Abstract
Neuraminidase inhibitors are agents used against influenza viruses; however, the emergence of drug-resistant strains is a major concern. Recently, the prevalence of oseltamivir-resistant seasonal influenza A (H1N1) virus increased globally and the emergence of oseltamivir-resistant pandemic influenza A (H1N1) 2009 viruses was reported. In this study, we developed a cycling probe real-time PCR method for the detection of oseltamivir-resistant seasonal influenza A (H1N1) and pandemic influenza A (H1N1) 2009 viruses. We designed two sets of primers and probes that were labeled with 6-carboxyfluorescein or 6-carboxy-X-rhodamine to identify single nucleotide polymorphisms (SNPs) that correspond to a histidine and a tyrosine at position 275 in the neuraminidase protein, respectively. These SNPs confer susceptibility and resistance to oseltamivir, respectively. In the 2007-2008 season, the prevalence of oseltamivir-resistant H1N1 viruses was 0% (0/72), but in the 2008-2009 season, it increased to 100% (282/282). In the 2009-2010 season, all of the pandemic influenza A (H1N1) 2009 viruses were susceptible to oseltamivir (0/73, 0%). This method is sensitive and specific for the screening of oseltamivir-resistant influenza A (H1N1) viruses. This method is applicable to routine laboratory-based monitoring of drug resistance and patient management during antiviral therapy.
The neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir are currently the antiviral drugs of choice for treatment and prophylaxis of influenza virus infections. NAIs prevent the release and spread of progeny virions from infected cells (16). A major concern is the emergence of drug-resistant strains during antiviral therapy. Oseltamivir-resistant viruses possessed a histidine-to-tyrosine amino acid substitution at position 275 in type N1 NA protein (His274Tyr in N2 numbering). This mutation was initially detected in patients who were infected with seasonal influenza A (H1N1) viruses after oseltamivir treatment (10). The prevalence of oseltamivir resistance was low in the 2007-2008 season, but a sudden increase was reported in the following season, when the His275Tyr mutants spread globally and were the predominant strain among seasonal H1N1 viruses (23).
In the spring of 2009, pandemic influenza A (H1N1) 2009 virus (H1N1pdm) emerged and circulated worldwide (4). Initial reports showed that all H1N1pdm viruses were sensitive to neuraminidase inhibitors, and recently, so far only 298 cases of oseltamivir-resistant H1N1pdm viruses possessing the His275Tyr mutation were reported by the Centers for Disease Control and Prevention and the World Health Organization (2, 3, 24). The majority of His275Tyr mutations in H1N1pdm viruses were detected after therapeutic or preventive administration of oseltamivir. Although the proportion of oseltamivir-resistant H1N1pdm viruses is low at the moment, continued monitoring for oseltamivir-resistant viruses is important because of the possibility that the prevalence of these resistant strains may increase, which happened among the contemporary seasonal H1N1 viruses (1, 20, 23).
Various high-throughput methods used in detecting the His275Tyr mutation among oseltamivir-resistant H1N1pdm viruses include pyrosequencing (7, 25), real-time PCR method using a TaqMan probe, and the rolling circle amplification (RCA) technology (12, 21, 22). Cycling probe real-time PCR is an alternative method that employs a sequence-specific chimeric probe in detecting single nucleotide polymorphisms (SNPs) (19). We previously applied this method to identify amantadine-resistant seasonal influenza A (H1N1) and A (H3N2) viruses with the Ser31Asn mutation in the M2 channel protein (19). We showed rapid detection of the Ser31Asn mutation from nasopharyngeal swabs in several hours by this method and demonstrated its high sensitivity and specificity, which are comparable to those of the gene sequencing method. In the study described in this report, we designed new sets of primers and probes to identify the His275Tyr mutation in NA which confers oseltamivir resistance, and we investigated the prevalence of the His275Tyr mutation among seasonal H1N1 viruses from the 2007-2008 and the 2008-2009 seasons and H1N1pdm viruses from the 2009-2010 season in Niigata, Japan.
