Abstract
The slow turnaround time for Mycobacterium tuberculosis drug susceptibility results is a barrier to care. We developed a rapid quantitative PCR (qPCR)-based phenotypic antimicrobial susceptibility test that utilizes amplification of the M. tuberculosis 16S rRNA gene after 3 days of incubation with antituberculosis drugs. To decrease background from killed organisms, we used propidium monoazide (PMA), a DNA-binding dye that penetrates damaged bacterial cells and renders DNA unamplifiable. M. tuberculosis was cultured in broth media containing PMA with or without drugs for 3 days prior to DNA extraction and real-time PCR amplification. 16S rRNA qPCR exhibited a significant decrease in threshold cycle (CT) time values (CT control − CT drug treated) with drug-susceptible strains compared with resistant strains. Susceptibility data were reported as ΔCT or as 2ΔCT and with appropriate cutoffs yielded an accuracy of 89 to 100% on 38 susceptible, multidrug-resistant, and extensively drug-resistant strains compared with conventional agar proportion susceptibility results for isoniazid, rifampin, ethambutol, streptomycin, amikacin, kanamycin, capreomycin, ofloxacin, moxifloxacin, ethionamide, para-aminosalicylic acid, linezolid, and cycloserine and compared with Bactec MGIT results for pyrazinamide. This PMA-qPCR assay is useful as a rapid 3-day first- and second-line drug susceptibility test for M. tuberculosis.
Diagnostics for drug-resistant tuberculosis (TB) have been identified by the World Health Organization as a key bottleneck in multidrug-resistant tuberculosis (MDR TB) control (20). Important gains have been made in rapid molecular diagnostics for isoniazid (INH) and/or rifampin (RIF) resistance, such as the commercially available INNO-LiPA Rif TB kit, the GenoType MTBDRplus assay, and the Cepheid MTB/RIF assay (4, 12). Clinically, these tests are helpful to quickly rule-in drug susceptibility. However, should MDR TB be detected, the clinician then immediately wants to know second-line drug susceptibilities as a basis for constructing a therapeutic regimen (9). However, there are limited diagnostic tools that address this need.
There are several drugs to treat MDR TB for which drug susceptibility testing (DST) is desired: pyrazinamide (PZA), ethambutol, levofloxacin, moxifloxacin, amikacin, capreomycin, streptomycin, kanamycin, ethionamide, para-aminosalicyclic acid, cycloserine, and linezolid. The standard DST method is culture based—either conventional agar proportion or liquid (1)—and completion requires 8 days to longer than a month, during which time the patient may be suboptimally treated. Furthermore, most laboratories only test a limited number of medications, and even among referral laboratories, there is inconsistency in the drugs offered for DST. Thus, isolates may be sent to several locations, augmenting delays.
Molecular methods for rapid second-line drug susceptibility testing are emerging and have promise (17). For instance, the GenoType MTBDRsl test offers rapid molecular information on the gyrA, rrs, and embB genes, which offers some predictive power for susceptibility to quinolones, aminoglycosides, and ethambutol, respectively (8, 13). However, these assays detect only the common mutations conferring resistance; therefore, their sensitivity is imperfect (5, 11). Other targets are emerging or unknown, such as those for para-aminosalicyclic acid, cycloserine, ethionamide, and linezolid (10, 16), and for some drugs, such as quinolones or linezolid, resistance may occur for reasons other than target mutation, such as efflux (7). The sum of these features limits molecular testing as a complete tool for second-line DST.
Finally, both molecular and conventional DST for TB yield a qualitative “susceptible” or “resistant” result for each drug, when in truth, bacterial resistance to antibiotics is a quantitative spectrum. Although typically MICs are not obtained, expert panels have recently advocated studying MIC benefits for TB (2). We agree with the desire for a quantitative metric of TB drug susceptibility, since as clinicians we face difficult and limited choices. In the setting of extensively drug-resistant (XDR) TB, we have few “susceptible” agents (19) and the choice becomes one of “least-resistant” drugs. Therefore, in this work we sought to develop a rapid quantitative and phenotypic DST assay that could accommodate any TB drug.
MATERIALS AND METHODS
Mycobacterial strains and culture conditions.
