Abstract
Due to the rise in methicillin-resistant Staphylococcus aureus (MRSA) infections and widespread use of vancomycin, MRSA isolates with reduced susceptibility to vancomycin are emerging (i.e., MIC creep). However, the prevalence of heterogeneous vancomycin-intermediate S. aureus (hVISA) is unknown due to the difficulty in detecting this phenotype. Recently, Etest glycopeptide resistance detection (GRD) strips have been developed to detect hVISA. This study assessed vancomycin susceptibility in MRSA isolates and determined the prevalence of hVISA by Etest GRD and population analysis profile-area under the curve ratio (PAP-AUC). The genetic backgrounds of 167 MRSA isolates collected from 2000 to 2008 were identified by pulsed-field gel electrophoresis. Vancomycin MICs were determined using Etest and two broth microdilution assays, MicroScan and Sensititre. Etest GRD was performed on all isolates, and those exhibiting a hVISA phenotype were further tested by PAP-AUC. The vancomycin MIC modes remained consistent at 1 μg/ml, as assessed by Sensititre and MicroScan. Etest reported a significant increase (mode MIC = 1.5 μg/ml) in the MIC between 2000 and 2008 (P < 0.01); however, this increase did not reflect a ≥2-fold change. In addition, the slight MIC increase did not increase linearly from 2000 to 2008, suggesting biological fluctuation, and is inconsistent with the concept of MIC creep. Etest GRD identified six hVISA isolates, two of which were confirmed to be hVISA by PAP-AUC. In conclusion, reduced vancomycin susceptibility was not detected in our hospital over a 9-year period using three different MIC methodologies, and the hVISA incidence was 1.2%, as determined by Etest GRD and PAP-AUC.
Vancomycin remains the antibiotic of choice to treat many methicillin-resistant Staphylococcus aureus (MRSA) infections. However, due to the dramatic rise in MRSA infections and widespread use of vancomycin, which is known to have marginal tissue penetration and slow bactericidal activity, MRSA strains with reduced susceptibility to vancomycin are emerging (10, 30, 44, 46). Although most of these strains have a vancomycin MIC within the susceptible range, according to the Clinical and Laboratory Standards Institute (CLSI), some reports have shown a generalized increase in vancomycin MIC over time (also known as MIC creep) (38, 41-43, 52).
Associated with this issue is the presence of heterogeneous vancomycin-intermediate S. aureus (hVISA). These organisms are described as being susceptible to vancomycin but contain a subpopulation that possesses a thicker cell wall and expresses resistance to vancomycin (5, 6, 49). With these characteristics, hVISA is considered the precursor of vancomycin-intermediate S. aureus (51). Infections caused by hVISA are a growing concern in hospitals, resulting in prolonged bacteremia, endocarditis, and osteomyelitis and ultimately leading to vancomycin treatment failure (4, 9, 16, 37). Contributing to this problem of hVISA is the fact that current diagnostic methods often fail to detect this resistance phenotype, and additional screening is required to detect hVISA (15, 24). The “gold standard” for identifying hVISA is the population analysis profile-area under the curve ratio (PAP-AUC), but this method is time-consuming, labor-intensive, and costly for clinical laboratories. Additional methods have been used to detect hVISA, but these are not standardized and have low predictability in hVISA detection (14). A new modified Etest, called Etest glycopeptide resistant detection (GRD), has recently been developed and demonstrated to have high sensitivity and specificity in detecting hVISA (24, 55).
The purpose of this study was to determine if the MIC to vancomycin had increased significantly among clinical MRSA isolates that were collected over a 9-year period and to determine the prevalence of hVISA within this population using the Etest GRD methodology and vancomycin population analysis profile.
MATERIALS AND METHODS
Bacterial strains.
A random selection of 167 clinical MRSA bloodstream isolates collected from 2000 to 2008 at a 689-bed academic medical center was used in this study. All isolates were processed at the time of collection and stored at −80°C until they were tested. S. aureus Mu3 (ATCC 700698; hVISA) and a non-methicillin-resistant S. aureus strain (ATCC 29213) were used as reference organisms for the antimicrobial susceptibility tests and PAP-AUC analysis. S. aureus strains NRS382 (USA100), NRS383 (USA200), NRS384 (USA300-0114), and NRS123 (USA400) were acquired from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA).