MATERIALS AND METHODS
Sample collection and virus isolation.
Nasopharyngeal swab specimens were collected from patients with influenza-like illness who visited a pediatric clinic in Niigata City, Japan, during three influenza seasons (2007-2008 season from January to March in 2008, the 2008-2009 season from January to March in 2009, and the 2009-2010 season in November and December in 2009). Samples were taken after a written informed consent was obtained. None of the patients had received anti-influenza virus drugs before samples were taken. The nasopharyngeal swabs were suspended in viral transport medium and kept at 4°C until transportation to the Division of Public Health, Department of Infectious Disease Control and International Medicine, Niigata University, within 1 week. Initial isolation of influenza viruses was performed using Madin-Darby canine kidney (MDCK) cells. One hundred-microliter aliquots of the supernatants of the nasopharyngeal swabs were inoculated onto MDCK cells, and the cells were then incubated at 34°C with 5% CO2 until a specific cytopathic effect was detected. Influenza virus isolates were typed and subtyped by hemagglutination inhibition assay using guinea pig red blood cells and commercially available influenza vaccine strain antisera (Denka Seiken Co., Ltd., Tokyo, Japan).
RNA extraction and reverse transcription.
Viral RNA was extracted from 100 μl of supernatants of nasopharyngeal swabs or virus culture supernatant using an Extragen II kit (Kainos, Tokyo, Japan), according to the manufacturer's instructions. Reverse transcription was performed using influenza A universal primer Uni12, as reported elsewhere (13). Preparation of RNA from other respiratory viruses was performed using random primers (Invitrogen Corp., Carlsbad, CA) (17).
Primers, probes, and PCR conditions.
Two PCR primer pairs were designed to amplify specifically the NA gene of seasonal H1N1 and H1N1pdm viruses (Table 1). Cycling probes for seasonal H1N1 viruses, sH1N1-His275 and sH1N1-Tyr275, were synthesized to detect the SNP at codon ATG/A, which corresponds to CAT (oseltamivir-sensitive His275 genotype) and TAT (oseltamivir-resistant Tyr275 genotype) in the reverse complement (TaKaRa Bio Inc., Japan) (Table 1). Likewise, the cycling probes for pandemic H1N1 viruses, H1N1pdm-His275 and H1N1pdm-Tyr275, were synthesized to detect the SNPs CAC (oseltamivir-sensitive His275 genotype) and TAC (oseltamivir-resistant Tyr275 genotype) (TaKaRa Bio Inc.) (Table 1). The underlined nucleotides indicate the RNA replacement in the chimeric probes used in the real-time PCR. The probes for seasonal H1N1 virus, sH1N1-His275 and sH1N1-Tyr275, were designed in the reverse-complement direction, and the probes for pandemic H1N1 virus, H1N1 pdm-His275 and H1N1 pdm-Tyr275, were designed such that the nucleotide replaced in the RNA sequence is adjacent to the SNP. Cycling probes were labeled with either 6-carboxyfluorescein (FAM) or 6-carboxy-X-rhodamine (ROX), which can detect the oseltamivir-sensitive genotype and the oseltamivir-resistant genotype, respectively.
TABLE 1.