The TB strains used in this study were either ATCC strains or clinical isolates confirmed by Mycobacterium tuberculosis complex (MTBC) gene probe. These included H37Rv (ATCC 27294) and 37 clinical isolates, including 10 susceptible strains, 25 MDR TB strains, and 2 XDR TB strains obtained from the Mycobacteriology Service Unit, Department of Microbiology, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand. All work was approved by the University of Virginia Institutional Biosafety Committee and Human Investigation Committees. TB isolates were cultured on Lowenstein-Jensen medium at 35°C for 3 weeks. Cell suspensions were prepared in Middlebrook's 7H9 broth supplemented with Middlebrook OADC (oleic acid-albumin-dextrose-catalase) enrichment (Difco, Livonia, MI) and adjusted to a 0.5 McFarland standard for the quantitative PCR (qPCR) experiments and Bactec MGIT assay and a 1.0 McFarland standard for the agar proportion method.
Antimicrobial agents.
The drugs used were isoniazid, rifampin, streptomycin sulfate, kanamycin sulfate, ofloxacin, ethionamide, para-aminosalicylic acid, d-cycloserine (Sigma-Aldrich, St. Louis, MO), ethambutol hydrochloride, amikacin, capreomycin sulfate (MP Biomedicals, Solon, OH), moxifloxacin, linezolid (injection form; University of Virginia Pharmacy, Charlottesville, VA), and pyrazinamide (BD Diagnostic System, Sparks, MD). Isoniazid, ethambutol, amikacin, kanamycin, streptomycin, capreomycin, cycloserine, and pyrazinamide were dissolved in sterile distilled water. Rifampin and ethionamide were dissolved in dimethyl sulfoxide (DMSO). Ofloxacin was dissolved in 0.1 N NaOH, and p-aminosalicylic acid was dissolved in ethanol. All stock solutions were stored in single-use aliquots at −80°C.
Traditional agar proportion method.
Antimicrobial susceptibility tests were carried out in Middlebrook 7H10 (M7H10) agar (Difco) according to standard procedures. Briefly, a 1.0 McFarland suspension was diluted 10-fold serially in sterile distilled water, and dilutions of 10−2 and 10−4 were inoculated onto M7H10 agar with and without drug and incubated at 35°C. Critical concentrations established by WHO (21), were used and several concentrations were tested to establish the MIC, defined as the lowest concentration of drug that inhibited more than 99% of the bacterial population. Results were read 21 days after inoculation of media. At the critical concentration of each drug, the bacterial growth was measured and the percentage of resistance was calculated, whereby ≥1% is defined as a resistance. M. tuberculosis H37Rv, susceptible to all drugs tested, was used as an internal quality control.
Bactec MGIT method.
Antimicrobial susceptibility to pyrazinamide was carried out in Bactec MGIT 960 PZA medium using the Bactec MGIT 960 PZA kit (BD Diagnostic System, Sparks, MD) according to the manufacturer's procedure. Briefly, two 7-ml Bactec MGIT 960 PZA tubes were used for each isolate—one with PZA and one as a growth control. Results were read by detection of mycobacterial growth during a 7- to 21-day incubation with a UV transilluminator (Cole-Parmer, Vernon Hills, IL).
Optimization of PMA concentrations and treatment conditions.
M. tuberculosis H37Rv was used as a strain to optimize conditions. For heat killing, a 0.5 McFarland suspension was diluted 1:100 (≈1.5 × 105 CFU/ml) in M7H9 broth and 500-μl aliquots (≈7.5 × 104 cells) were heated to 80°C for 20 min. For antimicrobial killing, cells were diluted in M7H9 broth plus 10% OADC with drug and incubated at 35°C for 3 days. The viability of cells was confirmed by subculturing 10 μl on M7H10 agar at 35°C for 6 weeks. The basic procedure of PMA treatment (14) was adapted as follows. PMA (Biotium, Inc., Hayward, CA) was dissolved in 20% DMSO and added to M. tuberculosis cells in concentrations from 5 to 100 μM. PMA-treated-cells were incubated in the dark for 10 min, followed by placing the tubes on ice and exposing them to light from 650-W halogen lamps (Britek 8061; Linco, Inc., Santa Fe Springs, CA) at a 20-cm distance for 2 min. M. tuberculosis cells were then harvested by centrifugation at 18,000 × g for 15 min and washed in phosphate-buffered saline (PBS), and the pellets were subjected to DNA extraction. Timing of PMA was examined by adding to culture cells at the beginning of drug treatment (day 0) versus at the end of incubation (day 3).
