Abstract
In normoxic cells the hypoxia-inducible factor-1α (HIF-1α) is rapidly degraded by the ubiquitin-proteasome pathway, and activation of HIF-1α to a functional form requires protein stabilization. Here we show that the product of the von Hippel-Lindau (VHL) tumor suppressor gene mediated ubiquitylation and proteasomal degradation of HIF-1α under normoxic conditions via interaction with the core of the oxygen-dependent degradation domain of HIF-1α. The region of VHL mediating interaction with HIF-1α overlapped with a putative macromolecular binding site observed within the crystal structure of VHL. This motif of VHL also represents a mutational hotspot in tumors, and one of these mutations impaired interaction with HIF-1α and subsequent degradation. Interestingly, the VHL binding site within HIF-1α overlapped with one of the minimal transactivation domains. Protection of HIF-1α against degradation by VHL was a multistep mechanism, including hypoxia-induced nuclear translocation of HIF-1α and an intranuclear hypoxia-dependent signal. VHL was not released from HIF-1α during this process. Finally, stabilization of HIF-1α protein levels per se did not totally bypass the need of the hypoxic signal for generating the transactivation response.
Keywords: hypoxia-inducible factor-1α/transcription/tumor suppression/ubiquitylation/von Hippel-Lindau protein
Introduction
von Hippel-Lindau (VHL) disease is caused by germ line mutations of the VHL gene. These mutations lead to the development of a variety of tumors including clear cell carcinomas of the kidney, pheochromocytomas and vascular tumors of the central nervous system and retina. VHL-associated neoplasms are typically hypervascular (reviewed by Kaelin and Maher, 1998), and under normoxic conditions VHL-deficient cells express vascular endothelial growth factor (VEGF) mRNA, which is normally expressed in a hypoxia-dependent fashion (Gnarra et al., 1996; Iliopoulos et al., 1996). Reintro duction of VHL into VHL-mutated renal carcinoma cells indicates that it functions as a negative regulator of VEGF mRNA levels by post-transcriptional (Gnarra et al., 1996; Iliopoulos et al., 1996) and/or transcriptional (Mukhopadhyay et al., 1997) mechanisms.
The VHL protein displays no sequence similarity to other known proteins, thus giving no clues about its function. Biochemical studies have shown that VHL is associated with elongins B and C, and cullin-2 (Cul-2) (Kaelin and Maher, 1998), forming the VHL–BC–Cul-2 complex. The crystal structure of the VHL–BC ternary complex shows two interfaces: one between VHL and elongin C and another between elongins B and C (Stebbins et al., 1999). The 35 residue-long elongin C binding domain of VHL represents one of the mutational hotspots in tumors (Kaelin and Maher, 1998), suggesting that VHL–BC complex formation is critical for tumor suppressor function. In addition, there is a mutational hotspot on a separate domain, the β domain of VHL (Kaelin and Maher, 1998), which overlaps with a putative macromolecular binding site identified in the VHL–BC crystal structure (Stebbins et al., 1999). Elongins B and C and Cul-2 all share homology to components of the SCF (Skp1–Cul-1–F-box protein) multiprotein complex, which targets cell cycle regulatory proteins for ubiquitin-mediated proteolysis (Ciechanover, 1998). Importantly, the structure of the VHL–BC complex extends these similarities to the SCF complex structure (Stebbins et al., 1999), indicating that these protein complexes may have similar functions. In excellent agreement with this model, VHL has recently been shown to be associated with an E3 ubiquitin ligase activity in cellular extracts (Iwai et al., 1999; Lisztwan et al., 1999).
In hypoxic cells, the hypoxia-inducible factor-1α (HIF-1α) mediates transcriptional activation of the VEGF gene (Iyer et al., 1998). HIF-1α mRNA is constitutively expressed in a number of mammalian cells (Kallio et al., 1997). In contrast, the HIF-1α protein is remarkably unstable in cells at normoxia, whereas hypoxia dramatically stabilizes the protein (Kallio et al., 1997). Under normoxic conditions HIF-1α protein degradation is mediated by the ubiquitin-proteasome pathway and a distinct oxygen-dependent degradation domain of HIF-1α (Huang et al., 1998; Kallio et al., 1999). Stabilization of HIF-1α initiates a multi-step pathway of activation of HIF-1α that includes hypoxia-dependent nuclear translocation and dimerization with a partner DNA binding factor, Arnt, to interact with cognate hypoxia-response elements of target promoters, followed by recruitment of transcriptional coactivators (Wenger and Gassmann, 1997; Kallio et al., 1998; Ema et al., 1999). Recently, two studies have indicated distinct roles of VHL in regulation of HIF-1α function: induction of a natural HIF-1α antisense transcript in VHL-deficient cells, resulting in negative regulation of HIF-1α function (Thrash-Bingham and Tartof, 1999). On the other hand, VHL has recently been reported to interact physically with HIF-1α, possibly targeting HIF-1α for protein degradation (Maxwell et al., 1999). Here we show that VHL directly mediated ubiquitylation and ensuing proteasomal degradation of HIF-1α at normoxia via physical interaction with the core of the oxygen-dependent degradation domain. This motif coincided with one of the minimal transactivation domains of HIF-1α. The domain mediating interaction with HIF-1α also represented one of the mutational hotspots of VHL. Protection against VHL-mediated degradation required both nuclear translocation of HIF-1α and an intranuclear hypoxia-dependent regulatory signal. Finally, stabilization of HIF-1α protein levels per se did not bypass the need of the hypoxic signal for generating the transactivation response.
Results
Regulation of HIF-1α protein stability by the VHL tumor suppressor protein
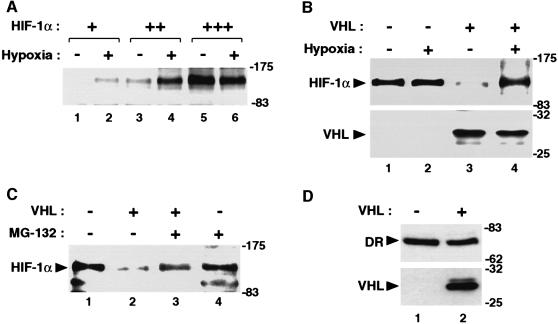
We and others have recently demonstrated that HIF-1α is regulated by the ubiquitin-proteasome pathway under normoxic conditions, resulting in very rapid turnover of the protein, and that one of the early responses to hypoxia is massive upregulation of HIF-1α protein levels (Kallio et al., 1997; Salceda and Caro, 1997; Huang et al., 1998; Kallio et al., 1999). Interestingly, VHL protein complexes have recently been demonstrated to harbor E3 ubiquitin-protein ligase activity, although the target protein for this activity has not yet been identified (Lisztwan et al., 1999; Iwai et al., 1999). Moreover, HIF-1α-regulated target genes such as VEGF are constitutively expressed at normoxia in VHL-deficient cells (Gnarra et al., 1996; Iliopoulos et al., 1996), indicating dysregulation of HIF-1α function in these cells. We were thus interested to investigate the potential mechanism of regulation of HIF-1α function by VHL. Due to the pronounced lability of the HIF-1α protein under normoxic conditions, HIF-1α is normally not detectable by immunoblot analysis of cellular extracts (Kallio et al., 1997, 1998). It was therefore not possible to investigate the effect of expression of VHL on endogenous HIF-1α protein levels. To establish experimental conditions to examine the effect of VHL on HIF-1α protein stability, we transiently transfected COS7 cells with FLAG epitope-tagged HIF-1α expression plasmids. As expected, at a low concentration (0.2 µg) of expression vector, HIF-1α was not detected at normoxia, and we observed potent stabilization of HIF-1α protein levels at hypoxia, as assessed by immunoblot analysis (Figure 1A). However, at higher concentrations (0.5–1 µg) of expression vector we could detect HIF-1α protein expression also under normoxic conditions (Figure 1A). At the highest concentrations of expression vector tested we observed significant HIF-1α expression levels both at normoxia and hypoxia (Figure 1A). Thus, these experiments suggest that the mechanism of degradation of HIF-1α had become saturated under these conditions and that one or several components of the degradation machinery were limiting.