Primers and probes for cycling probe real-time PCR method
| Subtype | Primer or probe | Sequence (5′-3′) | Locationa |
|---|---|---|---|
| Seasonal influenza A (H1N1) virus | sH1N1-His275Tyr forward primer | 5′-CAAGATCGAAAAGGGGAAG-3′ | 768-786 |
| sH1N1-His275Tyr reverse primer | 5′-GACACCCAAGGTCGATTTG-3′ | 896-914 | |
| sH1N1-His275b | 5′-(Eclipsec)-[ATG]dAAAATTGGGTG-(FAMe)-3′ | 812-825 | |
| sH1N1-Tyr275b | 5′-(Eclipse)-[ATA]AAAATTGGGTG-(ROXe)-3′ | 812-825 | |
| Influenza A pandemic (H1N1) 2009 | H1N1pdm-His275Tyr forward primer | 5′-TGGACAGGCCTCATACAAGA-3′ | 744-763 |
| H1N1pdm-His275Tyr reverse primer | 5′-GCCAGTTATCCCTGCACACA-3′ | 870-889 | |
| H1N1pdm-His275b | 5′-(Eclipse)-CCTAATTAT[CAC]T-(FAM)-3′ | 814-826 | |
| H1N1pdm-Tyr275b | 5′-(Eclipse)-AT[TAC]TATGAGGA-(ROX)-3′ | 821-833 |
Location of primers and probes in the NA-coding region (total, 1,413 bp), segment 6, of influenza A (H1N1) virus. Note that both cycling probes for seasonal H1N1 were designed as reverse complements.
Fluorescent dye and quencher-labeled DNA/RNA chimeric probe.
Quenching molecule.
Nucleotides inside brackets indicate the codon relevant to sequences for oseltamivir sensitivity (His) and resistance (Tyr). Boldface and italicized letters indicate the nucleotide replaced by RNA.
Fluorescent molecules.
Cycling probe real-time PCR was carried out using a CycleavePCRCore kit (TaKaRa Bio Inc.). Conditions of the PCR cycles were as follows: initial denaturation at 95°C for 10 s, followed by 40 cycles of denaturation at 95°C for 5 s, primer annealing at 55°C and 59°C for seasonal H1N1 and for H1N1pdm, respectively, for 10 s, and extension and subsequent detection of fluorescence at 72°C for 15 s. In each PCR run, one set of forward and reverse PCR primers and two (FAM- and ROX-labeled) cycling probes were used. Separate PCR runs are needed for seasonal H1N1 and H1N1pdm virus detection.
Human influenza A (H3N2) virus, influenza B virus, and other common human respiratory viruses, such as respiratory syncytial virus, parainfluenza virus, enterovirus, rhinovirus, human metapneumovirus, and adenovirus, were tested with the same cycling probes and primer sets to examine whether cross-reactions occur by the assay. No animal influenza virus strains were tested. All influenza viruses and other viruses used in this study were collected and isolated at the Division of Public Health, Department of Infectious Disease Control and International Medicine, Niigata University, and the Department of Virology, Niigata Prefectural Institute of Public Health and Environmental Sciences.
Control plasmids.
Four positive-control plasmids harboring the NA gene insert from a seasonal H1N1 oseltamivir-sensitive strain (sH1N1-OS), a seasonal H1N1 oseltamivir-resistant isolate with the His275Tyr mutation (sH1N1-OR), an H1N1pdm oseltamivir-sensitive strain (H1N1pdm-OS), or an H1N1pdm oseltamivir-resistant virus with the His275Tyr mutation (H1N1pdm-OR) were constructed. NA gene fragments were amplified using the same PCR primers designed in this study. NA gene inserts were cloned using a Mighty TA-cloning kit (TaKaRa Bio Inc.), according to the manufacturer's instructions.
NAI susceptibility assay.
Drug susceptibility testing was performed by the 50% inhibitory concentration (IC50) method in order to validate the results of the cycling probe real-time PCR assay (1). The susceptibility to oseltamivir carboxylate (Roche Products, Ltd., Basel, Switzerland) and zanamivir (GlaxoSmithKline, Brentford, United Kingdom) was examined by a previously described fluorescence-based NA inhibition assay using methylumbelliferone N-acetylneuraminic acid (MUNANA) as the substrate (14).
DNA sequencing.
The sequences of selected samples and control viruses used in this study were determined using previously reported primers (5, 26). The NA sequences were edited and assembled using the DNAStar Lasergene 7 program (Bioinformatics Pioneer DNAStar, Inc., WI).
