exp4 requires large parts of the eIF-5A molecule for recognition and does not just bind a short, contiguous export signal sequence.
Exp4 mediates export of eIF-5A from the nucleus
The way Exp4 interacts with eIF-5A closely resembles the interaction of other exportins with their respective export substrates in that binding is of high affinity in the presence of RanGTP, i.e. in a nuclear environment and weak without RanGTP, i.e. under cytoplasmic conditions (see Introduction). Also, just as is the case for other transport receptors, the trimeric eIF-5A–Exp4–RanGTP complex is disassembled in the simultaneous presence of cytoplasmic RanBP1 and RanGAP (data not shown). Taken together, this strongly suggests that Exp4 is the eIF-5A-specific exportin. However, we wanted to test this directly.
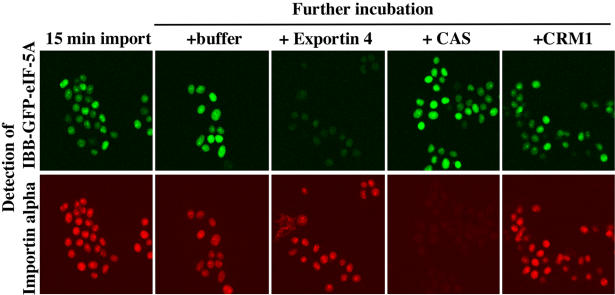
To target eIF-5A to the nucleus in the first place, we fused it to the IBB domain, a potent importin β-dependent nuclear import signal (Görlich et al., 1996a; Weis et al., 1996). The fusion also contained the green fluorescent protein (GFP) for detection by fluorescence microscopy. As an internal control, a second export substrate, Texas Red-labelled importin α, was also added and simultaneously detected in a separate channel. Import into nuclei of permeabilized cells was performed in the presence of a Xenopus egg extract that had been depleted by immobilized RanGTP of endogenous importins and exportins. Importin β was re-added to allow import of importin α and the IBB–eIF-5A fusion.
After 15 min incubation, importin α and the IBB–GFP–eIF-5A fusion had clearly accumulated in the nuclei (Figure 5). The mixture was then split into four, and either buffer, Exp4, CAS or CRM1 was added. Fifteen minutes later, each of the incubations was analysed by confocal microscopy. Exp4 specifically promoted export of the eIF-5A fusion, but had no effect on importin α export (Figure 5). Conversely, CAS promoted export of importin α as reported before (Kutay et al., 1997a), but left the eIF-5A fusion unaffected. The very small effect of CRM1 on nuclear localization of both importin α and eIF-5A is probably due to its more efficient NPC binding and competition of importin β-mediated import as compared with the other two exportins.
Fig. 5. Exp4 mediates nuclear export of an eIF-5A fusion protein. An IBB–GFP–eIF-5A fusion (0.4 µM) and Texas-Red-labelled Xenopus importin α (0.4 µM) were first allowed to accumulate in nuclei of permeabilized cells. Import was in the presence of an energy-regenerating system and a Xenopus egg extract that had been depleted of importin β-like transport receptors and replenished with importin β and RanBP1. Fifteen minutes later, the mixture was split into four and either buffer, 2 µM Exp4, CAS or CRM1 was added. After another 15 min, the distributions of the eIF-5A fusion and of importin α were recorded by confocal microscopy in the fluorescein and Texas Red channels, respectively. Note, Exp4 specifically promoted export of the eIF-5A fusion, but had no effect on importin α. Conversely, CAS promoted export of importin α but had no effect on eIF-5A localization.