Fig. 1. VHL mediates proteasomal degradation of HIF-1α under normoxic conditions. (A) Expression of HIF-1α under normoxic conditions. Increasing amounts [0.2 µg (+), 0.5 µg (++), 1.0 µg (+++)] of pFLAG CMV2/HIF-1α were transiently transfected into COS7 cells, and the cells were incubated for 12 h at normoxia (21% O2) or hypoxia (1% O2), as indicated. Whole-cell extracts were analyzed by immunoblotting using anti-FLAG antibodies. (B) Degradation of HIF-1α in the presence of VHL. pFLAG CMV2/HIF-1α was cotransfected into COS7 cells together with empty vector or wild-type VHL expression vector (pCMX/VHL). Whole-cell extracts were analyzed by immunoblotting using anti-FLAG or anti-VHL antibodies. (C) VHL mediates proteasomal degradation of HIF-1α. Cells were transfected with pFLAG CMV2/HIF-1α and pCMX/VHL, incubated in the absence or presence of 5 µM MG-132 for 6 h before harvesting, and cellular extracts were analyzed as in (B). (D) VHL does not affect dioxin receptor (DR) protein levels. COS7 cells were transfected with a FLAG-tagged dioxin receptor expression vector (pCMV/DR/FLAG) in the absence or presence of pCMX/VHL, and analyzed as in (B). The mobilities of molecular weight (kDa) markers are shown on the right hand side of the blots.
We next used the high level HIF-1α expression conditions for all subsequent experiments to examine the effect of VHL on HIF-1α protein levels. Transient coexpression of FLAG/HIF-1α and VHL resulted in reduction of the HIF-1α protein signal under normoxic conditions (Figure 1B, upper panel), indicating that VHL may have been limiting under the conditions of expression of HIF-1α alone. Interestingly, VHL failed to induce reduction of HIF-1α protein levels under hypoxic conditions (Figure 1B). In control experiments we detected similar levels of VHL expression in extracts from either normoxic or hypoxic cells (Figure 1B, lower panel). VHL-induced reduction of HIF-1α protein levels at normoxia was inhibited by treatment of the cells with the proteasome inhibitor MG-132 (Figure 1C). Taken together, these results strongly suggest that VHL mediates proteasomal degradation of HIF-1α. This effect of VHL was specific for HIF-1α, as transiently expressed VHL did not produce this effect on FLAG-tagged dioxin receptor (Figure 1D), a basic helix–loop–helix(bHLH)/PAS (Per/Arnt/Sim domain) protein belonging to the same class of transcription factors as HIF-1α.
Two domains of VHL are required for inducing protein degradation of HIF-1α
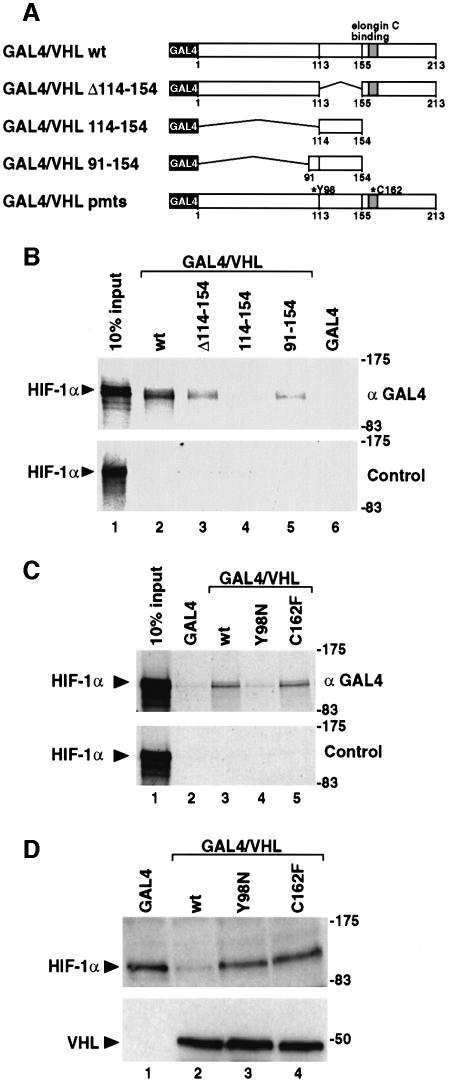
Given the potential role of VHL as an E3 ubiquitin ligase we examined whether VHL physically interacted with HIF-1α. 35S-labeled, in vitro translated HIF-1α was incubated with wild-type or mutant GAL4/VHL fusion proteins (schematically represented in Figure 2A) or the minimal GAL4 DNA binding domain alone prior to immunoprecipitation assays. In these experiments, 35S-labeled HIF-1α was co-immunoprecipitated in the presence of GAL4/VHL by anti-GAL4 specific antibodies, whereas no interaction was observed between HIF-1α and the minimal GAL4 DNA binding domain (Figure 2B, upper panel). Non-specific pre-immune rabbit antiserum did not precipitate HIF-1α protein in the presence of either VHL or GAL4 alone (Figure 2B, lower panel), indicating that wild-type VHL specifically interacted with HIF-1α in vitro.
Fig. 2. VHL requires two functional domains to induce HIF-1α degradation. (A) Schematic representation of GAL4-fused wild-type (wt), deletion or single amino acid point mutant (pmt) forms of VHL. (B) VHL directly interacts with HIF-1α. Equal concentrations of in vitro translated 35S-labeled full-length HIF-1α were incubated with in vitro translated wild-type GAL4/VHL or VHL deletion mutants or GAL4-DBD spanning the GAL4 DNA binding domain alone. Co-immunoprecipitation assays with anti-GAL4 antibody (upper panel) or control preimmune serum (lower panel) were carried out as described in Materials and methods. The precipitated material was analysed by SDS–PAGE and autoradiography. For loading controls, 10% of input 35S-labeled HIF-1α is shown in lane 1. (C) A tumor-derived point mutation impairs the interaction between VHL and HIF-1α. In vitro translated proteins were incubated and analyzed by co-immunoprecipitation as in (B). (D) Tumor-derived point-mutated forms of VHL fail to induce HIF-1α degradation. pFLAG CMV2/HIF-1α was transiently coexpressed in COS7 cells in the absence or presence of GAL4-fused wild-type or mutant forms of VHL as indicated. Cells were incubated at normoxia for 24 h, and whole cell extracts were prepared and analyzed by immunoblotting using anti-FLAG or anti-VHL antibodies. The mobilities of molecular weight (kDa) markers are shown on the right hand side of the blots.