RESULTS
LOD of cycling probe method.
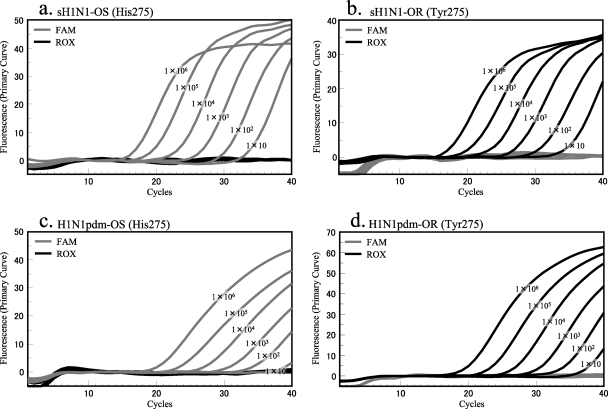
Control plasmids were used to determine the limit of detection (LOD) of each primer/probe set. All control plasmids were tested using a 10-fold dilution series from 1 × 101 to 1 × 107 copies (Fig. 1). The range of the threshold cycle (CT) values of 1 × 101 copies was from 35 to 39, and the range of CT values of 1 × 107 copies was from 15 to 17. The LOD for each of the four kinds of control plasmids was 10 copies.
FIG. 1.
Limit of detection of cycling probe real-time PCR with control plasmids. FAM fluorescence signals correspond to the oseltamivir-sensitive genotype (His275), and ROX fluorescence signals indicate the oseltamivir-resistant genotype (Tyr275). Control plasmids containing inserts of seasonal H1N1 sequences (sH1N1-OS and sH1N1-OR) reacted with probes sH1N1-His275 and sH1N1-Tyr275, respectively (a and b). Control plasmids harboring H1N1pdm sequences (H1N1pdm-OS and H1N1pdm-OR) reacted with H1N1pdm probes (c and d).
Specificity of cycling probe method.
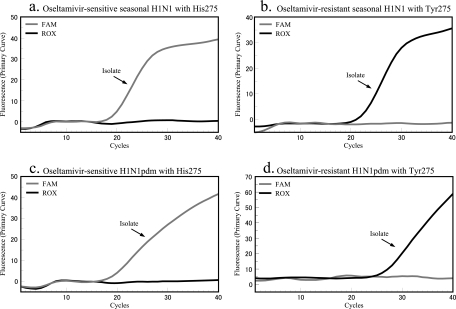
The specificity of the cycling probe real-time PCR assay was determined using previously characterized seasonal and pandemic H1N1 viruses. Using the seasonal H1N1 primer pair and probe set, all oseltamivir-sensitive seasonal H1N1 nasopharyngeal swabs and isolates tested positive, indicated by the presence of a FAM signal, and all oseltamivir-resistant seasonal H1N1 nasopharyngeal swabs and isolates tested positive, indicated by emission of a ROX fluorescent signal. Importantly, these probes did not show any cross-reactivity with oseltamivir-sensitive H1N1pdm or oseltamivir-resistant H1N1pdm samples (Fig. 2; Table 2). Likewise, when the pandemic H1N1 primers and probes were used, all oseltamivir-sensitive H1N1pdm samples yielded a corresponding FAM signal and all oseltamivir-resistant H1N1pdm samples gave a corresponding ROX signal. The pandemic H1N1 primers and probes did not exhibit cross-reactivity with seasonal H1N1 samples.
FIG. 2.
Detection of oseltamivir-sensitive and -resistant isolates with H275 and H275Y in NA gene of influenza A (H1N1) virus. Oseltamivir-sensitive and -resistant viruses of seasonal H1N1 virus reacted with the FAM probe and the ROX probe, respectively (a and b). Oseltamivir-sensitive and -resistant H1N1pdm viruses reacted with its specific corresponding probes (c and d).
TABLE 2.