DNA extraction.
PMA-treated cell pellets were resuspended in 200 μl nuclease-free water followed by boiling for 30 min and centrifugation at 18,000 × g for 3 min, with supernatant used for DNA template.
16S rRNA qPCR.
The primer MTB-F (5′-ACGGAAAGGTCTCTTCG-3′) and MTB-R (5′-CTTGGTAGGCCGTCAC-3′) (6) were used to amplify a 206-bp region within the 16S rRNA gene of M. tuberculosis complex (MTBC). PCR mixtures (25 μl) consisted of 12.5 μl of 2× iQSYBR green supermix (Bio-Rad, Hercules, CA), 0.25 μl of 50 μM forward primer, 0.25 μl of 50 μM reverse primer, 7 μl nuclease-free water, and 5 μl DNA template. Each set of samples included a nuclease-free water PCR control. PCR was performed on an iCycler (Bio-Rad, Hercules, CA) with initial denaturation at 95°C for 13.5 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 55°C for 20 s, and extension at 72°C for 20 s. Melting curve analysis was performed to confirm single amplicons.
Statistical analysis.
Means were compared by using the t test or Mann-Whitney test if data were nonparametric. All M. tuberculosis cultures were performed in duplicate. Data are shown as the mean or mean ± standard deviation (SD). The correlation between CT values (2ΔCT) and MICs or percentages of resistant colonies was calculated by Pearson correlation. Receiver-operating characteristic (ROC) analysis was performed with PASW Statistics software and was used to define a cutoff in the ΔCT and 2ΔCT values that was compared with agar proportion results as the “gold standard.” All P values were two tailed.
RESULTS
Development of the assay.
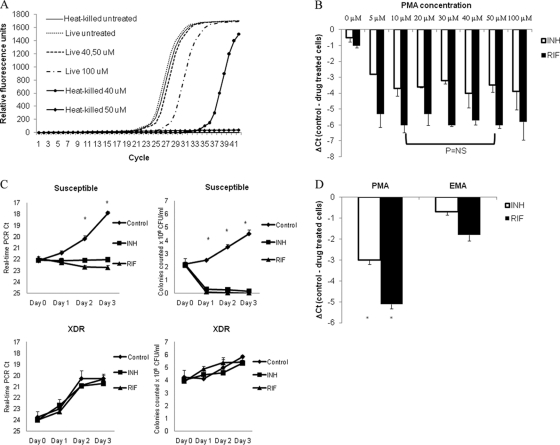
M. tuberculosis H37Rv was used to develop the protocol. We first performed qPCR of the 16S rRNA gene on M. tuberculosis cultures but found unacceptable background DNA in drug-treated cultures such that a difference in CT values between control and INH- or RIF-treated H37Rv could not be discerned (data not shown). We then evaluated RNA by reverse transcription (RT)-PCR, including the highly abundant 85B protein transcript, but again could not observe a difference in INH- or RIF-treated H37Rv cells (data not shown). We then used the DNA-binding chemical PMA which can penetrate dead or membrane-damaged cells and render cellular DNA unamplifiable (14). We tested the efficacy of several PMA concentrations on heat-killed cells, which indicated that PMA indeed decreased PCR amplification and increased the qPCR CT with heat-killed M. tuberculosis cells. Concentrations of 50 and 100 μM PMA completely inhibited PCR amplification (Fig. 1 A). At 100 μM PMA, the CT value of live cells was approximately 2 cycles higher than that of untreated cells (Fig. 1A), illustrating that excessive PMA can inhibit DNA amplification. Thus, 50 μM appeared to provide an optimal ΔCT for heat-killed cells.
FIG. 1.
Optimization of the PMA-PCR assay for TB drug susceptibility testing. The optimal PMA concentration was examined using cultured M. tuberculosis H37Rv, both heat killed (A) and antibiotic treated (B), followed by DNA extraction and real-time PCR. The data are shown as raw CT or ΔCT (CT control − CT drug), as indicated. (C) Real-time PCR CT was compared to colony counts as a function of incubation time over 1 to 3 days. The efficiency of PMA versus that of EMA was examined and compared by using drug-treated cells (D). *, P < 0.05.