The VHL Δ114–154 deletion mutant showed interaction with HIF-1α, whereas the VHL 114–154 fragment failed to do so (Figure 2B). A 23 amino acid-long N-terminal extension of this fragment generated VHL 91–154, which was able to interact with HIF-1α, indicating the importance of a structure located between residues 91 and 113 of VHL to interact with HIF-1α. Interestingly, this region of VHL is not only contained within the putative macromolecular binding site observed in the crystal structure of the VHL–BC complex (Stebbins et al., 1999), but also represents one of the mutational hotspots in tumors (Kaelin and Maher, 1998). This fact prompted us to examine whether tumor-derived mutations of VHL would affect its ability to interact with HIF-1α and/or to induce HIF-1α degradation. We performed these experiments using GAL4/VHL fusion proteins containing either a Y98N (the most frequent tumor mutation in this region; Kaelin and Maher, 1998) or a C162F single amino acid mutation. The C162F mutation has been demonstrated to render VHL unable to bind the elongin B–C complex (Lonergan et al., 1998; Lisztwan et al., 1999), and inhibit ubiquitin ligase activity in vitro (Lisztwan et al., 1999). In co-immunoprecipitation experiments, VHL Y98N was unable to interact with HIF-1α, whereas VHL C162F showed wild-type levels of interaction with HIF-1α (Figure 2C, upper panel). In our cellular degradation assay we transiently expressed at normoxia FLAG/HIF-1α in the presence or absence of wild-type or the individual point-mutated forms of VHL. Immunoblot analysis demonstrated that, in contrast to wild-type VHL, both the VHL Y98N and VHL C162F mutants failed to induce degradation of HIF-1α at normoxia (Figure 2D). These results demonstrate that both the HIF-1α interaction domain and the elongin C binding domain of VHL are necessary to mediate degradation of HIF-1α, and that regulation of HIF-1α may be involved in the tumor suppressor function of VHL.
The oxygen-dependent degradation domain of HIF-1α is targeted for regulation by VHL
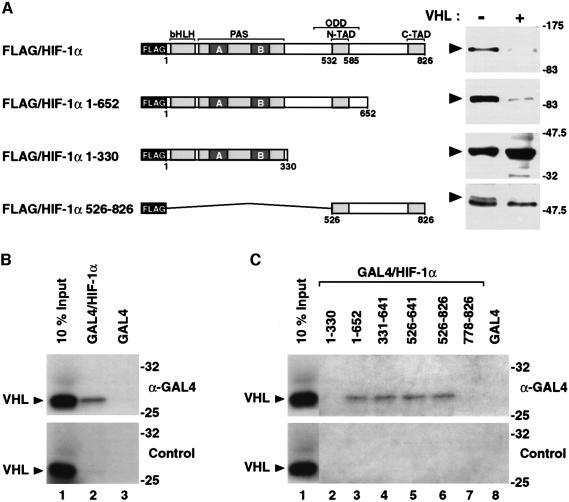
To identify the domain of HIF-1α that is targeted by VHL to mediate proteasomal degradation at normoxia, we transiently expressed in COS7 cells in the presence or absence of VHL either wild-type FLAG/HIF-1α or a series of FLAG-tagged HIF-1α deletion mutants. In analogy to wild-type HIF-1α, HIF-1α 1–652 lacking the C-terminus including the C-terminal transactivation domain (schematically represented in Figure 3A) was degraded in the presence of VHL. However, the protein levels of HIF-1α 1–330 lacking structures C-terminal of the PAS domain were not affected by VHL. HIF-1α 526–826 lacking N-terminal structures (including the bHLH and PAS domains) was also degraded upon exposure to VHL at normoxia (Figure 3A). In conclusion, these results indicate that a C-terminal region of HIF-1α spanning residues 526–652 mediated VHL-dependent degradation.
Fig. 3. VHL targets the oxygen-dependent degradation domain of HIF-1α. (A) FLAG-tagged wild-type HIF-1α or the indicated HIF-1α deletion mutants were transiently coexpressed in COS7 cells at normoxia in the absence or presence of VHL. Whole-cell extracts were prepared and analyzed as described in Figure 1. ODD, oxygen-dependent degradation domain; N- and C-TAD, N- and C-terminal transactivation domains. (B) 35S-labeled VHL was incubated with equal concentrations of in vitro translated GAL4-fusion proteins spanning full-length HIF-1α or GAL4-DBD alone. Co-immunoprecipitation assays were performed with anti-GAL4 antibodies (upper panel) or control non-specific rabbit antiserum (lower panel). The precipitated material was analyzed by SDS–PAGE and autoradiography. For loading controls, 10% of input 35S-labeled VHL is shown. (C) VHL, GAL4 fusion proteins spanning the indicated HIF-1α fragments or GAL4 alone were expressed by in vitro translation and analyzed as in (B).
We proceeded to map the domain of HIF-1α required to interact with VHL. To this end we used full-length HIF-1α or a set of HIF-1α deletion mutants fused to the GAL4 DNA binding domain, and performed co-immunoprecipitation assays following incubation with VHL. As expected, 35S-labeled VHL was specifically co-immunoprecipitated together with full-length HIF-1α (Figure 3B). In excellent agreement with the fact that the deletion mutant HIF-1α 1–330 was not degraded upon overexpression of VHL in COS7 cells under normoxic conditions (Figure 3A), GAL4/HIF-1α 1–330 failed to interact physically in vitro with 35S-labeled VHL (Figure 3C, upper panel). Moreover, GAL4/HIF-1α 778–826, spanning the C-terminal transactivation domain of HIF-1α, did not show any interaction with VHL (Figure 3C, upper panel). In contrast, the GAL4/HIF-1α fragments, which showed VHL-mediated degradation in normoxic cells, HIF-1α 1–652 and HIF-1α 526–826 (Figure 3A), clearly interacted with VHL (Figure 3C). In control reactions, the GAL4/HIF-1α fragments were expressed at similar levels (data not shown), and non-specific pre-immune rabbit antiserum did not precipitate VHL protein in the presence of any of the used fragments or GAL4 alone (Figure 3B and C, lower panels). In a more detailed analysis of the interaction of VHL with HIF-1α, VHL was co-immunoprecipitated by anti-GAL4 antibodies in the presence of GAL4/HIF-1α 331–641 or GAL4/HIF-1α 526–641. In fact, when compared with full-length HIF-1α, all these latter fragments of HIF-1α interacted with VHL with very similar efficacies (Figure 3C, upper panel). Taken together, these results indicate that a region of HIF-1α spanning residues 526–641 was essential for physical interaction with VHL. Interestingly, this region overlaps with the oxygen/redox-dependent degradation domain of HIF-1α, which has previously been demonstrated to mediate proteasomal degradation of HIF-1α in normoxic cells, and that has broadly been defined to be located between amino acid residues 401 and 603 of hHIF-1α (Huang et al., 1998; Kallio et al., 1999).