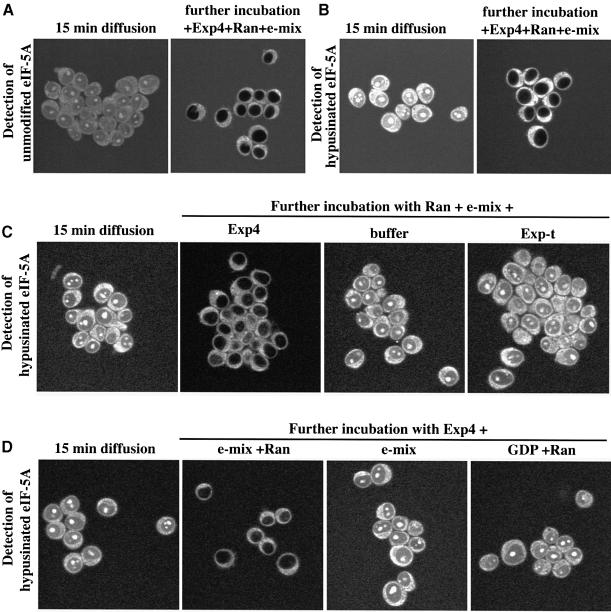
In Figure 5 we used the IBB domain to target eIF-5A to the nucleus artificially. We next wanted to know whether eIF-5A can also enter nuclei on its own and so we labelled untagged, recombinant eIF-5A at a 1:1 molar ratio with Alexa-maleimide to allow detection by fluorescence microscopy. When incubated with permeabilized cells, eIF-5A gave some cytoplasmic staining and also readily entered nuclei and accumulated in the nucleoli (Figure 6A). Consistent with the small size of eIF-5A (18 kDa), this accumulation occurred by passive diffusion as it was insensitive to dominant-negative importin β mutants (not shown) that are known to block facilitated NPC passage (Kutay et al., 1997b). When Exp4 was added along with Ran and an energy-regenerating system, the nucleolar and nuclear signals completely disappeared, indicating efficient nuclear export (Figure 6A). The recombinant eIF-5A lacks the hypusine modification and binds Exp4 ∼35 times more weakly than the fully modified, native protein. Export was nevertheless efficient, probably because Exp4 was added at 0.5 µM, a concentration above the KD (75 nM) for binding of unmodified eIF-5A. However, it was crucial to show that Exp4 would also export hypusinated eIF-5A and so we also performed in parallel the same assay with native eIF-5A. Native eIF-5A accumulated in the absence of Exp4 more strongly in the nucleoli than non-hypusinated eIF-5A (compare Figure 6A with B), indicating that the hypusine residue contributes to nucleolar retention. Also in this case, addition of Exp4, Ran and GTP resulted in very efficient export (Figure 6B). This effect was highly specific and occurred neither when Exp4, Ran or free GTP was omitted, nor when Exp4 was replaced by another exportin (Figure 6C and D). This is the expected result if eIF-5A was exported from the nucleus as a trimeric eIF-5A–Exp4–RanGTP complex.
Fig. 6. Characterization of eIF-5A export. (A) Alexa-labelled, non-hypusinated eIF5A was allowed to diffuse into nuclei of permeabilized cells, before a 15 min export reaction was started by addition of 0.5 µM Exp4, 1.5 µM Ran and an energy-regenerating system (‘e-mix’ containing 0.5 mM ATP and GTP). The distribution of eIF-5A was recorded before and after export by confocal fluorescence microscopy. Note, eIF-5A was efficiently depleted from nucleoli and the nucleoplasm during the export reaction. (B) Hypusinated eIF-5A was used for the export experiment, which was otherwise performed in parallel and under exactly identical conditions to those in (A). Note, nucleolar accumulation prior to export is stronger for hypusinated than for non-hypusinated eIF-5A. Nevertheless, addition of Exp4, Ran and e-mix caused essentially quantitative export. (C and D) Export of eIF-5A was performed as described for (B), using the combinations of 0.5 µM Exp4, 0.5 µM exportin-t (‘Exp-t’), 1.5 µM Ran, an energy-regenerating system and GDP indicated. Note, only the combination Exp4 plus Ran and e-mix allowed export of eIF-5A.
Discussion
Nuclear transport receptors of the importin β superfamily mediate many, although not all, transport pathways between nucleus and cytoplasm. Five family members from higher eukaryotes, namely importin β, importin 5, importin 7, transportin 1 and transportin SR, are known to function in import (Chi et al., 1995; Görlich et al., 1995; Imamoto et al., 1995; Pollard et al., 1996; Fridell et al., 1997; Jäkel and Görlich, 1998; Kataoka et al., 1999), while three receptors have been shown to function in export: CAS, which mediates the export of importin α (Kutay et al., 1997a); exportin-t, that of tRNA (Arts et al., 1998a; Kutay et al., 1998); and CRM1, which accounts for export of a variety of substrates (Fornerod et al., 1997a; Stade et al., 1997), many of which contain a leucine-rich NES. Here we identify a fourth mammalian exportin, Exp4, as an export receptor for eIF-5A.