Surprisingly, when we evaluated the ΔCT for INH- or RIF-treated cells, we found that 10 μM provided a statistically similar ΔCT for both drugs; thus, this lower concentration was pursued for subsequent studies (Fig. 1B). Addition of 10 μM PMA to INH- or RIF-treated M. tuberculosis cells did not completely inhibit real-time PCR: in fact, the CT remained in the 20s, even though all M. tuberculosis cells were killed (Fig. 1C), suggesting PMA did not completely penetrate all drug-treated M. tuberculosis cells. However, a discernible ΔCT of approximately 3 to 4 for INH and 4 to 6 for RIF was observed when H37Rv or other susceptible strains were used. In contrast, a drug-resistant strain exhibited little or no ΔCT after culture with INH or RIF, indicating little decrement in DNA replication (Fig. 1C). Of note, although we found statistically significant differences in CT after day 2 of culture, differences were more pronounced after 3 days of culture, and thus this time point was pursued. Next, we compared the similar compound ethidium monoazide (EMA) with propidium monoazide but found PMA offered statistically significantly greater ΔCT (Fig. 1D). Finally, we compared adding propidium monoazide for the duration of the culture versus adding it at the end of the 3-day culture and found the former was simpler to perform and yielded as good or better ΔCT (data not shown). Therefore, our final protocol utilized 10 μM PMA added at the beginning of 3-day culture prior to qPCR.
Evaluation of optimized PMA-qPCR assay with clinical M. tuberculosis isolates.
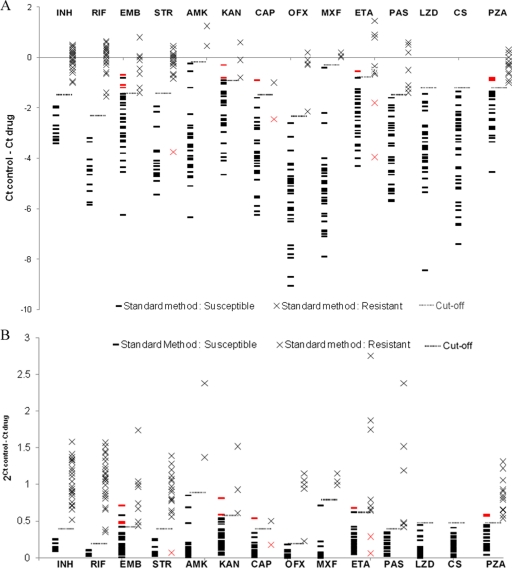
Thirty-eight M. tuberculosis isolates were tested for first- and second-line drug susceptibilities by the standard agar proportion method and MGIT method for pyrazinamide at the recommended critical concentrations (Table 1). These included 11 susceptible strains, 25 MDR TB strains, and 2 XDR TB strains, and 14 drugs. All strains were also subjected to the PMA-qPCR method in 7H9 broth at the standard liquid critical concentrations: therefore, for each isolate, a ΔCT (CT control − CT drug) was obtained for each drug. Figure 2 A shows the ΔCT values for all drugs on all strains and compares these results with the conventional agar proportion susceptibility results and MGIT results for pyrazinamide. This revealed that the average ΔCT for susceptible strains was significantly lower than that for resistant strains for each drug (P < 0.05). Indeed for some resistant strains, the ΔCT after drug treatment was above 0, indicating improved DNA replication in the presence of drug versus that in control media. Notably, the decrement in ΔCT in susceptible strains varied across drugs: for instance, it was relatively modest for INH and large for quinolones (−2.8 ± 0.5 versus −5.7 ± 1.6; P < 0.05). A cutoff ΔCT was ascribed by ROC analysis that yielded 89 to 100% accuracy versus agar proportion method and MGIT method results (Table 2), whereby a ΔCT above the cutoff indicated the strain was resistant, while a ΔCT below the cutoff indicated the strain was susceptible. There were 15 discrepant results (2.8% overall) where the PMA-qPCR assay differed from the agar proportion method: 4 false resistant for ethambutol, 1 false susceptible for streptomycin, 2 false resistant for kanamycin, 1 false susceptible and 1 false resistant for capreomycin, 2 false susceptible and 1 false resistant for ethionamide, and 3 false resistant for pyrazinamide.
TABLE 1.