The minimal N-terminal transactivation domain of HIF-1α is a target for ubiquitylation and proteasomal degradation by VHL
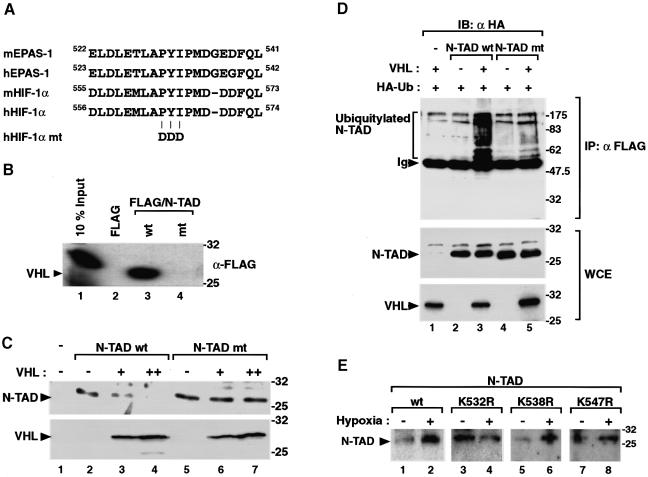
Interestingly, the VHL-interacting fragment GAL4/HIF-1α 526–641 contains not only the core of the oxygen-dependent degradation domain of HIF-1α but also the N-terminal transactivation domain N-TAD (Jiang et al., 1997; Pugh et al., 1997; Figure 3A). Within the N-TAD of HIF-1α a sequence motif of ∼19 amino acid residues (located between amino acids 556 and 574 of hHIF-1α) shows the strongest conservation between species and is also conserved in the related hypoxia-inducible factor EPAS-1/HLF (Figure 4A), which is expressed in a tissue-restricted fashion. This motif is also highly conserved in the hypoxia-responsive Drosophila Similar protein (reviewed by Taylor and Zhulin, 1999). Interestingly, this sequence motif has recently been reported to be important for the function of the oxygen-dependent degradation domain of HIF-1α since alanin substitutions within this domain impair hypoxia-dependent protein stabilization (Srinivas et al., 1999). We initially examined whether VHL could interact directly with the minimal N-TAD located between amino acids 532 and 585 of hHIF-1α (Jiang et al., 1997; Pugh et al., 1997). In co-immunoprecipitation assays, anti-FLAG antibodies could precipitate 35S-labeled VHL in the presence of the FLAG epitope-tagged minimal N-TAD of HIF-1α (Figure 4B), demonstrating that this region of HIF-1α is sufficient to mediate interaction with VHL. We next generated a point mutation within the context of the minimal N-TAD. As assessed in co-immunoprecipitation assays, substitution of the central PYI triplet with aspartic acid totally abolished interaction between VHL and the N-TAD (Figure 4B). In conclusion, these experiments demonstrate that the highly conserved core motif of the N-TAD is critical for interaction with VHL.
Fig. 4. The minimal N-TAD of HIF-1α is the target for VHL-mediated ubiquitylation and proteasomal degradation. (A) Alignment of N-TAD sequences of human (h) and mouse (m) HIF-1α and EPAS-1 revealed a conserved core sequence motif. The positions of amino acid substitutions are shown in mutant (mt) N-TAD. (B) Mutation of the N-TAD PYI motif abolishes interaction between HIF-1α and VHL. 35S-labeled VHL was incubated with equal concentrations of in vitro translated FLAG-tagged wild-type, mutant N-TAD or FLAG epitope alone. Co-immunoprecipitation assays were carried out using anti-FLAG antibodies and analyzed by SDS–PAGE and autoradiography. For loading controls, 10% of input 35S-labeled VHL is shown. (C) The PYI mutation confers resistance to VHL-mediated degradation. FLAG or FLAG-tagged wild-type or mutant N-TAD (1 µg/6-cm dish) were transiently coexpressed in COS7 cells in the absence or presence of increasing concentrations (1.0 µg, +; 2.0 µg/6-cm dish, ++) of VHL as indicated, and incubated at normoxia for 24 h. Whole-cell extracts were prepared and analyzed by immunoblotting using anti-FLAG or anti-VHL antibodies. (D) VHL mediates ubiquitylation of N-TAD. FLAG, or FLAG-tagged wild-type or mutant N-TAD and HA-tagged ubiquitin were transiently coexpressed in COS7 cells in the presence or absence of VHL under normoxic conditions in combination with MG132 for 6 h. After immunoprecipitation of whole-cell extracts by anti-FLAG, ubiquitylated forms of N-TAD were detected by anti-HA immunoblotting (upper panel). Ten percent of input whole-cell extracts are shown in the two lower panels. (E) Effect on protein stability following substitution of individual lysines to arginines within N-TAD. Wild-type or mutant N-TAD were transiently expressed in 293 cells and incubated at normoxia or hypoxia for 12 h. Whole-cell extracts were prepared and analyzed as in (C). The mobilities of molecular weight (kDa) markers are shown to the right of the blots.
We next transiently expressed under normoxic conditions the wild-type or mutant N-TAD in the absence or presence of increasing concentrations of VHL. Immunoblot analysis demonstrated that wild-type N-TAD protein was degraded in a dose-dependent manner by VHL (Figure 4C). In contrast, the stability of the mutant N-TAD was not affected by identical concentrations of VHL (Figure 4C).
To investigate the mechanism of VHL-mediated degradation of HIF-1α, we performed in vivo ubiquitylation experiments. We transiently coexpressed the FLAG-tagged wild-type or mutant N-TAD and HA-tagged ubiquitin in the presence or absence of VHL in COS7 cells under normoxic conditions in the presence of the proteasome inhibitor MG132. After anti-FLAG immunoprecipitation of whole-cell extracts, ubiquitylated N-TAD forms were detected by anti-HA immunoblotting analysis. Ubiquitylation of wild-type N-TAD was strongly enhanced by coexpression of VHL (Figure 4D). On the other hand, the mutant N-TAD showed very low levels of ubiquitylation even in presence of VHL. Importantly, these results clearly demonstrate that VHL mediates ubiquitylation of HIF-1α via physical interaction with the minimal N-TAD motif.
Given the ubiquitylation of the minimal N-TAD motif, we next examined which of the three lysines of N-TAD were targeted for degradation at normoxia. FLAG-tagged wild type or single lysine mutants (K532R, K538R and K547R, respectively) of N-TAD were transiently expressed in 293 cells. The cells were incubated at normoxia or hypoxia for 12 h, and whole-cell extracts were analyzed by immunoblotting. Analogous to full-length HIF-1α, the minimal wild-type GAL4/N-TAD fusion protein showed significant degradation under normoxic conditions, and was stabilized by hypoxia. Interestingly, mutation of K532 stabilized the protein at normoxia, whereas expression of the two other lysine mutants was hardly detectable at normoxia (Figure 4E). These results suggest that K532 is critical for degradation of HIF-1α.
Subcellular localization of VHL at normoxia and hypoxia
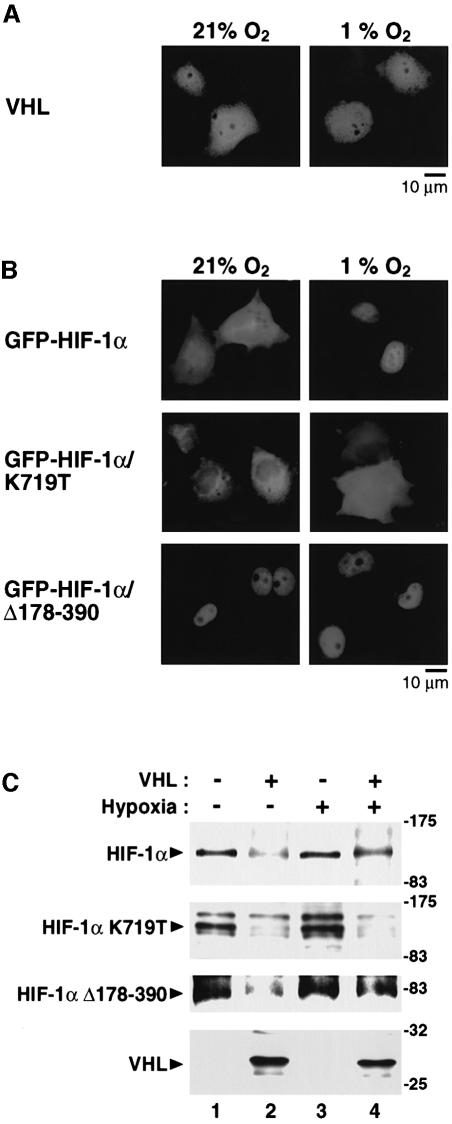
We have previously demonstrated that hypoxia induces nuclear translocation of HIF-1α (Kallio et al., 1998). In the case of VHL, nuclear-cytoplasmic trafficking has been suggested to be required for VHL function (Lee et al., 1999). To study the intracellular localization of VHL in relation to its function in normoxic versus hypoxic cells we transiently expressed VHL in COS7 cells. At normoxia, immunofluorescence by an anti-VHL antibody was detected throughout the cells with some preference toward localization in the cytoplasmic compartment of the cells (Figure 5A). A very similar distribution of VHL immunoreactivity was observed under hypoxic conditions (Figure 5A). Semiquantitative analysis of the subcellular localization of VHL immunoreactivity (Kallio et al., 1998) under normoxic conditions revealed that 47% of the transfected cells had equal distribution of fluorescence in the cytoplasm and the nucleus, whereas in 45% of the transfected cells cytoplasmic fluorescence predominated over that detected in the nucleus. No transfected cell showed exclusive nuclear staining. Hypoxic treatment of the cells had no effect on the intracellular distribution of VHL since under these conditions 47, 51 and 0% of the transfected cells fell into the three different categories, respectively.