Exp4 is the most distant family member identified so far. Nevertheless, it functions according to the same principles as the other exportins i.e. it binds its export cargo preferentially in a nuclear environment in the presence of RanGTP, forming a trimeric eIF-5A–Exp4–RanGTP export complex. This complex is subsequently transferred out of the nucleus. Cytoplasmic disassembly of the complex and concomitant cargo release are brought about by the concerted action of RanGAP and RanBP1 (or RanBP2, respectively), and also result in the hydrolysis of the Ran-bound GTP. The ‘empty’ exportin can then re-enter the nucleus and participate in another round of export.
Available expressed sequence tags and genomic sequences indicate that Exp4 orthologues exist in vertebrates (human, cattle, rat, pig, frog and zebrafish), invertebrates (Halocynthia roretzi and Caenorhabditis elegans) and plants (cotton). It is remarkable, however, that the yeast S.cerevisiae has no identifiable Exp4 orthologue. It remains to be seen whether yeast also shows the phenomenon of active nuclear export of eIF-5A and if so, by which components it is mediated.
Another study has suggested that eIF-5A export is mediated by CRM1 (Rosorius et al., 1999). Our data clearly argue against this scenario and instead demonstrate that CRM1 binds eIF-5A at least 1000 times more weakly than Exp4 (Figure 4). The concentration of eIF-5A in HeLa cells is ∼6 µM (data not shown; see Hershey, 1994), i.e. similarly abundant to Ran. This high abundance might explain why higher eukaryotic cells employ a specialized pathway for eIF-5A export.
The identification of a novel export pathway poses the question for the corresponding export signal. The hypusine modification is apparently part of the signal that allows eIF-5A to access the Exp4 pathway. Recombinant eIF-5A that lacks the modification binds to Exp4 35 times more weakly than the fully modified protein, i.e. the recognition of the modification appears to contribute ∼8 KJ/mol binding energy. Deoxyhypusine can partially, but not fully, substitute for hypusine (Table I), indicating that the hydroxyl moiety of hypusine also contributes to Exp4 binding. Our data also indicate that Exp4 recognizes more than just the hypusine and the flanking residues, and for stable binding requires large parts of the eIF-5A molecule (see Table I). This resembles certain other export substrates such as tRNA (Arts et al., 1998b; Lipowsky et al., 1999), importin α (Kutay et al., 1997a; Herold et al., 1998) or snurportin 1 (Huber et al., 1998; Paraskeva et al., 1999), where the identity of the export substrate per se, and not some short ‘transplantable’ export signal, appears to be recognized by their cognate exportins.
Does Exp4 also have export substrates other than eIF-5A? Given that hypusine is part of the export signal and that eIF-5A is apparently the only hypusine-containing protein, it appears unlikely that Exp4 recognizes any other export substrate in an identical way. However, it is known that a given transport pathway can be accessed by very different signals. Transportin, for example, can bind and import proteins containing an M9 or a BIB domain (Pollard et al., 1996; Jäkel and Görlich, 1998) and even has separate binding sites for these two domains (Jäkel and Görlich, 1998). By analogy, Exp4 might then also recognize and export different types of cargoes, and we are currently testing this possibility.
A central question is why do higher eukaryotic cells need to export eIF-5A from their nuclei? This question is intimately linked to the question of the function of eIF-5A, which is still open at present. eIF-5A has been suggested to recognize the NES in the HIV-Rev protein and to mediate its export (Ruhl et al., 1993; Bevec et al., 1996). This assumption has already been contradicted by Henderson and Percipalle (1997), who reported that recombinant eIF-5A is unable to bind Rev specifically. We can confirm and extend this conclusion in that native, hypusinated eIF-5A also does not specifically interact with Rev. We used immobilized Rev to bind Rev-interacting factors from a complete HeLa extract and observed efficient CRM1 binding in the expected RanGTP-regulated manner (see Supplementary figure), confirming CRM1 as the receptor and export mediator for proteins with a Rev-type NES (Fornerod et al., 1997a; Fukuda et al., 1997; Stade et al., 1997). In contrast, eIF-5A was undetectable amongst the Rev-interacting proteins.