Critical concentrations for susceptibility testing using agar proportion and PMA-qPCRa
| Antimicrobial agent | Critical concn (μg/ml) for susceptibility testing by: |
|
|---|---|---|
| M7H10 agar proportion (MIC range) | PMA-qPCR with M7H9 broth | |
| Isoniazid | 0.2 (0.03-32) | 0.1 |
| Rifampin | 1 (0.03-32) | 1 |
| Ethambutol | 5 (0.31-10) | 5 |
| Streptomycin | 2 (0.31-10) | 1 |
| Amikacin | 5 (0.31-10) | 1 |
| Kanamycin | 5 (0.31-10) | 1 |
| Capreomycin | 10 (0.31-10) | 2.5 |
| Ofloxacin | 2 (0.125-8) | 2 |
| Moxifloxacin | 2 (0.125-8) | 0.25 |
| Ethionamide | 5 (0.31-10) | 5 |
| p-Aminosalicylic acid | 2 (0.125-8) | 2 |
| Linezolid | 1 (0.125-8) | 1 |
| Cycloserine | 30 (0.47-30) | 30 |
| Pyrazinamideb | 100 (ND)c | 100 |
FIG. 2.
Correlation between PMA-qPCR results and standard agar proportion method and between PMA-qPCR results and the MGIT method for pyrazinamide. Thirty-eight M. tuberculosis isolates were tested by PMA-qPCR, agar proportion, and the Bactec MGIT assay. Shown is the relationship between standard method results versus ΔCT (A) and 2ΔCT (B). Cutoff threshold values were determined by ROC analysis. The red symbols (red × and red bars) indicate a standard assay result that was discrepant from PMA-qPCR. INH, isoniazid; RIF, rifampin; EMB, ethambutol; STR, streptomycin; AMK, amikacin; KAN, kanamycin; CAP, capreomycin; OFX, ofloxacin; MXF, moxifloxacin; ETA, thionamide; PAS, para-aminosalicylic acid; LZD, linezolid; CS, cycloserine; PZA, pyrazinamide.
TABLE 2.
Accuracy of PMA-qPCR compared with agar proportion
| Antimicrobial agent | PMA-qPCR |
No. of samples with result by agar proportion |
Accuracy (%) | |||
|---|---|---|---|---|---|---|
| Cutoff ΔCT | 2ΔCT | Result | Susceptible | Resistant | ||
| Isoniazid | −1.5 | 0.4 | Susceptible | 11 | 0 | 100 |
| Resistant | 0 | 27 | ||||
| Rifampin | −2.4 | 0.2 | Susceptible | 11 | 0 | 100 |
| Resistant | 0 | 27 | ||||
| Ethambutol | −1.4 | 0.4 | Susceptible | 25 | 0 | 89 |
| Resistant | 4 | 9 | ||||
| Streptomycin | −1.4 | 0.4 | Susceptible | 15 | 1 | 97 |
| Resistant | 0 | 22 | ||||
| Amikacin | −0.1 | 0.9 | Susceptible | 36 | 0 | 100 |
| Resistant | 0 | 2 | ||||
| Kanamycin | −0.9 | 0.6 | Susceptible | 33 | 0 | 95 |
| Resistant | 2 | 3 | ||||
| Capreomycin | −1.5 | 0.4 | Susceptible | 35 | 1 | 95 |
| Resistant | 1 | 1 | ||||
| Ofloxacin | −2.4 | 0.2 | Susceptible | 32 | 0 | 100 |
| Resistant | 0 | 6 | ||||
| Moxifloxacin | −0.2 | 0.8 | Susceptible | 34 | 0 | 100 |
| Resistant | 0 | 4 | ||||
| Ethionamide | −0.7 | 0.6 | Susceptible | 27 | 2 | 92 |
| Resistant | 1 | 8 | ||||
| p-Aminosalicylic acid | −1.5 | 0.4 | Susceptible | 31 | 0 | 100 |
| Resistant | 0 | 7 | ||||
| Linezolid | −1.2 | 0.5 | Susceptible | 38 | 0 | 100 |
| Resistant | 0 | 0 | ||||
| Cycloserine | −1.2 | 0.5 | Susceptible | 38 | 0 | 100 |
| Resistant | 0 | 0 | ||||
| Pyrazinamidea | −1.2 | 0.5 | Susceptible | 21 | 0 | 92 |
| Resistant | 3 | 14 | ||||
MGIT.