Fig. 5. Protection of HIF-1α against VHL-mediated degradation requires nuclear translocation of HIF-1α and a hypoxia-dependent intranuclear signal. (A) Subcellular localization of VHL. COS7 cells were transfected with pCMX/VHL and after 24 h of expression, the cells were incubated for 6 h at normoxia or hypoxia. The subcellular localization was determined by indirect immunofluorescence using anti-VHL antibodies. (B) Subcellular distribution of GFP–HIF-1α chimeric proteins. GFP fusion proteins spanning wild-type or mutant forms of HIF-1α were transiently expressed in COS7 cells and incubated as above. Photographs were taken using a Zeiss fluorescent microscope. (C) Effect of VHL on degradation of wild-type and mutant forms of HIF-1α showing constitutively cytoplasmic or nuclear localization. FLAG-tagged wild-type or mutant forms of HIF-1α were transiently expressed in the absence or presence of VHL and incubated for 12 h at normoxia or hypoxia. Whole-cell extracts were prepared and assayed as in Figure 1.
Protection of HIF-1α from VHL-dependent degradation is a multi-step pathway requiring hypoxia-induced nuclear translocation of HIF-1α and a hypoxia-dependent regulatory signal
We next compared the effect of VHL on the stability of wild-type HIF-1α versus the stability of a HIF-1α single amino acid point mutant, HIF-1α K719T, which fails to enter the nucleus at hypoxia and is constitutively localized in the cytoplasm (Kallio et al., 1998; Figure 5B). Under normoxic conditions, both wild-type HIF-1α and HIF-1α K719T were degraded in the presence of VHL (Figure 5C). Interestingly, whereas wild-type HIF-1α showed significant (albeit not total) resistance to VHL under hypoxic conditions (Figures 1B and 5C), the HIF-1α K719T mutant was potently degraded upon exposure to VHL in hypoxic cells (Figure 5C). In conclusion, these results suggest that hypoxia-induced nuclear translocation of HIF-1α protects HIF-1α from VHL-mediated proteasomal degradation.
To test whether nuclear localization is sufficient to protect HIF-1α against VHL-induced proteolysis we examined the effect of VHL on the stability of a HIF-1α mutant, HIF-1α Δ178–390, which shows constitutive nuclear localization (Kallio et al., 1998; Figure 5B). Strikingly, this protein was degraded at normoxia upon exposure to VHL and showed hypoxia-dependent protein stabilization (Figure 5C). In biochemical experiments, cytosolic or nuclear extracts were prepared from COS7 cells transiently expressing wild-type or mutant HIF-1α in the absence or presence of VHL. HIF-1α and VHL co-immunoprecipitation was detected only in nuclear extracts of hypoxic cells; whereas VHL was constitutively associated with the nuclear HIF-1α Δ178–390 mutant, and no interaction was observed between HIF-1α K719T and VHL in nuclear extracts from either hypoxic or normoxic cells (data not shown). In summary, nuclear localization per se cannot explain protection of HIF-1α against degradation by VHL. Our results suggest that, in addition to nuclear translocation, a distinct hypoxia-dependent intranuclear event or signal is required for stabilization of HIF-1α. Given the striking overlap within the N-TAD between the structures that mediate degradation, physical interaction with VHL, and hypoxia-inducible transactivation, it is possible that this putative intranuclear stabilizing signal may be linked to the transactivation function of the protein.
VHL is not released from HIF-1α in hypoxic cells
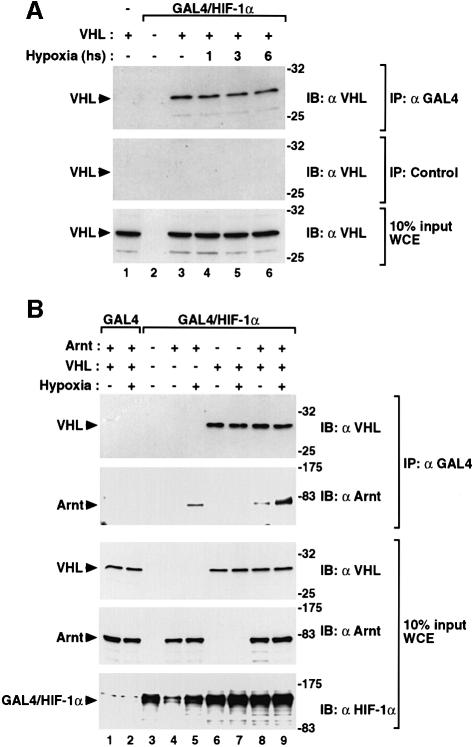
Since we found that both nuclear translocation of HIF-1α and a hypoxia-induced activation signal were necessary to protect HIF-1α from VHL-mediated degradation, we next examined the mechanistically important question of whether VHL was released from HIF-1α under hypoxic conditions. We transiently expressed GAL4/HIF-1α or the minimal GAL4 DNA binding domain in COS7 cells in the absence or presence of VHL under normoxic or hypoxic conditions for increasing periods of time. In co-immunoprecipitation experiments using whole-cell extracts, VHL was detected by immunoblotting using anti-VHL antibodies. As expected, VHL was specifically co-immunoprecipitated together with GAL4/HIF-1α under normoxic conditions. However, VHL was also co-immunoprecipitated under hypoxic conditions at levels similar to those observed at normoxia (Figure 6A). These results suggest that dissociation of the HIF-1α–VHL complex is not necessary for protection of HIF-1α from VHL-mediated degradation, and that there exists a mechanism for hypoxia-dependent inactivation of VHL function when remaining associated with HIF-1α.
Fig. 6. VHL, Arnt and HIF-1α form a ternary nuclear complex. (A) VHL is not released from HIF-1α in hypoxic cells. GAL4, GAL4/HIF-1α and/or VHL were transiently coexpressed in COS7 cells in the presence of MG132 at normoxia or hypoxia for the indicated periods of time. Following immunoprecipitation of whole cell extracts by anti-GAL4 (upper panel) or control (middle panel) antibodies, VHL was detected by immunoblotting using anti-VHL antibodies. Ten percent of input whole-cell extracts is shown in the lower panel. (B) Association with Arnt. GAL4 or GAL4/HIF-1α were transiently expressed in COS7 cells in the absence or presence of Arnt or VHL at normoxia or hypoxia for 6 h, and analyzed as above.