Probe reaction performance with various virus samples
| Sample type | Virusa | Subtypeb | Susceptibility to oseltamivirc | No. of samples | No. of samples positive with: |
|||
|---|---|---|---|---|---|---|---|---|
| Seasonal H1N1 probe set |
H1N1pdm probe set |
|||||||
| sH1N1-His275 | sH1N1-Tyr275 | H1N1pdm-His275 | H1N1pdm-Tyr275 | |||||
| Isolate | Influenza A virus | Seasonal H1N1 | Sensitive | 10 | 10 | 0 | 0 | 0 |
| Influenza A virus | Seasonal H1N1 | Resistant | 10 | 0 | 10 | 0 | 0 | |
| Influenza A virus | H1N1pdm | Sensitive | 10 | 0 | 0 | 10 | 0 | |
| Influenza A virus | H1N1pdm | Resistant | 3 | 0 | 0 | 0 | 3 | |
| Influenza A virus | H3N2 | NAd | 10 | 0 | 0 | 0 | 0 | |
| Influenza B virus | NA | NA | 10 | 0 | 0 | 0 | 0 | |
| Respiratory syncytial virus | NA | NA | 5 | 0 | 0 | 0 | 0 | |
| Parainfluenza virus | NA | NA | 1 | 0 | 0 | 0 | 0 | |
| Enterovirus | NA | NA | 1 | 0 | 0 | 0 | 0 | |
| Rhinovirus | NA | NA | 2 | 0 | 0 | 0 | 0 | |
| Adenovirus | NA | NA | 1 | 0 | 0 | 0 | 0 | |
| Nasopharyngeal swab | Influenza A virus | Seasonal H1N1 | Sensitive | 15 | 15 | 0 | 0 | 0 |
| Influenza A virus | Seasonal H1N1 | Resistant | 15 | 0 | 15 | 0 | 0 | |
| Influenza A virus | H1N1pdm | Sensitive | 15 | 0 | 0 | 15 | 0 | |
| Influenza A virus | H3N2 | NA | 15 | 0 | 0 | 0 | 0 | |
| Influenza B virus | NA | NA | 10 | 0 | 0 | 0 | 0 | |
| Respiratory syncytial virus | NA | NA | 5 | 0 | 0 | 0 | 0 | |
| Human metapneumovirus virus | NA | NA | 5 | 0 | 0 | 0 | 0 | |
| Negative sample | NA | NA | 10 | 0 | 0 | 0 | 0 | |
Viruses were initially detected by virus isolation and PCR using specific primers.
Typed and subtyped by hemagglutinin inhibition assay with vaccine strain antisera and PCR using specific primers.
Resistant strains of both subtypes had a histidine-to-tyrosine change in residue 275 of the NA gene.
NA, not addressed.
The cycling probe method was tested on human influenza A (H3N2) and influenza B viruses and other common respiratory viruses. Results showed that none of these viruses tested positive using the same set of primers and probes (Table 2).
Validation of cycling probe method by NAI susceptibility assay.
The median IC50s of oseltamivir carboxylate for oseltamivir-sensitive seasonal H1N1 and H1N1pdm viruses were 2.34 ± 0.70 nM (n = 15) and 2.06 ± 0.99 nM (n = 22), respectively. Oseltamivir-resistant seasonal H1N1 and H1N1pdm viruses exhibited a 300- to 400-fold increase in IC50 (982.76 ± 421.47 nM, n = 24) compared to the IC50s of the oseltamivir-sensitive seasonal H1N1 and oseltamivir-sensitive H1N1pdm strains. For zanamivir, the median IC50s were 1.91 ± 0.60 nM, 1.10 ± 1.61 nM, and 0.99 ± 0.49 nM for oseltamivir-sensitive seasonal H1N1, oseltamivir-resistant seasonal H1N1, and oseltamivir-sensitive H1N1pdm viruses, respectively. None of the viruses demonstrated reduced susceptibility to zanamivir.