eIF-5A appears to be an RNA-binding protein (Liu et al., 1997), with probably the hypusine modification and the C-terminal domain contributing to the interaction (Liu et al., 1997; Kim et al., 1998; Peat et al., 1998). Although we do not yet know which RNAs eIF-5A normally interacts with, it would be an attractive possibility that eIF-5A functions as an export adapter for these RNAs. There is no definitive evidence against this hypothesis; however, a number of complications need to be considered. First, assuming that the essential function of eIF-5A is conserved between eukaryotes and archaea, and given that archaea have no nuclei, one has to conclude that nuclear export of other macromolecules cannot be the primary function of eIF-5A. Of course, eukaryotic eIF-5A might be a multifunctional protein that took over a role in RNA export later in evolution. Secondly, if eIF-5A was an export adapter, then one should expect it to be actively imported into nuclei. However, nuclear accumulation of eIF-5A appears to occur solely by passive diffusion, as it is insensitive to reagents that block facilitated NPC passage. Thirdly, the hypusine modification is essential for eIF-5A function, probably involved in the interaction with its functional targets and required for the RNA-binding activity of eIF-5A. Our data indicate that the hypusine is also involved in the interaction with Exp4. It remains to be seen whether the hypusine in an eIF-5A–Exp4–RanGTP complex is then still available for RNA binding (and export).
Alternatively, Exp4 might mask the hypusine and other crucial parts of the eIF-5A molecule in order to prevent an interaction of eIF-5A with potential targets in the nucleus. Thus, eIF-5A might be actively exported from nuclei, not because it is a transporter for other macromolecules, but instead because its function must be restricted to the cytoplasm. The bets for eIF-5A’s function are still open and its elucidation will require the identification of the macromolecules with which eIF-5A interacts in a physiological context. The available data would be consistent with eIF-5A being involved in some aspect of translation or cytoplasmic degradation of mRNA or with eIF-5A functioning as an RNA chaperonin or in some other facet of RNA metabolism. We found that eIF-5A accumulates in the absence of Exp4 in nucleoli, the sites where ribosomes are assembled. Such untimely binding of eIF-5A to pre-ribosomal particles might interfere with ribosome biogenesis. Exp4-mediated export of eIF-5A would certainly be an elegant solution to avoid such a problem.
Materials and methods
Protein identification
The Exp4 band (see Figure 1) was subjected to a tryptic digest. Resulting peptides were separated by HPLC and analysed by Edman degradation, resulting in the peptide sequences depicted in Figure 2. eIF-5A (DDBJ/EMBL/GenBank accession No. P10159) was identified through partial sequence of two tryptic peptides, VHLVGIDIFTGK and NGFVVLK, and by mass spectrometry. For thymidylate synthase (DDBJ/EMBL/GenBank accession No. P04818), the following tryptic peptides were identified by mass spectrometry: PVAGSELPR, RPLPPAAQER and TGTGTLSVF GMQAR.
Molecular cloning
Database searches identified a mouse expressed sequence tag that matched some of our Exp4 partial sequence information, but lacked the 5′ and 3′ ends. The missing sequence information was obtained through several rounds of 3′ and 5′ RACE using mRNA from mouse 3T3 cells as a template. The full-length cDNA was finally amplified by RT–PCR, cloned into the TOPO TA vector and several clones were sequenced to generate a master sequence. N-His6-tagged Exp4 expression construct (pQE9-Exp4) was generated by inserting the coding sequence as a SalI–HindIII fragment into pQE9 (Qiagen). The zz-tagged Exp4 expression construct (zz Exp4) was derived from pQE9-Exp4 by replacing the His tag for a zz tag. The expression construct for untagged eIF-5A (pQE60-eIF-5A) was generated by cloning the coding sequence of the human protein into the NcoI–HindIII sites of pQE60. The IBB–GFP–eIF-5A-His6 construct is a pQE60 derivative, generated by fusing residues 1–63 from Rch1 (human importin α1) with GFP and human eIF-5A. To generate the N-His6-tagged human deoxyhypusine synthase expression construct, the coding sequence (DDBJ/EMBL/GenBank accession No. L39068) was cloned from HeLa cDNA into the BamHI–HindIII sites of pQE30. All constructs were verfied by DNA sequencing.
Recombinant protein expression in E.coli
All recombinant proteins were expressed in the E.coli BLR strain. Purification of importin β, CAS, Ran, RanQ69L, zz-tagged Ran wild type and RanQ69L, NTF2, CRM1 and importin α have been previously described (Kutay et al., 1997a, 1998; Paraskeva et al., 1999).
His-tagged Exp4 was purified on nickel NTA agarose, followed by ammonium sulfate precipitation and gel filtration. zz-tagged proteins were directly absorbed from the E.coli lysates to IgG Sepharose for the binding experiments.