Next, we examined whether Arnt was associated with the HIF-1α–VHL complex. GAL4/HIF-1α or the minimal GAL4 DNA binding domain transiently expressed in COS7 cells in the absence or presence of VHL and/or Arnt at normoxia or hypoxia. As expected, Arnt was specifically co-immunoprecipitated together with GAL4/HIF-1α under hypoxic conditions in the absence of VHL (Figure 6B). Moreover, in the presence of VHL, Arnt and GAL4/HIF-1α were also co-immunoprecipitated in a hypoxia-dependent fashion (Figure 6B), demonstrating that these proteins formed a ternary complex in hypoxic cells.
Role of protein stabilization in regulation of the hypoxia-dependent transactivation function of the HIF-1α N-TAD domain
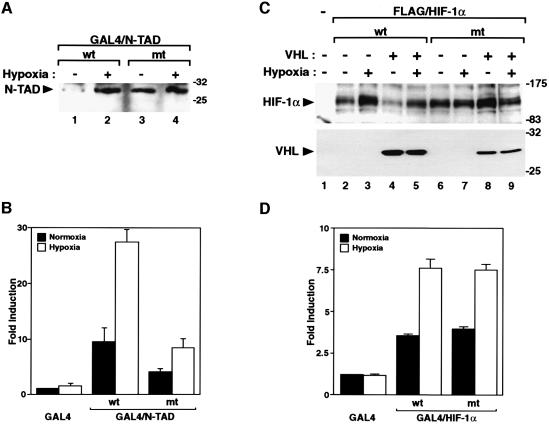
We and others have observed that transcriptional activation by HIF-1α in hypoxic cells is mediated by two distinct transactivation domains, N- and C-TAD (Jiang et al., 1997; Pugh et al., 1997; Ema et al., 1999; Carrero et al., 2000). Given the fact that VHL interacted with the minimal N-TAD structure, we were interested to compare under normoxic and hypoxic conditions both the stability and transactivation functions of GAL4 fusion proteins harboring either the wild-type and/or the PYI mutant N-TAD motifs (Figure 4A). Whereas wild-type GAL4/N-TAD showed hypoxia-dependent protein stabilization (Figures 4E and 7A), PYI mutant GAL4/N-TAD protein levels were readily detectable in extracts from normoxic cells, and not significantly increased following exposure of the cells to hypoxia (Figure 7A). Thus, the failure of this mutant to interact with VHL (Figure 4B) correlated with constitutively stable protein expression levels similar to those generated by wild-type GAL4/N-TAD under hypoxic conditions. In transactivation assays using a GAL4-driven luciferase reporter gene, we observed, as expected (Carrero et al., 2000), rather modest (∼3-fold) activation of the function of the minimal wild-type GAL4/N-TAD by hypoxia (Figure 7B). In the context of full-length HIF-1α, mutation of the PYI motif rendered the protein stable under normoxic conditions and resistant to VHL-mediated degradation (Figure 7C), demonstrating that this motif is the critical determinant for VHL-dependent degradation of HIF-1α. Although the mutant protein showed an increased constitutive transcriptional activity in comparison with wild-type HIF-1α, it was still hypoxia-inducible (Figure 7D). Within the minimal N-TAD domain, mutation of the PYI motif reduced the transactivation function with regard to both the activity observed at normoxia and hypoxia (Figure 7B). These results indicate that the mutation may generally have altered the structure important for transactivation, possibly impairing some of the protein contacts that may be required for the full activation response. Nevertheless, this mutated construct was inducible by hypoxia, yielding an ∼2-fold increase in transcriptional activity (Figure 7B). These data suggest that the stabilized protein resistant to degradation by VHL is still capable of mediating a hypoxia-dependent activation response, and that protein stabilization per se does not bypass the need of the hypoxic signal for transactivation.
Fig. 7. Mutant N-TAD maintains an hypoxia-inducible transactivation response. (A) Constitutive stabilization of PYI mutant N-TAD. GAL4-fused wild-type and mutant N-TAD were transiently expressed in COS7 cells at normoxia or hypoxia for 12 h. Whole-cell extracts were prepared and assayed as in Figure 1. (B) Hypoxia-inducible transactivation function of mutant N-TAD. The transcriptional activation function by GAL4-fused wild-type or mutant N-TAD proteins was monitored using a GAL4-responsive luciferase reporter gene as described in Materials and methods. Six hours after transfection, cells were incubated for 36 h at either normoxia or hypoxia. Reporter gene activities are expressed relative to the activity of the GAL4 DNA binding domain alone at normoxia, and normalized against internal control β-galactosidase activities. Values represent the mean ± SD of three independent experiments. (C) PYI mutant stabilizes full-length HIF-1α in normoxic cells. FLAG-tagged wild-type and mutant HIF-1α were transiently expressed in absence or presence of VHL and analyzed as above. (D) Transcription activation functions of wild-type and mutant HIF-1α proteins were assayed as in (B).
Discussion
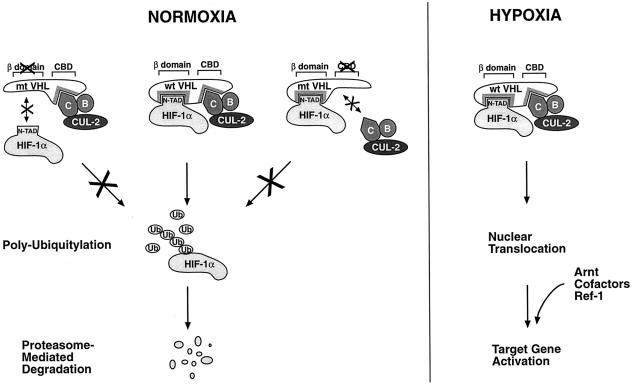
We have demonstrated that VHL interacts with HIF-1α to mediate ubiquitin-proteasomal degradation of HIF-1α under normoxic conditions. The HIF-1α-binding domain of VHL coincides with one of the hotspots of cancer-causing mutations in the VHL gene (Kaelin and Maher, 1998). In fact, a frequent tumor mutation occurring within this region of VHL abrogated VHL–HIF-1α complex formation, suggesting the importance of VHL–HIF-1α interaction for the tumor suppressor function of VHL. HIF-1α is among the most short-lived proteins currently known. The half-life of HIF-1α after exposure of cells to hypoxia and subsequent return to normoxia is in the range of a few minutes (Wang et al., 1995). The present data indicate that VHL functions by recruiting HIF-1α to the VHL–BC–Cul-2 complex. Importantly, the naturally occurring C162F mutation of VHL fails to bind the elongin B and C complex, and has recently been demonstrated to abrogate VHL-dependent ubiquitylation activity in vitro (Lisztwan et al., 1999). As schematically summarized in Figure 8, this mutant also failed to mediate degradation of HIF-1α, strongly supporting the notion that VHL regulates HIF-1α function via targeting it for ubiquitylation.
Fig. 8. Model of conditional regulation of HIF-1α function. Under normoxic conditions (left panel) HIF-1α is targeted for ubiquitin-proteasomal degradation by VHL. The shaded areas in VHL indicate mutational hotspots in tumors that coincide with the HIF-1α or the elongin C binding domain (CBD) of VHL, respectively. Mutations in either of these two domains stabilize HIF-1α protein. Hypoxia (right panel) leads to inhibition of degradation of HIF-1α by induction of nuclear translocation and an as yet unidentified nuclear regulatory signal that may be linked to recruitment of a partner DNA binding factor, Arnt, transcriptional coactivators and/or the redox regulator Ref-1. See text for details.
Degradation of a protein by the ubiquitin system involves two critical steps: covalent attachment of multiple ubiquitin molecules to the target protein, and degradation of the ubiquitin-tagged substrate by the 26S proteasome (reviewed by Ciechanover, 1998). Although the cascade of enzymatic pathways that mediate conjugation of ubiquitin to its substrates has been rather well characterized (for recent reviews see Ciechanover, 1998; Hershko and Ciechanover, 1998), one of the central questions remains how proteins are selected for degradation (reviewed by Laney and Hochstrasser, 1999). Obviously, this process must be highly specific since short-lived proteins need to be identified and differentiated from more stable proteins within the cells.