DNA sequencing.
Sequencing results were consistent with the findings from the cycling probe real-time PCR assay and NAI susceptibility test. All oseltamivir-resistant viruses had the His275Tyr mutation in the NA gene.
Prevalence of oseltamivir-resistant influenza viruses.
A total of 427 influenza A (H1N1) virus isolates that were collected during three epidemic seasons between January 2008 and December 2009 in Niigata in Japan were screened for the prevalence of the His275Tyr mutation that confers resistance to oseltamivir (Table 3). A nasopharyngeal swab specimen was collected from each patient during the individual's first visit to the medical facility, before any anti-influenza drug was administered. In the 2007-2008 influenza season, none of 72 (0%) seasonal H1N1 isolates were oseltamivir resistant; however, in the 2008-2009 season, all (282 of 282, 100%) of the seasonal H1N1 isolates were oseltamivir resistant. In the 2009-2010 season, seasonal H1N1 viruses were not detected and none of 73 (0%) H1N1pdm isolates were oseltamivir-resistant strains (Table 3).
TABLE 3.
Numbers of oseltamivir-sensitive and -resistant strains of influenza A/H1N1 viruses during 2007-2008, 2008-2009, and 2009-2010 seasons in Niigata
| Season | Virus subtype | No. of influenza A (H1N1) virus-positive samplesa | No. of oseltamivir-resistant virusesb | Proportion of oseltamivir-resistant viruses (%) |
|---|---|---|---|---|
| 2007-2008 | Seasonal influenza A (H1N1) | 72 | 0 | 0.0 |
| 2008-2009 | Seasonal influenza A (H1N1) | 282 | 282 | 100.0 |
| 2009-2010 | Seasonal influenza A (H1N1) | 0 | 0 | |
| Influenza A pandemic (H1N1) 2009 | 73 | 0 | 0.0 |
Samples were collected at a pediatric clinic in Niigata Prefecture in Japan.
Oseltamivir-resistant viruses with His275Tyr mutation in NA.
DISCUSSION
This study demonstrated the application of the cycling probe real-time PCR method in detecting the His275Tyr mutation in NA. This method correctly identified the oseltamivir-sensitive (His275) and oseltamivir-resistant (His275Tyr) genotypes of both seasonal and pandemic H1N1 viruses. We previously reported on a cycling probe real-time PCR method for detecting the Ser31Asn mutation in the M2 channel protein which confers resistance to amantadine (19). Our results suggest that the cycling probe real-time PCR method is applicable to detecting drug-resistant viruses by SNP genotyping.
This method showed high specificity in identifying the His275Tyr mutation in NA among human seasonal H1N1 and pandemic H1N1 viruses. The results of this assay were in agreement with the results of the IC50 method and gene sequencing. The mutation was detected in both nasopharyngeal swab samples and virus isolates, despite the difference in the virus concentration between the two types of samples. In addition, the method did not show any false-positive reactions with the other influenza A, influenza B, or other respiratory viruses. Thus, our method is very specific, and it is suitable for the detection of the His275Tyr mutation among human influenza A viruses. However, we could not perform this method on classical swine, triple-swine reassortant, or avian influenza viruses because we can handle only human influenza virus strains, as regulated by law. Although the sequence of the amplified NA gene segment in our cycling probe method showed variations compared to the sequences of nonhuman influenza viruses, further study is needed in order to evaluate the specificity of this assay with nonhuman influenza viruses.