Recombinant human full-length eIF-5A and the N-terminal domain from eIF-5A (residues 1–83) were purified on SP–Sepharose FF, followed by ammonium sulfate precipitation (70% saturation) and gel filtration on Superdex 75. The IBB–GFP–eIF-5A-His6 fusion and deoxyhypusine synthase were purified on nickel NTA agarose.
Purification of native eIF-5A from HeLa cells
Fully hypusinated eIF-5A was purified from 350 ml of cytoplasmic HeLa extract by a series of chromatographic steps on SP–Sepharose, MonoQ, Superdex 75, and yielded ∼3 mg of >95% pure protein. For details see Supplementary material.
Deoxyhypusination of recombinant eIF-5A
For enzymatic deoxyhypusination, we adapted a published protocol (Yan et al., 1996; Liu et al., 1997) as follows: 100 µM untagged eIF-5A was incubated for 3 h at 37°C with 5 µM deoxyhypusine synthase, 200 mM glycine pH 9.0, 2 mM NAD, 2 mM spermidine, 2 mM dithiothreitol. The efficiency of modification was between 80 and 90%. eIF-5A was separated from the enzyme by gel filtration. Deoxyhypusination introduces an additional positive charge into eIF-5A that causes eIF-5A to bind more tightly to cation exchangers. We could therefore use SP–Sepharose to separate the non-modified and deoxyhypusinated forms of eIF-5A at a preparative scale.
Antibodies
Anti-human eIF-5A antibodies were the kind gift of Dr J.Hauber. Anti-CRM1 antibodies were raised against the 12 C-terminal residues of human CRM1 and used after affinity purification.
Kinetic measurement of the RanGTPase
The assay was as described before (Bischoff et al., 1995; Kutay et al., 1997b), with the modification that the measurements were performed at 15°C in 20 mM HEPES–KOH pH 7.5, 120 mM potassium acetate, 1 mM magnesium acetate.
Permeabilized cell assays
These were essentially performed as described previously (Kutay et al., 1997a). Further details are given in the figure legends.
DDBJ/EMBL/GenBank accession Nos
The nucleotide sequence of Exp4 will be available from the DDBJ/EMBL/GenBank under the accession No. AF145021.
Supplementary material
Supplementary material to this paper is available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank Dr J.Hauber for the kind gift of anti-eIF-5A antibodies, Dr R.Lührmann for the kind gift of cytoplasmic HeLa cell extract, K.Ribbeck for supplying permeabilized cells and Dr M.Pool for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB352) and the Human Frontier Science Programme Organisation (grant RG0198/1998M).
References
- Aoki H., Dekany,K., Adams,S.L. and Ganoza,M.C. (1997) The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J. Biol. Chem., 272, 32254–32259. [DOI] [PubMed] [Google Scholar]
- Arts G.J., Fornerod,M. and Mattaj,I.W. (1998a) Identification of a nuclear export receptor for tRNA. Curr. Biol., 8, 305–314. [DOI] [PubMed] [Google Scholar]
- Arts G.J., Kuersten,S., Romby,P., Ehresmann,B. and Mattaj,I.W. (1998b) The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J., 17, 7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartig D., Schümann,H. and Klink,F. (1990) The unique post-translational modification leading to deoxyhypusine or hypusine is a general feature of the archaebacterial kingdom. Syst. Appl. Microbiol., 13, 112–116. [Google Scholar]
- Bartig D., Lemkemeier,K., Frank,J., Lottspeich,F. and Klink,F. (1992) The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur. J. Biochem., 204, 751–758. [DOI] [PubMed] [Google Scholar]
- Benne R., Brown-Luedi,M.L. and Hershey,J.W. (1978) Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D and eIF-5 from rabbit reticulocytes. J. Biol. Chem., 253, 3070–3077. [PubMed] [Google Scholar]
- Bevec D. et al. (1996) Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science, 271, 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. and Görlich,D. (1997) RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett., 419, 249–254. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe,C., Kretschmer,J., Wittinghofer,A. and Ponstingl,H. (1994) RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc. Natl Acad. Sci. USA, 91, 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Krebber,H., Smirnova,E., Dong,W.H. and Ponstingl,H. (1995) Coactivation of RanGTPase and inhibition of GTP dissociation by Ran GTP binding protein RanBP1. EMBO J., 14, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N.C., Adam,E.J. and Adam,S.A. (1995) Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol., 130, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N.C., Adam,E.J.H., Visser,G.D. and Adam,S.A. (1996) RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J. Cell Biol., 135, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper H.L., Park,M.H., Folk,J.E., Safer,B. and Braverman,R. (1983) Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc. Natl Acad. Sci. USA, 80, 1854–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg J.E. and Lund,E. (1998) Functions of the GTPase Ran in RNA export from the nucleus. Curr. Opin. Cell Biol., 10, 400–408. [DOI] [PubMed] [Google Scholar]
