Abstract
The epidemiology of Acinetobacter baumannii emerging in combat casualties is poorly understood. We analyzed 65 (54 nonreplicate) Acinetobacter isolates from 48 patients (46 hospitalized and 2 outpatient trainees entering the military) from October 2004 to October 2005 for genotypic similarities, time-space relatedness, and antibiotic susceptibility. Clinical and surveillance cultures were compared by amplified fragment length polymorphism (AFLP) genomic fingerprinting to each other and to strains of a reference database. Antibiotic susceptibility was determined, and multiplex PCR was performed for OXA-23-like, -24-like, -51-like, and -58-like carbapenemases. Records were reviewed for overlapping hospital stays of the most frequent genotypes, and risk ratios were calculated for any association of genotype with severity of Acute Physiology and Chronic Health Evaluation II (APACHE II) score or injury severity score (ISS) and previous antibiotic use. Nineteen genotypes were identified; two predominated, one consistent with an emerging novel international clone and the other unique to our database. Both predominant genotypes were carbapenem resistant, were present at another hospital before patients' admission to our facility, and were associated with higher APACHE II scores, higher ISSs, and previous carbapenem antibiotics in comparison with other genotypes. One predominated in wound and respiratory isolates, and the other predominated in wound and skin surveillance samples. Several other genotypes were identified as European clones I to III. Acinetobacter genotypes from recruits upon entry to the military, unlike those in hospitalized patients, did not include carbapenem-resistant genotypes. Acinetobacter species isolated from battlefield casualties are diverse, including genotypes belonging to European clones I to III. Two carbapenem-resistant genotypes were epidemic, one of which appeared to belong to a novel international clone.
Acinetobacter baumannii infections are well-described complications of severe combat-related injuries suffered in Iraq or Afghanistan by military service members (42). Acinetobacter species are notorious for multiantibiotic resistance and survival in the hospital, making eradication difficult or impossible (6, 27). Origin and transmission mechanisms both in wounded service members and in other nosocomial settings remain ambiguous, however. Presumptions of an environmental source (12) are based on literature prior to 1986 before any species differentiation within the genus and are probably incorrect. Hypotheses of Acinetobacter acquisition from the battlefield environment or skin colonization prior to injury are not supported by cultures of healthy service members' skin, nor wound cultures taken within 48 h of injury, nor soil cultures around field hospitals (13, 30, 41).
A. baumannii strains differ in antimicrobial susceptibility and transmissibility. Several reports have shown that multidrug-resistant (MDR) Acinetobacter strains were frequently associated with numerous outbreaks throughout Europe and beyond. Many of these strains were found to belong to three major clonal lineages (EU clones I to III) (2, 4, 22, 37). It is unknown whether these or other epidemic or sporadic strains influence patient outcomes.
The National Naval Medical Center (NNMC) in Bethesda, MD, is a referral site for all wounded U.S. Marine Corps (USMC) and Navy service members, in addition to all U.S. service members with neurotrauma. Prior to admission, patients followed a complex aeromedical evacuation/hospitalization process as previously described (29, 30), except that forward hospitals were in Iraq and Landstuhl, Germany (Landstuhl Regional Medical Center [LRMC]), for this study.
Antibiogram data indicate that the rate of Acinetobacter isolations at NNMC among U.S. service members injured in Southwest Asia has been declining since 2004. Acinetobacter strains obtained during peak prevalence (October 2004 to October 2005) were investigated to determine genetic diversity, relatedness to global strains and clones, incidence origin based on time and space, and clinical impact using amplified fragment length polymorphism (AFLP) analysis. AFLP is a well-validated high-resolution genomic fingerprinting method based on selective amplification of restriction fragments (5, 22, 36) that allows for organismal differentiation at strain level and enables comparison with a large well-established worldwide Acinetobacter AFLP database (5). In addition to AFLP, Acinetobacter strains were also characterized by antibiotic susceptibility phenotypes and the occurrence of OXA genes encoding carbapenem resistance (40).