Phenotypic assay, such as IC50 method, is the “gold standard” for identifying oseltamivir resistance. However, this method is time-consuming because it requires virus culture. Thus, several rapid detection methods were developed, including pyrosequencing, TaqMan probe real-time PCR assay, and RCA, for screening samples for the His275Tyr mutation, which confers resistance to oseltamivir (7, 12, 21, 22, 25). These methods showed high specificities and sensitivities in detecting the drug-resistant influenza virus. Of these methods, pyrosequencing is well-established and provides a definitive identification of the His275Tyr mutation, as well as other novel mutations that are associated with reduced drug susceptibility (6-8). However, not all laboratories can perform pyrosequencing as a routine assay for influenza virus surveillance because the machine and reagents are expensive and the procedures involved are complex. Thus, we developed the cycling probe real-time PCR assay as a low-cost alternative for screening for the His275Tyr mutation. This method has a high specificity and sensitivity in detecting SNPs which are comparable to those of the TaqMan and RCA methods. In addition, the probes that were used in this study can easily be synthesized by various manufacturers, and the cost of reagents is comparable to that of the reagents for the TaqMan method.
We utilized the cycling probe real-time PCR assay in determining the prevalence of the His275Tyr mutation among H1N1 viruses in three influenza seasons in Niigata, Japan. Our results showed that the prevalence of oseltamivir-resistant seasonal H1N1 strains had increased dramatically from the 2007-2008 season (0%) to the 2008-2009 season (100%), as reported in other studies (1, 15, 19, 20). In the 2009-2010 season, all of our H1N1pdm samples were oseltamivir sensitive. This result suggested that the oseltamivir-resistant H1N1pdm virus has not yet gained the genetic fitness to spread like the oseltamivir-resistant seasonal H1N1 viruses in the 2008-2009 season. It was observed that oseltamivir-resistant H1N1pdm viruses emerged after oseltamivir treatment and prophylaxis (2, 3, 24). One possible explanation as to why we were not able to detect oseltamivir-resistant H1N1pdm strains was that we collected samples from patients only prior to antiviral drug treatment. We are aware that during NAI treatment it is important to collect and examine time series samples from immunocompromised patients and from patients who manifested long-term clinical symptoms for monitoring for the emergence of drug-resistant viruses. Our method, which is capable of obtaining results within 3 h after receipt of nasopharyngeal swabs, is applicable in screening of clinical samples for resistance to oseltamivir during antiviral therapy.
One disadvantage of this method is that a new set of primers and probes has to be developed in the event that a novel drug-resistant strain would emerge during treatment with either oseltamivir, zanamivir, peramivir, or laninamivir (9, 11, 18). However, so far, all of the currently circulating oseltamivir-resistant seasonal H1N1 viruses had the His275Tyr mutation (1, 15, 19, 20), and the oseltamivir-resistant H1N1pdm viruses also possessed the same mutation (2, 3, 24). This mutation is very common among the contemporary oseltamivir-resistant viruses that belonged to the N1 group (1, 2, 3, 15, 19, 20, 24). Thus, should these viruses continue to persist in the future, the cycling probe real-time PCR assay can provide a fast, simple, and low-cost alternative for the laboratory-based surveillance of oseltamivir-resistant viruses.
In summary, we developed a highly sensitive and specific method of detecting the His275Tyr mutation in NA among seasonal H1N1 and H1N1pdm viruses by cycling probe real-time PCR assay. We clarified the prevalence of the His275Tyr mutation in three influenza seasons using this method. We demonstrated that the cycling probe method is applicable in monitoring of drug resistance as part of routine influenza virus surveillance work, and this method may provide information useful to clinicians during antiviral therapy.