PFGE.
To determine the epidemiological relatedness of the MRSA isolates, pulsed-field gel electrophoresis (PFGE) was performed using SmaI DNA digestion as previously described (32). All gels were analyzed using BioNumerics software (Applied Maths, Austin, TX) and normalized with NCTC 8325 (19). Seventeen to 20 unique isolates, as assessed by PFGE, were selected from each year (2000 to 2008) for vancomycin susceptibility testing and evaluation for the presence of hVISA. Some isolates that were indistinguishable by PFGE and that appeared in multiple years were included in the analysis. S. aureus isolates NRS382, NRS383, NRS384, and NRS123 were used as standard controls for PFGE types USA100, USA200, USA300, and USA400, respectively.
Antimicrobial susceptibility tests.
Vancomycin MICs were measured and compared by Etest (AB bioMérieux, Solna, Sweden) and two broth microdilution tests, Sensititre (TREK Diagnostic Systems, Cleveland, OH) and MicroScan (Siemens Healthcare Diagnostics, Deerfield, IL). Each of the susceptibility tests utilized a 0.5 McFarland standard inoculum in sterile water and was performed according to the manufacturers' instructions. Results from the broth microdilution tests were read manually.
Etest GRD.
Etest GRD was performed on all isolates according to the manufacturer's instructions (AB bioMérieux). Briefly, a bacterial suspension of a 0.5 McFarland standard inoculum in sterile water was spread on a Mueller-Hinton agar plate with 5% sheep blood (Remel, Lenexa, KS). Next, a GRD strip consisting of a double-sided gradient with vancomycin and teicoplanin was applied to the plate. The plate was incubated at 37°C, and the elliptical zones of the Etest GRD strip were read at 24 and 48 h. Isolates defined to be positive for hVISA had a GRD value of ≥8 μg/ml for either vancomycin or teicoplanin and a standard vancomycin Etest MIC of <4 μg/ml. Two or more microcolonies present in the zone of inhibition at ≥8 μg/ml also were an indicator for a positive GRD test (55).
PAP-AUC.
PAP-AUC was done as described by Wootton et al. (53). Briefly, overnight cultures grown in Trypticase soy broth (BD Diagnostics, Sparks, MD) on an orbital shaker at 37°C and 250 rpm were serially diluted onto brain heart infusion agar (BD Diagnostics) plates containing 0, 0.5, 1, 1.5, 2, 4, and 8 μg/ml of vancomycin. The numbers of CFU were counted after 48 h of incubation at 37°C. The number of CFU/ml was plotted against the aforementioned vancomycin concentrations using GraphPad Prism software (San Diego, CA). The AUC was computed by the trapezoidal method for each test isolate and S. aureus Mu3. A ratio was calculated by dividing the AUC of the test isolate by the AUC of Mu3; a ratio of between ≥0.9 and <1.3 was considered positive for hVISA.
Statistical analysis.
Statistical software (SAS, version 9.2; SAS Institute, Inc., Cary, NC) was utilized to analyze the MICs over time for each antimicrobial susceptibility test and to compare the MICs among the different tests for each year. The summary statistics for the MICs from the different tests for each year were presented by the geometric mean values. For comparison, the MIC values were sorted into four categories: ≤0.75, 1, 1.5, and ≥ 2. The generalized linear mixed models (GLIMMIX) procedure was used to fit a multinomial model with fixed effects for the type of test, year, and the interaction of the two and a random effect for different bacterial isolates nested within each year. A P value of <0.05 was considered significant for all statistical tests.
RESULTS
PFGE profiles of MRSA isolates.
PFGE was utilized to identify the genetic backgrounds of 167 MRSA isolates, and the pulsed-field profiles were compared to those of standard USA100 to USA400 strains (see Fig. S1 in the supplemental material). Forty-four (26%), 1 (0.6%), 12 (7%), and 8 (5%) of the isolates had profiles that were within two band differences of those for USA100, USA200, USA300, and USA400, respectively. The remaining 102 (61%) isolates were distinguishable from the USA100 to USA400 strains. Fifty-four percent of the isolates appeared more than once between 2000 and 2008, including one USA100-like isolate that was present in 7 of the 9 years.
Vancomycin MICs from Sensititre, MicroScan, and Etest.