Relationship of PMA-qPCR and MIC values or percentage resistant.
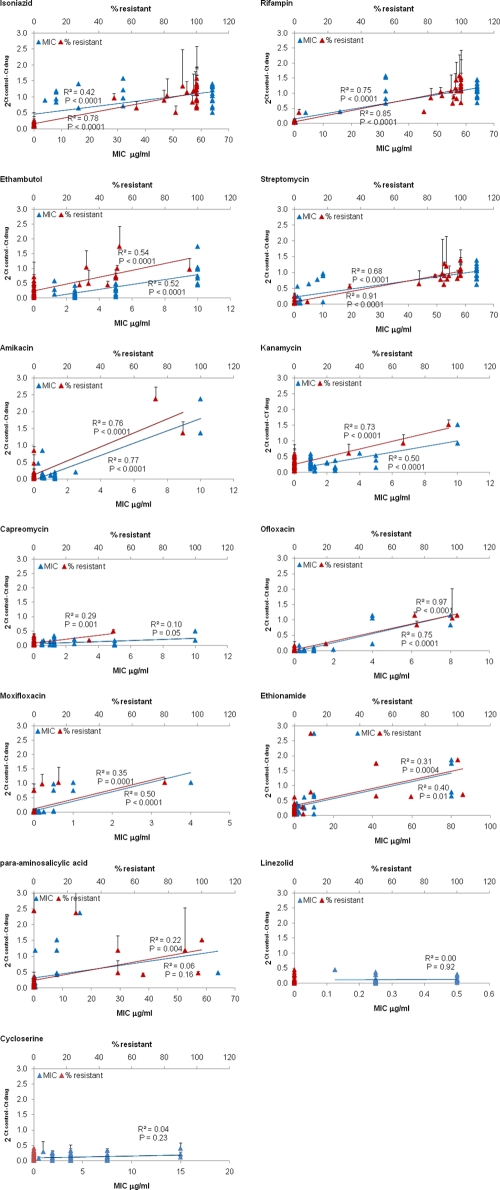
Since every unit decrease of real-time PCR CT represents a doubling in DNA, a potentially more accurate way to report the ΔCT (control − drug) data is as 2ΔCT, whereby a value of 1 represents no change in DNA between control and drug (20), values >1 reflect an increase in DNA with drug versus control, and values approaching 0 reflect decreases in DNA (e.g., 2−5 = 0.03). The 2ΔCT data versus agar proportion results are shown in Fig. 2B. We examined whether these 2ΔCT values thus offer a quantitative metric of drug susceptibility that mirrored conventional quantitative metrics of the percentage of resistant colonies at a critical concentration as well as the M. tuberculosis MIC value (Fig. 3). The 2ΔCT metric statistically correlated with both the percentage of resistant colonies and the MIC for each drug (P < 0.05), except capreomycin, para-aminosalicylic acid, linezolid, and cycloserine. The R2 values were on average higher for the 2ΔCT correlation with percentage of resistance than with the 2ΔCT correlation with MIC, but this difference was not statistically significant (0.61 ± 0.28 versus 0.46 ± 0.27; P = 0.09). The correlation with the MIC was highest for rifampin, ofloxacin, and amikacin (R2 ≥ 0.75). We could not analyze the 2ΔCT correlation with the percentage of resistance for linezolid and cycloserine because all isolates were susceptible, with 0% resistant colonies. Of note, the four false-susceptible PMA-qPCR results for streptomycin, capreomycin, and ethionamide were isolates with a low proportion of resistant colonies (7%, 34%, and 5%, respectively).
FIG. 3.
Correlation between PMA-qPCR (2ΔCT) and MIC or percentage of resistant colonies. Thirty-eight M. tuberculosis strains were tested by conventional agar proportion method critical concentration (where the percentages of resistant colonies were counted [upper x axis, red symbols]) and for MIC (lower x axis, blue symbols). All cultures were performed in duplicate, and therefore mean ± SD values are shown. The correlation of these values with 2ΔCT was tested for isoniazid, rifampin, ethambutol, streptomycin, amikacin, kanamycin, capreomycin, ofloxacin, moxifloxacin, ethionamide, para-aminosalicylic acid, linezolid, and cycloserine. Pyrazinamide MICs were not obtained. Best-fit lines and Pearson regression R2 values are shown.