MATERIALS AND METHODS
Patient investigation.
All positive A. baumannii cultures isolated at NNMC and 3 from LRMC during the period October 2004 to September 2005 were identified. The average daily number of inpatients at NNMC for this study period was 128. The isolates were collected when clinically indicated as follows: wound cultures in the operating room either as debrided tissue or swabs of the wound bed, sterile urine or blood cultures, and respiratory cultures via tracheal aspirate or bronchoalveolar lavage. Skin surveillance cultures were generally obtained from nares, axilla, and groin of all patients admitted from Iraq within 24 h of admission per the infection control policy initiated in 2003 (38).
Two electronic medical records and a USMC database detailing patient movement after injury provided age; gender; time, severity, and mechanism of injury; and length of hospital and intensive care unit (ICU) stay. Illness severity was assessed by both the objective composite trauma injury severity score (ISS) (1) and Acute Physiology and Chronic Health Evaluation II score (APACHE II) (19) at time of culture. This study was approved by the National Naval Medical Center Institutional Review Board (protocol number B06-136) in compliance with all federal regulations governing the protection of human subjects.
Provisional species identification and strain storage.
Samples from wounds, blood, sputum, urine, and skin surveillance were cultured using standard microbiological methods. Suspect colonies were presumptively identified using the Vitek system (bioMérieux, Durham, NC) to species level, deidentified as to patient with a unique code, and placed in fastidious broth (sucrose, glycerol) in vials (Fisher Scientific) and frozen at −70°C.
AFLP genomic fingerprint analysis for identification and typing.
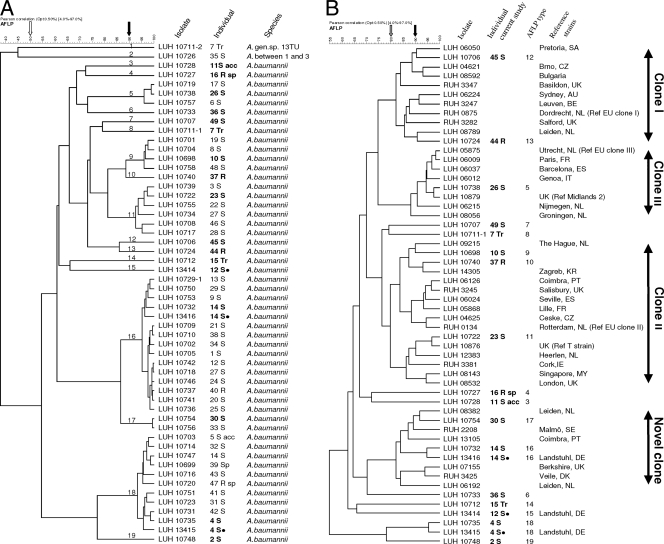
AFLP DNA fingerprinting was performed as previously described (21). Restriction fragments generated with EcoRI and MseI were selectively amplified using a Cy5-labeled Eco and an Mse primer; resulting fragments were separated by electrophoresis. Digitized electrophoretic profiles were grouped according to similarity using Bionumerics 4.5 (Applied Maths, Sint-Martens-Latem, Belgium), resulting in a dendrogram depicting the pattern of relatedness (Fig. 1). Isolates were also compared to an AFLP database comprising >2,000 reference strains of all Acinetobacter species and including epidemiologically defined strains from multiple cities and countries and the major European clones. Cutoffs of 50% similarity were used for species identification (5, 24), cutoffs of ≥80% similarity were used for clone identification (23), and cutoffs of ≥90% similarity were used for strain identification (34, 35) per recently established guidelines (34).
FIG. 1.