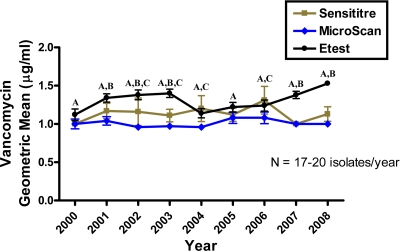
The vancomycin geometric mean MICs from the three different antimicrobial susceptibility tests ranged from 1.00 to 1.31 μg/ml for Sensititre, 0.96 to 1.08 μg/ml for MicroScan, and 1.12 to 1.53 μg/ml for Etest (Fig. 1). The Sensititre MIC results fluctuated the most from year to year, yet there were no significant differences between the years due to the large variability among the isolates from each year. MicroScan displayed the lowest MICs and remained stable throughout the study. When the MIC values from Etest were compared, there was a significant increase between 2000 and 2008 (P < 0.01); however, this was not a gradual increase over time (Fig. 1). There was a significant increase in the MIC between 2000 and 2001 (P = 0.04), but the MIC did not significantly change for the next 2 years. In 2003 and 2004, the MIC significantly dropped (P = 0.01) and did not rise again until 2007 (P = 0.02).
FIG. 1.
Vancomycin MICs of MRSA, displayed as geometric means ± standard errors of the means, were measured by Sensititre, MicroScan, and Etest for isolates recovered from 2000 to 2008. Etest reported significantly higher MICs than MicroScan (indicated by A) and Sensititre (indicated by B). Sensititre results were also significantly higher than MicroScan results for certain years (indicated by C) (P < 0.05).
The vancomycin MIC modes remained fixed throughout the study period, with overall modes of 1 μg/ml for Sensititre and MicroScan and 1.5 μg/ml for Etest (Table 1). Utilizing the Sensititre and MicroScan methodologies, 81% and 94% of the isolates, respectively, had a MIC of 1 μg/ml, while using Etest, 31% and 63% of the isolates had MICs of 1 μg/ml and 1.5 μg/ml, respectively. The overall frequency of MRSA isolates for which the MIC was ≥2 μg/ml varied from 17% by Sensititre to 4% by MicroScan and 2% by Etest. Etest results were significantly elevated compared with those of MicroScan for every year, and this was also true when the Etest MICs were compared to the Sensititre MICs for 5 of the 9 years (Fig. 1; P < 0.05). For 2002 to 2004 and 2006, the Sensititre results were statistically higher than the MicroScan results (Fig. 1; P < 0.05).
TABLE 1.
Annual frequencies and percentages for each test for vancomycin
| Yr (no. of isolates) and test | No. (%) of isolates for which vancomycin MIC (μg/ml) was: |
|||
|---|---|---|---|---|
| ≤0.75 | 1 | 1.5 | ≥2 | |
| 2000 (18) | ||||
| Sensititre | 1 (6) | 16 (88) | 1 (6) | |
| MicroScan | 1 (6) | 16 (88) | 1 (6) | |
| Etest | 4 (22) | 6 (33) | 8 (44) | 0 (0) |
| 2001 (18) | ||||
| Sensititre | 0 (0) | 14 (78) | 4 (22) | |
| MicroScan | 0 (0) | 17 (94) | 1 (6) | |
| Etest | 0 (0) | 5 (28) | 13 (72) | 0 (0) |
| 2002 (19) | ||||
| Sensititre | 2 (11) | 12 (63) | 5 (26) | |
| MicroScan | 1 (5) | 18 (95) | 0 (0) | |
| Etest | 1 (5) | 3 (16) | 14 (74) | 1 (5) |
| 2003 (20) | ||||
| Sensititre | 0 (0) | 17 (85) | 3 (15) | |
| MicroScan | 1 (5) | 19 (95) | 0 (0) | |
| Etest | 0 (0) | 4 (20) | 15 (75) | 1 (5) |
| 2004 (19) | ||||
| Sensititre | 0 (0) | 15 (79) | 4 (21) | |
| MicroScan | 1 (5) | 18 (95) | 0 (0) | |
| Etest | 1 (5) | 11 (58) | 7 (37) | 0 (0) |
| 2005 (19) | ||||
| Sensititre | 0 (0) | 16 (84) | 3 (16) | |
| MicroScan | 0 (0) | 17 (89) | 2 (11) | |
| Etest | 1 (5) | 8 (42) | 10 (53) | 0 (0) |
| 2006 (18) | ||||
| Sensititre | 0 (0) | 12 (67) | 6 (33) | |
| MicroScan | 0 (0) | 16 (89) | 2 (11) | |
| Etest | 0 (0) | 10 (56) | 7 (39) | 1 (5) |
| 2007 (19) | ||||
| Sensititre | 0 (0) | 19 (100) | 0 (0) | |
| MicroScan | 0 (0) | 19 (100) | 0 (0) | |
| Etest | 0 (0) | 4 (21) | 15 (79) | 0 (0) |
| 2008 (17) | ||||
| Sensititre | 0 (0) | 14 (82) | 3 (18) | |
| MicroScan | 0 (0) | 17 (100) | 0 (0) | |
| Etest | 0 (0) | 0 (0) | 16 (94) | 1 (6) |
| Total (167) | ||||
| Sensititre | 3 (2) | 135 (81) | 29 (17) | |
| MicroScan | 4 (2) | 157 (94) | 6 (4) | |
| Etest | 7 (4) | 51 (31) | 105 (63) | 4 (2) |
Etest GRD for detecting hVISA.