DISCUSSION
We report a 3-day qPCR-based drug susceptibility test for tuberculosis that yielded highly accurate results compared with the conventional 21-day agar proportion method for all drugs evaluated (and the MGIT method for pyrazinamide). The protocol is inexpensive (costing ∼$2 in reagents per drug tested) and can serve as an important adjunct to confirm molecular susceptibility test results or as an early indicator of the final culture-based DST. We think the most useful setting for this assay is when MDR TB has been detected by molecular or first-line susceptibility testing and the clinician needs to know the results, quickly, of second-line susceptibility tests for the entire list of possibilities. Even under the most efficient circumstances, our experience in treating MDR TB in Virginia has shown that the time period between first-line results and complete second-line results can exceed 2 months. An early 3-day second-line drug panel would be highly valuable in this interim, and indeed this was the impetus for developing this test.
The levels of agreement between our assay and agar proportion were higher than that typically appreciated for new TB susceptibility tests (15), with an accuracy of 89 to 100% for each drug. There were only 15 instances in which the PMA-qPCR assay was discrepant from the conventional methods. The agreement may in part owe to the ability to tune the ΔCT or 2ΔCT cutoff to fit the agar proportion and MGIT results optimally. Also our lab had relatively resistant isolates in terms of the percentage of resistant colonies; therefore, the separation between susceptible and resistant may have been more pronounced. Prospective evaluation of the assay on a larger number of isolates is warranted, including isolates with a low percentage of resistant colonies, in order to ascertain the reliability of these cutoffs and whether they need to be adjusted. This process is now ongoing by our collaborators in Thailand and Tanzania. However, in the meantime, it appears secure to assume that an isolate after 3 days of drug treatment that exhibits a decrement in CT of 2 (ΔCT = −2, 2ΔCT = 0.25) can be considered susceptible since 99% (333/337) of isolates were susceptible, regardless of the drug. Likewise an isolate that exhibits an increase in CT (ΔCT > 0, 2ΔCT > 1) can safely be considered resistant, since 100% (62/62) were resistant by conventional means. The zone between a ΔCT of 0 and a ΔCT of −2 deserves more evaluation, but this was a minority of isolates overall (133/532 = 25%). Our assay was reproducible, however, in that the average standard deviation for all isolates was 0.11, including for the ΔCT of the zone from 0 to −2. Finally, we chose 3 days in an attempt to balance turnaround time with performance. We speculate that extending the assay to 4, 5, or 6 days would only enhance separation of susceptible and resistant ΔCT values.
The reliability of conventional DST for most second-line anti-XDR TB drugs and the appropriate critical concentrations are still questioned by the field. In this context, we think that a quantitative result could be valuable compared with the qualitative one of both conventional and molecular methods. For instance, it is possible that some isolates are clearly resistant, and some are clearly susceptible, and the difficulty lies with those that are intermediate. Such is standard antibiogram nomenclature for most bacteria, with cutoffs ascribed from the isolate's MIC. Remarkably, we found that the 2ΔCT statistically correlated with MIC for each drug.
We find it interesting that several isolates actually experienced more DNA replication in the setting of the drug than the control (ΔCT > 0, 2ΔCT > 1). This also has been observed with culture-based methods; in fact, some strains can grow preferentially amid supraphysiologic concentrations of drug (22). The finding may have profound clinical ramifications, in terms of deliberately avoiding particular drugs. However, by conventional reporting these strains will only be termed “resistant.”
We found that the decrements in ΔCT in susceptible isolates varied across drugs, being relatively modest for INH and large for quinolones. This may relate to the fact that quinolones inhibit DNA gyrase and thus halt DNA replication directly, while other antibiotics inhibit other cellular processes and thus DNA replication indirectly. Alternately, differences could be due to the effect of certain antibiotics on the cellular membrane and thus the efficiency of PMA permeability. Regardless of these interdrug differences, a particular ΔCT or 2ΔCT could be ascribed for each agent. We chose the housekeeping gene 16S rRNA since it is of high copy number and is the basis of many commercial assays. Since the assay is performed on pure M. tuberculosis culture, we did not need the encumbrance of the expense of an internal probe, and SYBR green was sufficient for measuring amplification, which was always of a single amplicon by melt curve analysis.