Acknowledgments
We thank Junko Yamamoto and Kazuhide Okazawa of TaKaRa Bio Inc. for technical assistance in developing the cycling probe assay. We are grateful to Akinori Miyashita and Ryozo Kuwano in the Department of Molecular Genetics, Bioresource Science Branch, Center for Bioresources, Brain Research Institute, Niigata University, for utilization of the DNA sequencer. We thank Akemi Watanabe for technical assistance in virus isolation and Yoshiko Kato for intensive secretarial work.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Baranovich, T., et al. 2010. Emergence of H274Y oseltamivir-resistant A(H1N1) influenza viruses in Japan during the 2008-2009 season. J. Clin. Virol. 47:23-28. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2009. Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:969-972. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2009. Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients—Seattle, Washington, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:893-896. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2009. Update: novel influenza A (H1N1) virus infections—worldwide, May 6, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:453-458. [PubMed] [Google Scholar]
- 5.Dapat, C., et al. 2009. Epidemiology of human influenza A and B viruses in Myanmar from 2005 to 2007. Intervirology 52:310-320. [DOI] [PubMed] [Google Scholar]
- 6.Deyde, V., and L. Gubareva. 2009. Influenza genome analysis using pyrosequencing method: current applications for a moving target. Expert Rev. Mol. Diagn. 9:493-509. [DOI] [PubMed] [Google Scholar]
- 7.Deyde, V., et al. 2010. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob. Agents Chemother. 54:1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deyde, V., et al. 2007. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 196:249-257. [DOI] [PubMed] [Google Scholar]
- 9.Gubareva, L., L. Kaiser, and F. Hayden. 2000. Influenza virus neuraminidase inhibitors. Lancet 355:827-835. [DOI] [PubMed] [Google Scholar]
- 10.Gubareva, L., L. Kaiser, M. Matrosovich, Y. Soo-Hoo, and F. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 11.Hayden, F. 2009. Developing new antiviral agents for influenza treatment: what does the future hold? Clin. Infect. Dis. 48(Suppl. 1):S3-S13. [DOI] [PubMed] [Google Scholar]
- 12.Hindiyeh, M., et al. 2010. Rapid detection of influenza A pandemic (H1N1) 2009 virus neuraminidase resistance mutation H275Y by real-time reverse transcriptase PCR. J. Clin. Microbiol. 48:1884-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann, E., J. Stech, Y. Guan, R. Webster, and D. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 14.Hurt, A., I. Barr, G. Hartel, and A. Hampson. 2004. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res. 62:37-45. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami, C., et al. 2009. Isolation of oseltamivir-resistant influenza A/H1N1 virus of different origins in Yokohama City, Japan, during the 2007-2008 influenza season. Jpn. J. Infect. Dis. 62:83-86. [PubMed] [Google Scholar]
- 16.McKimm-Breschkin, J., et al. 2003. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 47:2264-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shobugawa, Y., et al. 2009. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J. Clin. Microbiol. 47:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugaya, N., and Y. Ohashi. 2010. Long-acting neuraminidase inhibitor laninamivir octanoate (CS-8958) versus oseltamivir as treatment for children with influenza virus infection. Antimicrob. Agents Chemother. 54:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki, Y., et al. 2010. Rapid and specific detection of amantadine-resistant influenza A viruses with a Ser31Asn mutation by the cycling probe method. J. Clin. Microbiol. 48:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ujike, M., et al. 2010. Oseltamivir-resistant influenza viruses A (H1N1) during 2007-2009 influenza seasons, Japan. Emerg. Infect. Dis. 16:926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Vries, E., et al. 2010. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J. Clin. Virol. 47:34-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, B., et al. 2010. Detection of the rapid emergence of the H275Y mutation associated with oseltamivir resistance in severe pandemic influenza virus A/H1N1 09 infections. Antiviral Res. 87:16-21. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization. 2009, posting date. Influenza A(H1N1) virus resistance to oseltamivir. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/influenza/oseltamivir_summary_south_2008/en /index.html. Accessed 13 January 2010.
- 24.World Health Organization. 2010, posting date. Pandemic (H1N1) 2009—update 105. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/don/2010_06_18/en/index.html. Accessed 24 June 2010.
- 25.World Health Organization. 2009, posting date. Protocol for antiviral susceptibility testing by pyrosequencing. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/resources/publications/swineflu/pyrosequencing_protocol/en/index.html. Accessed 27 May 2009.
- 26.World Health Organization. 2009, posting date. Sequencing primers and protocol. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf. Accessed 15 July 2009.