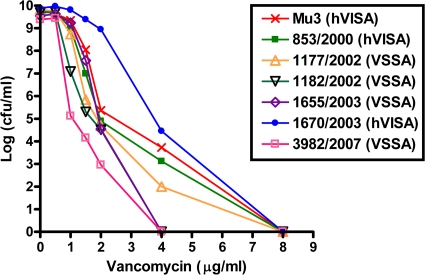
Of the 167 isolates tested by Etest GRD, 6 were positive for hVISA by having a teicoplanin MIC value of ≥8 μg/ml and a standard Etest vancomycin MIC of <4 μg/ml (Table 2). No consistent relationship existed for the year that the isolate was obtained or the clonal pattern and the presence of hVISA.
TABLE 2.
Isolates positive for hVISA by Etest GRD
| Isolate | Yr | Clonal pattern | GRD values (μg/ml) for VAN/TECa | Vancomycin MIC (μg/ml) by Sensititre, MicroScan, Etest | PAP-AUC result |
|---|---|---|---|---|---|
| 853 | 2000 | Not defined | 1/16 | 2, 1, 1.5 | hVISA |
| 1177 | 2002 | Not defined | 2/32 | 2, 1, 2 | VSSAb |
| 1182 | 2002 | USA400 | 1.5/24 | 1, 1, 1.5 | VSSA |
| 1655 | 2003 | USA100 | 1.5/8 | 1, 1, 1.5 | VSSA |
| 1670 | 2003 | Not defined | 2/12 | 2, 1, 2 | hVISA |
| 3982 | 2007 | Not defined | 1/≥32 | 1, 1, 1.5 | VSSA |
VAN, vancomycin; TEC, teicoplanin.
VSSA, vancomycin-susceptible S. aureus.
PAP-AUC for detecting hVISA.
PAP-AUC was performed to confirm the positive results from the GRD Etest, and two isolates were identified to be hVISA using this analysis (Fig. 2). The two hVISA isolates from 2000 and 2003 had PAP-AUC ratios of 0.91 and 1.23, respectively. Both hVISAs had vancomycin MICs of 1.5 to 2 μg/ml according to Sensititre and Etest, while MicroScan reported a 1-μg/ml MIC for both isolates (Table 2).
FIG. 2.
Population analysis profile curves of the six isolates that were positive for hVISA by Etest GRD. Two isolates were identified to be hVISA and four isolates were defined to be vancomycin-susceptible S. aureus (VSSA) compared to the susceptibility of the Mu3 reference strain.