- Dou Q.P. and Chen,K.Y. (1990) Characterization and reconstitution of a cell free system for NAD(+)-dependent deoxyhypusine formation on the 18 kDa eIF-4D precursor. Biochim. Biophys. Acta, 1036, 128–137. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Lührmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Floer M. and Blobel,G. (1996) The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem., 271, 5313–5316. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel,G. and Rexach,M. (1997) Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J. Biol. Chem., 272, 19538–19546. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997a) Crm1 is an export receptor for leucine rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fornerod M., van Deursen,J., van Baal,S., Reynolds,A., Davis,D., Murti,K.G., Fransen,J. and Grosveld,G. (1997b) The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J., 16, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell R.A., Truant,R., Thorne,L., Benson,R.E. and Cullen,B.R. (1997) Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J. Cell Sci., 110, 1325–1331. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano,S., Nakamura,T., Adachi,M., Yoshida,M., Yanagida,M. and Nishida,E. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Gordon E.D., Mora,R., Meredith,S.C., Lee,C. and Lindquist,S.L. (1987) Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved among eukaryotes. J. Biol. Chem., 262, 16585–16589. [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Görlich D., Kostka,S., Kraft,R., Dingwall,C., Laskey,R.A., Hartmann,E. and Prehn,S. (1995) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol., 5, 383–392. [DOI] [PubMed] [Google Scholar]
- Görlich D., Henklein,P., Laskey,R.A. and Hartmann,E. (1996a) A 41 amino acid motif in importin α confers binding to importin β and hence transit into the nucleus. EMBO J., 15, 1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Pante,N., Kutay,U., Aebi,U. and Bischoff,F.R. (1996b) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J., 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski,M., Bischoff,F.R., Kutay,U., Bork,P., Hartmann,E., Prehn,S. and Izaurralde,E. (1997) A novel class of RanGTP binding proteins. J. Cell Biol., 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B.R. and Percipalle,P. (1997) Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-β. J. Mol. Biol., 274, 693–707. [DOI] [PubMed] [Google Scholar]
- Herold A., Truant,R., Wiegand,H. and Cullen,B.R. (1998) Determination of the functional domain organization of the importin α nuclear import factor. J. Cell Biol., 143, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J.W. (1994) Expression of initiation factor genes in mammalian cells. Biochimie, 76, 847–852. [DOI] [PubMed] [Google Scholar]
- Hieda M., Tachibana,T., Yokoya,F., Kose,S., Imamoto,N. and Yoneda,Y. (1999) A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J. Cell Biol., 144, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J., Cronshagen,U., Kadokura,M., Marshallsay,C., Wada,T., Sekine,M. and Lührmann,R. (1998) Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J., 17, 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N., Shimamoto,T., Kose,S., Takao,T., Tachibana,T., Matsubae,M., Sekimoto,T., Shimonishi,Y. and Yoneda,Y. (1995) The nuclear pore targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett., 368, 415–419. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay,U., von Kobbe,C., Mattaj,I.W. and Görlich,D. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J., 16, 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S. and Görlich,D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson B.P., Malandrin,L. and Johansson,H.E. (2000) Cell cycle arrest in archaea by the hypusination inhibitor N(1)-guanyl-1,7-diamino heptane. J. Bacteriol., 182, 1158–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Hou,Y. and Inouye,M. (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem., 272, 196–202. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank,N.M., O’Neill,E.M., Huang,L.S. and O’Shea,E.K. (1998) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature, 396, 482–486. [DOI] [PubMed] [Google Scholar]
- Kang H.A. and Hershey,J.W. (1994) Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem., 269, 3934–3940. [PubMed] [Google Scholar]
- Kataoka N., Bachorik,J.L. and Dreyfuss,G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol., 145, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper W.M., Berry,K.W. and Merrick,W.C. (1976) Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem., 251, 5551–5557. [PubMed] [Google Scholar]