To our knowledge, this is the first report of PMA and PCR to determine antibiotic susceptibility for M. tuberculosis. One group evaluated ethidium monoazide with M. tuberculosis: however, this was in an assay that required a flow cytometer (18). The protocol is straightforward and amenable to high throughput and miniaturization. The assay can be performed by any TB lab with culture and real-time PCR capabilities. Another advantage is that the short incubation period should be accommodating to strains that grow poorly, a major problem for the conventional method. Finally, the short growth period also minimizes the concerns of drug stability during the weeks required for conventional assays.
There were limitations to this work. As mentioned, our resistant isolates were generally of high-level resistance; thus, the performance and cutoff values among low-level-resistant isolates are less clear. We had relatively few isolates resistant to moxifloxacin and other second-line drugs and none resistant to linezolid and cycloserine: therefore, additional testing is needed.
Acknowledgments
This work was supported by National Institutes of Health grant R41AI073072, the University of Virginia Department of Medicine and Center for Global Health, and the Virginia Department of Health.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Ardito, F., B. Posteraro, M. Sanguinetti, S. Zanetti, and G. Fadda. 2001. Evaluation of BACTEC Mycobacteria Growth Indicator Tube (MGIT 960) automated system for drug susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 39:4440-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Association of Public Health Laboratories. 2007. Proceedings of the 5th National Conference on Laboratory Aspects of Tuberculosis. Association of Public Health Laboratories, Silver Spring, MD.
- 3.Reference deleted.
- 4.Blakemore, R., et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brossier, F., N. Veziris, A. Aubry, V. Jarlier, and W. Sougakoff. 2010. Detection by GenoType MTBDRsl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 48:1683-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burggraf, S., et al. 2005. Comparison of an internally controlled, large-volume LightCycler assay for detection of Mycobacterium tuberculosis in clinical samples with the COBAS AMPLICOR assay. J. Clin. Microbiol. 43:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escribano, I., et al. 2007. Importance of the efflux pump systems in the resistance of Mycobacterium tuberculosis to fluoroquinolones and linezolid. Chemotherapy 53:397-401. [DOI] [PubMed] [Google Scholar]
- 8.Giannoni, F., et al. 2005. Evaluation of a new line probe assay for rapid identification of gyrA mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis J. Curry National Tuberculosis Center and California Department of Public Health. 2008. Drug-resistant tuberculosis: a survival guide for clinicians, 2nd ed. Francis J. Curry National Tuberculosis Center, San Francisco, CA.
- 10.Huang, W. L., H. Y. Chen, Y. M. Kuo, and R. Jou. 2009. Performance assessment of the GenoType MTBDRplus test and DNA sequencing in detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 47:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiet, V. S., et al. 2010. Evaluation of the MTBDRsl test for detection of second-line-drug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 48:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makinen, J., H. J. Marttila, M. Marjamaki, M. K. Viljanen, and H. Soini. 2006. Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 44:350-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nocker, A., C. Y. Cheung, and A. K. Camper. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310-320. [DOI] [PubMed] [Google Scholar]
- 15.Park, W. G., W. R. Bishai, R. E. Chaisson, and S. E. Dorman. 2002. Performance of the microscopic observation drug susceptibility assay in drug susceptibility testing for Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4750-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter, E., S. Rusch-Gerdes, and D. Hillemann. 2007. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 51:1534-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiguchi, J., et al. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45:179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soejima, T., K. Iida, T. Qin, H. Taniai, and S. Yoshida. 2009. Discrimination of live, anti-tuberculosis agent-injured, and dead Mycobacterium tuberculosis using flow cytometry. FEMS Microbiol. Lett. 294:74-81. [DOI] [PubMed] [Google Scholar]
- 19.Velayati, A. A., et al. 2009. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136:420-425. [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization. 2009. Key bottlenecks in M/XDR-TB control and patient care. World Health Organization, Geneva, Switzerland.
- 21.World Health Organization. 23 July 2007, posting date. Policy guidance on TB drug-susceptibility testing (DST) of second-line drugs. WHO/HTM/TB/2008. World Health Organization, Geneva, Switzerland. [PubMed]
- 22.Zhong, M., et al. 2010. An interesting case of rifampicin-dependent/-enhanced multidrug-resistant tuberculosis. Int. J. Tuber. Lung Dis. 14:40-44. [PubMed] [Google Scholar]