DISCUSSION
Reduced susceptibility to vancomycin in S. aureus has been a major medical concern for over a decade. Several studies have reported elevated vancomycin MICs in MRSA isolates where the MICs are at the upper end of the susceptibility range (18, 27, 38, 40-43, 50, 52). However, not all medical institutions are reporting an increase in vancomycin MICs (1, 11, 20, 33). One study measured vancomycin MICs in MRSA isolates collected from 2002 to 2006 from nine different U.S. medical centers using a reference broth microdilution assay with a broad range of precise incremental dilutions (39). Their results showed that they did not detect a reduction in vancomycin susceptibility at any of the test sites, which are similar to our results using three separate antimicrobial susceptibility tests. Although the MIC by Etest did increase from 2000 to 2008, it fluctuated over time and was clinically insignificant to support a consistent decline in vancomycin susceptibility. More specifically, the majority of the isolates in this study had a vancomycin MIC of 1 or 1.5 μg/ml, and the difference between the two Etest MICs from 2000 and 2008 was <1.5-fold, whereas the other studies have detected a ≥1.5-fold MIC increase or a significant increase in the frequency of isolates having vancomycin MICs of ≥1 μg/ml (41, 43, 52). None of these trends were found in our study.
The conflicting results in the literature documenting elevated vancomycin MICs in MRSA may partly be due to failure of epidemiologically typing the MRSA isolates. Some MRSA clones have higher levels of dissemination than others and possess elevated vancomycin MICs, which may cause false perceptions of an overall reduction in vancomycin susceptibility for all strains of MRSA (8, 26, 31, 48). The purpose of our study was to assess vancomycin susceptibility in the population of clinically encountered MRSA clonal types and not to define the overall MRSA susceptibility profile for clinical purposes in the associated patient population. PFGE was performed to eliminate clonal strains within each year that could otherwise influence the results. This study identified a highly diverse sample of MRSA isolates. Twenty-six percent of the isolates were USA100-like, a common MRSA strain found in hospitals, whereas 7% and 5% were similar to the community-acquired strains USA300 and USA400, respectively.
The type of antimicrobial susceptibility test that is performed to measure the vancomycin MICs may also affect the assessment of whether susceptibility within the population has changed. In this study, three standard susceptibility tests that are used in the clinical setting were chosen to measure the MICs. Although elevated vancomycin MICs could not be demonstrated by the three different tests, there were differences among the assays. In agreement with a previous study, Sensititre panels had the highest variability (47). It is unclear why MicroScan reported the lowest vancomycin MICs. Other studies found MicroScan to report vancomycin MICs in MRSA similar to or higher than those that Etest and Sensititre report (17, 45, 47). Etest reported vancomycin MICs higher than those reported by the two broth microdilution assays. This was expected and has been described elsewhere (33, 36). Etest strips contain a gradient of vancomycin concentrations, including an intermediate reading of 1.5 μg/ml, as opposed to the broth microdilution assays, where the concentrations are in 2-fold increments. Most of the isolates had a MIC of 1.5 μg/ml, causing the Etest results to be more elevated than the results of the other two assays. Etest also measures the MIC for a larger proportion of bacteria at ∼108 CFU than the broth microdilution assays at ∼104 CFU, making Etest more sensitive in detecting resistant mutants and MIC increases (11, 33, 43).
In addition to assessing a reduction in vancomycin susceptibility, the prevalence of hVISA was determined by the newly developed Etest GRD. Two studies have verified that the Etest GRD has high sensitivity (93 to 94%) in detecting hVISA compared to PAP-AUC but variable specificity (82% and 95%) (24, 55). Nonetheless, the specificity of the Etest GRD is similar to or higher than the specificities of other screening tests, such as the Etest macrodilution method, the vancomycin screening plate method with brain heart infusion agar, and the teicoplanin screening plate method with Mueller-Hinton agar (14). According to the Etest GRD, 3.5% of the MRSA isolates in our sample were positive for hVISA. All of the positive isolates had teicoplanin MIC values ≥8 μg/ml. Teicoplanin is more sensitive in the detection of the hVISA phenotype than vancomycin (55). It was revealed by PAP-AUC that only two of the six isolates were confirmed to be hVISA. The small sample size precludes any conclusion regarding the disparity observed between Etest GRD and PAP-AUC but does suggest that additional study is warranted. The two hVISA isolates had vancomycin MICs of 1.5 to 2 μg/ml by Etest and Sensititre. Vancomycin MICs of >1 μg/ml have been associated with hVISA (2, 12, 33), but this is not a strong indicator for hVISA because Etest and Sensititre reported equally high MICs in the vancomycin-susceptible isolates. The incidence of hVISA in other hospitals has varied, ranging from 0 to 50% (3, 4, 7, 13, 21, 22, 23, 25, 28, 29, 34, 35, 50, 52, 54). The type of screening test used to detect hVISA and the number of isolates screened are responsible for this wide range. A standard screening test with high sensitivity and specificity for detecting hVISA needs to be implemented to better understand the prevalence and clinical impact of these organisms.