Earlier experiments have indicated that regulation of HIF-1α protein levels by the proteasome pathway is mediated by a C-terminal structure of HIF-1α spanning PEST sequence motifs (Huang et al., 1998; Kallio et al., 1999). This structure has been termed the oxygen-dependent degradation domain (Huang et al., 1998), and notably, this region harbors the N-TAD domain of HIF-1α. What is the degradation motif within the C-terminal region of HIF-1α that mediates regulation by VHL? Transplantable sequence elements, destruction boxes recognized and targeted by a proteolytic apparatus, have been identified in many short-lived proteins (Laney and Hochstrasser, 1999) but there is still limited information with regard to the structural characteristics of these elements. Here we demonstrate that VHL directly interacts with the N-TAD of HIF-1α, and specifically ubiquitylates this structure. More specifically, point mutagenesis indicates that the highly conserved central PYI motif of the N-TAD was critical for interaction with VHL and for VHL-induced ubiquitylation and proteasomal degradation.
We have recently observed that exposure of cells to hypoxia or a hypoxia-mimicking agent results in marked reduction in recovery of ubiquitylated HIF-1α complexes. This effect of hypoxia, in turn, correlates with the hypoxia-induced stabilization of HIF-1α protein levels (Kallio et al., 1999). Thus, HIF-1α is differentially regulated by ubiquitylation, and inhibition of ubiquitylation constitutes an early and critical step in hypoxia-dependent activation of HIF-1α function. It is likely that these effects are correlated with protection of HIF-1α against regulation by VHL. If this is the case, what is the mechanism that renders VHL-mediated degradation of HIF-1α inactive in hypoxic cells? Obviously, this is a key question to understanding the mechanism of conditional regulation of HIF-1α function. As schematically illustrated in Figure 8, we show here that hypoxia-induced protection of HIF-1α against regulation by VHL involves two distinct and successive steps: nuclear translocation of HIF-1α and an intranuclear event or signal required for protection of HIF-1α against VHL-induced proteolysis. Given the striking overlap within the N-TAD of HIF-1α between the structures that mediate oxygen-dependent degradation, physical interaction with VHL and hypoxia-inducible transactivation, it is possible that the putative intranuclear stabilizing signal is linked to the transactivation function of the protein. We and others have previously observed that the hypoxia-inducible function of both the N-TAD and C-TAD is critically dependent on the recruitment of the transcriptional coactivator CBP/p300 and SRC-1/p160 (Arany et al., 1996; Kallio et al., 1998; Ema et al., 1999; Carrero et al., 2000). This recruitment appears to be facilitated by the redox regulator Ref-1 (Ema et al., 1999; Carrero et al., 2000). Thus, the functional architecture of the N-TAD encompasses overlapping structures that have two different, in fact opposing functions, i.e. protein degradation versus activation of gene transcription, creating an important ‘switch’ in regulation of HIF-1α protein function. The hypoxia-dependent intranuclear mechanism of protection may involve dimerization with Arnt, recruitment of coactivators and/or recruitment of Ref-1. Importantly, the present data indicate that protein stabilization per se does not provide the sole basis for rendering HIF-1α transcriptionally active. It is possible that covalent modification of the C-terminus of HIF-1α may play a role in determining this regulatory effect. For instance, it is also an attractive scenario that the hypoxic signal may determine a conformational change in HIF-1α, which facilitates recruitment of the coactivators and inactivates VHL function.
Materials and methods
Plasmid constructs
pFLAG CMV2/HIF-1α1–826, pCMX-GAL4/HIF-1α and deletion mutants thereof, and pCMV/Arnt have been described elsewhere (Kallio et al., 1997, 1998; Carrero et al., 2000), or were constructed by inserting relevant fragments of hHIF-1α into pFLAG-CMV2 (Kodak). Amino acid substitutions in pFLAG or GAL4/HIF-1α or NTAD were generated using a QuikChange site-directed mutagenesis kit (Stratagene). Expression plasmids for green fluorescent protein (GFP) fused to wild-type or mutant forms of HIF-1α have been described previously (Kallio et al., 1998). Wild-type or mutant forms of VHL and VHL GAL4 fusion proteins were constructed by inserting a blunted NheI–EcoRI fragment of pCI/VHL wild-type or mutant/FLAG (generously provided by Dr Joan W.Conaway, Howard Hughes Medical Institute, USA) into EcoRV-digested pCMX or pCMX-GAL4. The hemagglutinin (HA)-tagged ubiquitin expression plasmid, pMT123, was kindly provided by Dr D.Bohmann (European Molecular Biology Laboratory, Germany). The FLAG-tagged dioxin receptor expression plasmid, pCMV/DR/FLAG, was obtained from Dr J.McGuire (Karolinska Institutet, Sweden).
Cell culture and transient transfection experiments
COS7 and 293 cells (obtained from ATCC) were routinely maintained in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum plus penicillin (50 IU/ml) and streptomycin (50 µg/ml). For analysis of protein expression, we transiently transfected using the FuGene6 (Boehringer Mannheim) reagent wild-type or mutant forms of FLAG-tagged HIF-1α (1 µg) and VHL (2 µg) into COS7 cells in 6-cm diameter plastic dishes. After 12 h of incubation, cells were treated for 12 h at hypoxia (1% O2) or normoxia (21% O2). GAL4 reporter gene assays were performed as described using a β-galactosidase expression plasmid as an internal control (Carrero et al., 2000). After 6 h of transfection, cells were incubated for 36 h under hypoxic or normoxic conditions prior to analysis of reporter gene activity.
Immunoblotting and immunoprecipitation assays
For the detection of HIF-1α or VHL fusion protein expression whole-cell extracts were prepared as previously described (Kallio et al., 1997). Nuclear and cytosolic extracts were prepared as previously described (Gradin et al., 1996). Fifty micrograms of protein were blotted onto nitrocellulose filters following SDS–PAGE. Anti-FLAG M2 (Kodak), anti-VHL (PharMingen) or anti-Arnt (Kallio et al., 1997) antibodies were used as primary antibodies, diluted 1:500 in TBS containing 0.1% Tween-20 (TBS-T) and 1% non-fat milk for 1 h. After several washes, a 1:1000 dilution of anti-mouse Ig-horseradish peroxidase conjugate (Amersham Life Science) in TBS-T buffer containing 1% non-fat milk was used as a secondary antibody and incubated with the sample for 1 h at room temperature. After extensive washing with TBS-T buffer, immunocomplexes were visualized using enhanced chemiluminescence (Amersham Pharmacia Biotech).
For in vivo immunoprecipitation experiments, we transiently expressed wild-type or mutant forms of FLAG-tagged or GAL4-fused HIF-1α, VHL and HA-tagged ubiquitin or Arnt (4 µg each) in COS7 cells in 10-cm diameter plastic dishes. After 12 h of transfection, cells were treated for 1, 3 or 6 h under hypoxic or normoxic conditions before harvesting the cells. The cell pellet was resuspended in 200 µl of whole-cell extract buffer (Kallio et al., 1997) supplemented with 20 µM N-ethylmaleimide, followed by centrifugation for 30 min at 14 000 r.p.m. For detection of co-immunoprecipitated proteins, cells were treated with 5 µM MG-132 (Calbiochem) for 6 h prior to harvest. Subsequent to protein extraction, 900 µg of total cell proteins were incubated with anti-FLAG antibodies at room temperature for 1 h. Thirty microliters of a 50% slurry of protein G–Sepharose in TEG buffer (20 mM Tris–HCl pH 7.4, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol), containing 150 mM NaCl and 0.1% Triton X-100, were then added to the reaction mixtures and incubated for 12 h at 4°C under rotation. After rapid centrifugation, the resulting Sepharose pellets were washed three times with supplemented TEG buffer, and co-immunoprecipitated proteins were analyzed by SDS–PAGE followed by immunoblotting using anti-HA (Santa Cruz Biotechnology), anti-VHL or anti-Arnt antibodies diluted 1:500 in TBS-T and 1% non-fat milk.