- Kim K.K., Hung,L.W., Yokota,H., Kim,R. and Kim,S.H. (1998) Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 Å resolution. Proc. Natl Acad. Sci. USA, 95, 10419–10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C., Bischoff,F.R., Ponstingl,H. and Wittinghofer,A. (1995) Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry, 34, 639–647. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997a) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde,E., Bischoff,F.R., Mattaj,I.W. and Görlich,D. (1997b) Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J., 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Lipowsky,G., Izaurralde,E., Bischoff,F.R., Schwarzmaier,P., Hartmann,E. and Görlich,D. (1998) Identification of a tRNA-specific nuclear export receptor. Mol. Cell, 1, 359–369. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C. and Woese,C.R. (1998) Universally conserved translation initiation factors. Proc. Natl Acad. Sci. USA, 95, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff,F.R., Izaurralde,E., Kutay,U., Schäfer,S., Gross,H.J., Beier,H. and Görlich,D. (1999) Coordination of tRNA nuclear export with processing of tRNA. RNA, 5, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.P., Nemeroff,M., Yan,Y.P. and Chen,K.Y. (1997) Interaction of eukaryotic initiation factor 5A with the human immunodeficiency virus type 1 Rev response element RNA and U6 snRNA requires deoxyhypusine or hypusine modification. Biol. Signals, 6, 166–174. [DOI] [PubMed] [Google Scholar]
- Lounsbury K.M. and Macara,I.G. (1997) Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J. Biol. Chem., 272, 551–555. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Paraskeva E., Izaurralde,E., Bischoff,F.R., Huber,J., Kutay,U., Hartmann,E., Lührmann,R. and Görlich,D. (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol., 145, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H., Cooper,H.L. and Folk,J.E. (1982) The biosynthesis of protein-bound hypusine (Nε-(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (Nε-(4-aminobutyl)lysine). J. Biol. Chem., 257, 7217–7222. [PubMed] [Google Scholar]
- Peat T.S., Newman,J., Waldo,G.S., Berendzen,J. and Terwilliger,T.C. (1998) Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 Å resolution. Structure, 6, 1207–1214. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Michael,W.M., Nakielny,S., Siomi,M.C., Wang,F. and Dreyfuss,G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell, 86, 985–994. [DOI] [PubMed] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Rosorius O., Reichart,B., Kratzer,F., Heger,P., Dabauvalle,M.C. and Hauber,J. (1999) Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J. Cell Sci., 112, 2369–2380. [DOI] [PubMed] [Google Scholar]
- Ruhl M. et al. (1993) Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell Biol., 123, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Abid,M.R. and Miyazaki,M. (1996) Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett., 384, 151–154. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G., Smirnova,E., Deane,R., Solsbacher,J., Kutay,U., Görlich,D., Ponstingl,H. and Bischoff,F.R. (1997) Yrb4p, a yeast ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J., 16, 6237–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J., Schwelberger,H.G., Smit-McBride,Z., Kang,H.A. and Hershey,J.W. (1991) Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.P., Yin,K.C. and Waxman,L. (1997) Effects of inhibitors of RNA and protein synthesis on the subcellular distribution of the eukaryotic translation initiation factor, eIF-5A and the HIV-1 Rev protein. Biol. Signals, 6, 143–149. [DOI] [PubMed] [Google Scholar]
- Shiba T., Mizote,H., Kaneko,T., Nakajima,T. and Kakimoto,Y. (1971) Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim. Biophys. Acta, 244, 523–531. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Eder,P.S., Kataoka,N., Wan,L., Liu,Q. and Dreyfuss,G. (1997) Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol., 138, 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Weis K., Ryder,U. and Lamond,A.I. (1996) The conserved amino-terminal domain of hSRP1 α is essential for nuclear protein import. EMBO J., 15, 1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Wöhl T., Klier,H., Ammer,H., Lottspeich,F. and Magdolen,V. (1993) The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol. Gen. Genet., 241, 305–311. [DOI] [PubMed] [Google Scholar]
- Wolff E.C., Park,M.H. and Folk,J.E. (1990) Cleavage of spermidine as the first step in deoxyhypusine synthesis. The role of NAD. J. Biol. Chem., 265, 4793–4799. [PubMed] [Google Scholar]
- Yan Y.P., Tao,Y. and Chen,K.Y. (1996) Molecular cloning and functional expression of human deoxyhypusine synthase cDNA based on expressed sequence tag information. Biochem. J., 315, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk D. and Jacobson,A. (1998) A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J., 17, 2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]