In conclusion, a decrease in vancomycin susceptibility in MRSA isolates from our institution over a 9-year period was not detected by three standard antimicrobial susceptibility tests. The vancomycin MICs differed among the three susceptibility tests, in which Etest resulted in the highest MICs and MicroScan reported the lowest MICs. Finally, the GRD Etest and PAP-AUC defined a low prevalence of hVISA at 1.2%.
Supplementary Material
Acknowledgments
We thank Valerie Shostrom for her assistance with the statistical analysis of the data. We also acknowledge Laura Orwe and Elsie Forsung for their help with performing PFGE analysis.
Footnotes
Published ahead of print on 20 October 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alós, J. I., A. García-Cañas, P. García-Hierro, and F. Rodríguez-Salvanés. 2008. Vancomycin MICs did not creep in Staphylococcus aureus isolates from 2002 to 2006 in a setting with low vancomycin usage. J. Antimicrob. Chemother. 62:773-775. [DOI] [PubMed] [Google Scholar]
- 2.Bae, I. G., J. J. Federspiel, J. M. Miró, C. W. Woods, L. Park, M. J. Rybak, T. H. Rude, S. Bradley, S. Bukovski, C. G. de la Maria, S. S. Kanj, T. M. Korman, F. Marco, D. R. Murdoch, P. Plesiat, M. Rodriguez-Creixems, P. Reinbott, L. Steed, P. Tattevin, M. F. Tripodi, K. L. Newton, G. R. Corey, and V. G. Fowler, Jr. 2009. Heterogeneous vancomycin-intermediate susceptibility phenotype in bloodstream methicillin-resistant Staphylococcus aureus isolates from an international cohort of patients with infective endocarditis: prevalence, genotype, and clinical significance. J. Infect. Dis. 200:1355-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benquan, W., T. Yingchun, Z. Kouxing, Z. Tiantuo, Z. Jiaxing, and T. Shuqing. 2002. Staphylococcus heterogeneously resistant to vancomycin in China and antimicrobial activities of imipenem and vancomycin in combination against it. J. Clin. Microbiol. 40:1109-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 5.Cui, L., A. Iwamoto, J. Q. Lian, H. M. Neoh, T. Maruyama, Y. Horikawa, and K. Hiramatsu. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado, A. J., T. Riordan, R. Lamichhane-Khadka, D. C. Winnett, J. Jimenez, K. Robinson, F. G. O'Brien, S. A. Cantore, and J. E. Gustafson. 2007. Hetero-vancomycin-intermediate methicillin-resistant Staphylococcus aureus isolate from a medical center in Las Cruces, New Mexico. J. Clin. Microbiol. 45:1325-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, and F. C. Tenover. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, R. L., and J. H. Jorgensen. 2008. Inhibitory activities of 11 antimicrobial agents and bactericidal activities of vancomycin and daptomycin against invasive methicillin-resistant Staphylococcus aureus isolates obtained from 1999 through 2006. Antimicrob. Agents Chemother. 52:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horne, K. C., B. P. Howden, E. A. Grabsch, M. Graham, P. B. Ward, S. Xie, B. C. Mayall, P. D. Johnson, and M. L. Grayson. 2009. Prospective comparison of the clinical impacts of heterogeneous vancomycin-intermediate methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-susceptible MRSA. Antimicrob. Agents Chemother. 53:3447-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howden, B. P. 2005. Recognition and management of infections caused by vancomycin-intermediate Staphylococcus aureus (VISA) and heterogenous VISA (hVISA). Intern. Med. J. 35:S136-S140. [DOI] [PubMed] [Google Scholar]
- 14.Howden, B. P., J. K. Davies, P. D. Johnson, T. P. Stinear, and M. L. Grayson. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howden, B. P., P. D. Johnson, P. B. Ward, T. P. Stinear, and J. K. Davies. 2006. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 50:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howden, B. P., P. B. Ward, P. G. Charles, T. M. Korman, A. Fuller, P. du Cros, E. A. Grabsch, S. A. Roberts, J. Robson, K. Read, N. Bak, J. Hurley, P. D. Johnson, A. J. Morris, B. C. Mayall, and M. L. Grayson. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521-528. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, D. I., L. K. Hidayat, R. Quist, J. Hindler, A. Karlsson, A. Yusof, and A. Wong-Beringer. 2008. Comparison of method-specific vancomycin minimum inhibitory concentration values and their predictability for treatment outcome of meticillin-resistant Staphylococcus aureus (MRSA) infections. Int. J. Antimicrob. Agents 32:378-385. [DOI] [PubMed] [Google Scholar]
- 18.Hussain, F. M., S. Boyle-Vavra, P. B. Shete, and R. S. Daum. 2002. Evidence for a continuum of decreased vancomycin susceptibility in unselected Staphylococcus aureus clinical isolates. J. Infect. Dis. 186:661-667. [DOI] [PubMed] [Google Scholar]
- 19.Iandolo, J. J., J. P. Bannantine, and G. C. Stewart. 1997. Genetic and physical map of the chromosome of Staphylococcus aureus, p. 39-53. In K. B. Crossley and G. L. Accher (ed.), The staphylococci in human disease. Churchill Livingstone, New York, NY.