In in vitro immunoprecipitation assays, GAL4 and FLAG fusion proteins containing full-length or various deletion mutants of HIF-1α, and/or wild-type or mutant VHL proteins were translated either in the presence or absence of [35S]methionine in rabbit reticulocyte lysate (Promega). Labeled translation products were separated by SDS–PAGE and analysed by phosphoimager (Fuji) for calculation of the protein concentration on the basis of incorporated [35S]methionine. Immunoprecipitation experiments were performed as previously described (Gradin et al., 1996) using GAL4 (Upstate Biotech) or FLAG antibodies or pre-immune rabbit serum, and precipitated proteins were analyzed by SDS–PAGE followed by autoradiography.
Immunostaining of VHL and visualization of intracellular trafficking of GFP-tagged proteins in living cells
COS7 cells grown on cover slips were transiently transfected with VHL expression plasmid and immunostained, and analyzed as described (Kallio et al., 1998). COS7 cells were transiently transfected with GFP-tagged protein expression plasmids, and after 24 h of expression the cells were incubated under either normoxic or hypoxic conditions for 6 h. Subcellular distribution of fluorescence activity was examined using a Zeiss Axiovert 135 microscope with an FITC-filter set, and epifluorescence with illumination from a Gixenon burner (Carl Zeiss Jena GmbH, Jena, Germany).
Acknowledgments
Acknowledgements
We thank Dr Joan W.Conaway for kindly providing wild-type and mutant VHL constructs, and Dr Dirk Bohmann for supplying the HA-tagged ubiquitin expression vector. This study was supported by grants from the Swedish Medical Research Council, the JSPS Research Fellowships for Young Scientists (to K.T.), the Swedish Cancer Society (to Y.M.) and the Fundacão para a Ciencia e a Technologia (to T.P.).
References
- Arany Z., Huang,L.E., Eckner,R., Bhattacharya,S., Jiang,C., Goldberg,M.A., Bunn,H.F. and Livingston,D.M. (1996) An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl Acad. Sci. USA, 93, 12969–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero P., Okamoto,K., Coumailleau,P., O’Brien,S., Tanaka,H. and Poellinger,L. (2000) Redox-regulated recruitment of the transcriptional coactivators CBP and SRC-1 to the hypoxia-inducible factor-1α. Mol. Cell. Biol., 20, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. (1998) The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J., 17, 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M., Hirota,K., Mimura,J., Abe,H., Yodoi,J., Sogawa,K., Poellinger,L. and Fujii-Kuriyama,Y. (1999) Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J., 18, 1905–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra J.R., Zhou,S., Merrill,M.J., Wagner,J.R., Krumm,A., Papavassiliou,E., Oldfield,E.H., Klausner,R.D. and Linehan,W.M. (1996) Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl Acad. Sci. USA, 93, 10589–10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradin K., McGuire,J., Wenger,R.H., Kvietikova,I., Whitelaw,M.L., Toftgård,R., Tora,L., Gassmann,M. and Poellinger,L. (1996) Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol. Cell. Biol., 16, 5221–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Huang L.E., Gu,J., Schau,M. and Bunn,H.F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl Acad. Sci. USA, 95, 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos O., Levy,A.P., Jiang,C., Kaelin,W.G.,Jr and Goldberg,M.A. (1996) Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl Acad. Sci. USA, 93, 10595–10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K., Yamanaka,K., Kamura,T., Minato,N., Conaway,R.C., Conaway,J.W., Klausner,R.D. and Pause,A. (1999) Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA, 96, 12436–12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer N.V. et al. (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev., 12, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.H., Zheng,J.Z., Leung,S.W., Roe,R. and Semenza,G.L. (1997) Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem., 272, 19253–19260. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. Jr and Maher,E.R. (1998) The VHL tumour-suppressor gene paradigm. Trends Genet., 14, 423–426. [DOI] [PubMed] [Google Scholar]
- Kallio P.J., Pongratz,I., Gradin,K., McGuire,J. and Poellinger,L. (1997) Activation of hypoxia-inducible factor 1α: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc. Natl Acad. Sci. USA, 94, 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio P.J., Okamoto,K., O’Brien,S., Carrero,P., Makino,Y., Tanaka,H. and Poellinger,L. (1998) Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J., 17, 6573–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio P.J., Wilson,W.J., O’Brien,S., Makino,Y. and Poellinger,L. (1999) Regulation of the hypoxia-inducible transcription factor 1α by the ubiquitin-proteasome pathway. J. Biol. Chem., 274, 6519–6525. [DOI] [PubMed] [Google Scholar]
- Laney J.D. and Hochstrasser,M. (1999) Substrate targeting in the ubiquitin system. Cell, 97, 427–430. [DOI] [PubMed] [Google Scholar]
- Lee S., Neumann,M., Stearman,R., Stauber,R., Pause,A., Pavlakis,G.N. and Klausner,R.D. (1999) Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol., 19, 1486–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J., Imbert,G., Wirbelauer,C., Gstaiger,M. and Krek,W. (1999) The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev., 13, 1822–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan K.M., Iliopoulos,O., Ohh,M., Kamura,T., Conaway,R.C., Conaway,J.W. and Kaelin,W.G.,Jr (1998) Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol., 18, 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.H. et al. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–275. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Knebelmann,B., Cohen,H.T., Ananth,S. and Sukhatme,V.P. (1997) The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol. Cell. Biol., 17, 5629–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh C.W., O’Rourke,J.F., Nagao,M., Gleadle,J.M. and Ratcliffe,P.J. (1997) Activation of hypoxia-inducible factor-1; definition of regulatory domains within the α subunit. J. Biol. Chem., 272, 11205–11214. [DOI] [PubMed] [Google Scholar]
- Salceda S. and Caro,J. (1997) Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem., 272, 22642–22647. [DOI] [PubMed] [Google Scholar]
- Srinivas V., Zhang,L.P., Zhu,X.H. and Caro,J. (1999) Characterization of an oxygen/redox-dependent degradation domain of hypoxia-inducible factor 1α (HIF-1α) proteins. Biochem. Biophys. Res. Commun., 260, 557–561. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Kaelin,W.G.,Jr and Pavletich,N.P. (1999) Structure of the VHL–ElonginC–ElonginB complex: implications for VHL tumor suppressor function. Science, 284, 455–461. [DOI] [PubMed] [Google Scholar]
- Taylor B.L. and Zhulin,I.B. (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev., 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash-Bingham C.A. and Tartof,K.D. (1999) aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J. Natl Cancer Inst., 91, 143–151. [DOI] [PubMed] [Google Scholar]
- Wang G.L., Jiang,B.H., Rue,E.A. and Semenza,G.L. (1995) Hypoxia-inducible factor 1 is a basic-helix–loop–helix–PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA, 92, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger R.H. and Gassmann,M. (1997) Oxygen(es) and the hypoxia-inducible factor-1. Biol. Chem., 378, 609–616. [PubMed] [Google Scholar]