- 20.Jones, R. N. 2006. Microbiological features of vancomycin in the 21st century: minimum inhibitory concentration creep, bactericidal/static activity, and applied breakpoints to predict clinical outcomes or detect resistant strains. Clin. Infect. Dis. 1:S13-S24. [DOI] [PubMed] [Google Scholar]
- 21.Khosrovaneh, A., K. Riederer, S. Saeed, M. S. Tabriz, A. R. Shah, M. M. Hanna, M. Sharma, L. B. Johnson, M. G. Fakih, and R. Khatib. 2004. Frequency of reduced vancomycin susceptibility and heterogeneous subpopulation in persistent or recurrent methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 38:1328-1330. [DOI] [PubMed] [Google Scholar]
- 22.Kim, H. B., W. B. Park, K. D. Lee, Y. J. Choi, S. W. Park, M. D. Oh, E. C. Kim, and K. W. Choe. 2003. Nationwide surveillance for Staphylococcus aureus with reduced susceptibility to vancomycin in Korea. J. Clin. Microbiol. 41:2279-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, M. N., S. H. Hwang, Y. J. Pyo, H. M. Mun, and C. H. Pai. 2002. Clonal spread of Staphylococcus aureus heterogeneously resistant to vancomycin in a university hospital in Korea. J. Clin. Microbiol. 40:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard, S. N., K. L. Rossi, K. L. Newton, and M. J. Rybak. 2009. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J. Antimicrob. Chemother. 63:489-492. [DOI] [PubMed] [Google Scholar]
- 25.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, C., C. J. Graber, M. Karr, B. A. Diep, L. Basuino, B. S. Schwartz, M. C. Enright, S. J. O'Hanlon, J. C. Thomas, F. Perdreau-Remington, S. Gordon, H. Gunthorpe, R. Jacobs, P. Jensen, G. Leoung, J. S. Rumack, and H. F. Chambers. 2008. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004-2005. Clin. Infect. Dis. 46:1637-1646. [DOI] [PubMed] [Google Scholar]
- 27.Lodise, T. P., J. Graves, A. Evans, E. Graffunder, M. Helmecke, B. M. Lomaestro, and K. Stellrecht. 2008. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 52:3315-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maor, Y., M. Hagin, N. Belausov, N. Keller, D. Ben-David, and G. Rahav. 2009. Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J. Infect. Dis. 199:619-624. [DOI] [PubMed] [Google Scholar]
- 29.Maor, Y., G. Rahav, N. Belausov, D. Ben-David, G. Smollan, and N. Keller. 2007. Prevalence and characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia in a tertiary care center. J. Clin. Microbiol. 45:1511-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maree, C. L., R. S. Daum, S. Boyle-Vavra, K. Matayoshi, and L. G. Miller. 2007. Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerg. Infect. Dis. 13:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musta, A. C., K. Riederer, S. Shemes, P. Chase, J. Jose, L. B. Johnson, and R. Khatib. 2009. Vancomycin MIC plus heteroresistance and outcome of methicillin-resistant Staphylococcus aureus bacteremia: trends over 11 years. J. Clin. Microbiol. 47:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunes, A. P., R. P. Schuenck, C. C. Bastos, M. M. Magnanini, J. B. Long, N. L. Iorio, and K. R. Santos. 2007. Heterogeneous resistance to vancomycin and teicoplanin among Staphylococcus spp. isolated from bacteremia. Braz. J. Infect. Dis. 11:345-350. [DOI] [PubMed] [Google Scholar]
- 35.Plipat, N., G. Livni, H. Bertram, and R. B. Thomson, Jr. 2005. Unstable vancomycin heteroresistance is common among clinical isolates of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:2494-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash, V., J. S. Lewis II, and J. H. Jorgensen. 2008. Vancomycin MICs for methicillin-resistant Staphylococcus aureus isolates differ based upon the susceptibility test method used. Antimicrob. Agents Chemother. 52:4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rybak, M. J., and R. L. Akins. 2001. Emergence of methicillin-resistant Staphylococcus aureus with intermediate glycopeptide resistance: clinical significance and treatment options. Drugs 61:1-7. [DOI] [PubMed] [Google Scholar]
- 38.Rybak, M. J., S. N. Leonard, K. L. Rossi, C. M. Cheung, H. S. Sader, and R. N. Jones. 2008. Characterization of vancomycin-heteroresistant Staphylococcus aureus from the metropolitan area of Detroit, Michigan, over a 22-year period (1986 to 2007). J. Clin. Microbiol. 46:2950-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sader, H. S., P. D. Fey, D. N. Fish, A. P. Limaye, G. Pankey, J. Rahal, M. J. Rybak, D. R. Snydman, L. L. Steed, K. Waites, and R. N. Jones. 2009. Evaluation of vancomycin and daptomycin potency trends (MIC creep) against methicillin-resistant Staphylococcus aureus isolates collected in nine U.S. medical centers from 2002 to 2006. Antimicrob. Agents Chemother. 53:4127-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakoulas, G., P. A. Moise-Broder, J. Schentag, A. Forrest, R. C. Moellering, Jr., and G. M. Eliopoulos. 2004. Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 42:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwaber, M. J., S. B. Wright, Y. Carmeli, L. Venkataraman, P. C. DeGirolami, A. Gramatikova, T. M. Perl, G. Sakoulas, and H. S. Gold. 2003. Clinical implications of varying degrees of vancomycin susceptibility in methicillin-resistant Staphylococcus aureus bacteremia. Emerg. Infect. Dis. 9:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soriano, A., F. Marco, J. A. Martínez, E. Pisos, M. Almela, V. P. Dimova, D. Alamo, M. Ortega, J. Lopez, and J. Mensa. 2008. Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 46:193-200. [DOI] [PubMed] [Google Scholar]
- 43.Steinkraus, G., R. White, and L. Friedrich. 2007. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001-05. J. Antimicrob. Chemother. 60:788-794. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, D. L. 2006. The role of vancomycin in the treatment paradigm. Clin. Infect. Dis. 1:S51-S57. [DOI] [PubMed] [Google Scholar]
- 45.Swenson, J. M., K. F. Anderson, D. R. Lonsway, A. Thompson, S. K. McAllister, B. M. Limbago, R. B. Carey, F. C. Tenover, and J. B. Patel. 2009. Accuracy of commercial and reference susceptibility testing methods for detecting vancomycin-intermediate Staphylococcus aureus. J. Clin. Microbiol. 47:2013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabó, J. 2009. hVISA/VISA: diagnostic and therapeutic problems. Expert Rev. Anti Infect. Ther. 7:1-3. [DOI] [PubMed] [Google Scholar]
- 47.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, S. K. McAllister, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover, F. C., and R. C. Moellering, Jr. 2007. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin. Infect. Dis. 44:1208-1215. [DOI] [PubMed] [Google Scholar]
- 50.Walsh, T. R., A. Bolmström, A. Qwärnström, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 52.Wang, G., J. F. Hindler, K. W. Ward, and D. A. Bruckner. 2006. Increased vancomycin MICs for Staphylococcus aureus clinical isolates from a university hospital during a 5-year period. J. Clin. Microbiol. 44:3883-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]
- 54.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yusof, A., A. Engelhardt, A. Karlsson, L. Bylund, P. Vidh, K. Mills, M. Wootton, and T. R. Walsh. 2008. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J. Clin. Microbiol. 46:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.