Dendrogram of Acinetobacter strains grouped according to their similarity in AFLP genotyping generated with Pearson's product-moment correlation coefficient as similarity measure and UPGMA (unweighted-pair group method using average linkages) as clustering algorithm. The horizontal scale denotes similarities among strains and groups of strains. Black dots indicate strains isolated from the same patient at Landstuhl Regional Medical Center (LRMC). (A) Comparison of the isolates of the current study. Numbers 1 to 49 designate the patients. Tr, trainee; S, soldier; S acc, soldier with accident; R, retiree; Sp, spouse; R sp, retiree spouse. For each individual, each unique genotype was included, as were the three isolates obtained from Landstuhl. Arrows denote cutoff levels for species (50%, white) and strains (90%, black). (B) Comparison of A. baumannii types (bold) with those of strains and clones of the Leiden University Medical Center (LUMC) AFLP database. For clone allocation, the cutoff level is 80% (gray arrow). SA, South Africa; CZ, Czech Republic; UK, United Kingdom; AU, Australia; BE, Belgium; NL, Netherlands; FR, France; ES, Spain; IT, Italy; KR, Croatia; PT, Portugal; IE, Ireland; MY, Malaysia; SE, Sweden; DE, Germany; DK, Denmark.
Antibiotic susceptibility testing.
Resistance was determined by disk diffusion using 12 antimicrobial agents/combinations with known activity against susceptible Acinetobacter strains following Clinical and Laboratory Standards Institute (CLSI) guidelines (39). Despite overlapping in drug classes, the 12 antibiotics tested reflect the most common resistance mechanisms found in Acinetobacter species. Zone sizes were read and interpreted using the semiautomated Biomic image analysis system (Giles Scientific, Inc., New York, NY). Agents (Oxoid) (content in μg/disk; susceptibility breakpoint in mm; resistance breakpoint in mm) included gentamicin (10; ≥15; ≤12), netilmicin (30; ≥15; ≤12), tobramycin (10; ≥15; ≤12), amikacin (30; ≥17; ≤14), ampicillin plus sulbactam (10 plus 10; ≥15; ≤11), piperacillin (100; ≥21; ≤17), ceftazidime (30; ≥18; ≤14), meropenem (10; ≥16; ≤13), imipenem (10; ≥16; ≤13), ofloxacin (5; ≥16; ≤12), sulfamethoxazole plus trimethoprim (23.75 plus 1.25; ≥16; ≤10), and tetracycline (30; ≥19; ≤14). Inhibition zone sizes were characterized as susceptible, intermediate, or resistant.
Colistin susceptibility testing was done using Etest (AB Biodisk, Solna, Sweden). A 0.5 McFarland standard of the organism was streaked to confluence on a 150-mm Muller-Hinton agar plate; e-strips were applied to the plate and read at 18 to 24 h of incubation. An organism was considered MDR if it was resistant to ≥3 antibiotics (23).
Detection of genes encoding OXA carbapenemases and ISAba1.
A multiplex PCR for detection of OXA-23-like, -24-like, -51-like, and -58-like (OXA-23, -24, -51, and -58) carbapenemases was performed as described by Woodford et al. (40). PCR for ISAba1 was performed according to the work of Turton et al. (32).
Statistical analysis.
Means and ranges were calculated to describe the patient clinical characteristics. Comparison of AFLP type and site of culture for the 4 major types was performed using Fisher's exact test. Risk ratios (RRs) with 95% confidence intervals (CIs) were calculated to compare the distribution of the most frequent AFLP genotypes to that of the other genotypes with respect to APACHE II score (≥10), ISS (≥25), and both total hospital and ICU stays.
RESULTS
Patient demographics.
We analyzed 46 inpatients with cultured Acinetobacter spp., 38 wounded service members, 6 retirees/family members hospitalized for nontrauma indications on the same wards or ICU (Table 1), and 2 service members who were noncombat traumas. One (patient 5 S, Fig. 1) had been injured on a ship, and one (patient 11 S, Fig. 1) had been injured in a motor vehicle accident. Among all patients, 42 (91%) were male, and mean age was 29 years (range, 19 to 84 years). Among combatants, the predominant injury was from improvised explosive devices (n = 24, 63%); the majority had multiple wounds (27, 56%), and injuries were predominantly on the extremities (30, 71%), and head and neck (18, 47%). Injuries were moderate to severe: the average ISS was 21 and the average APACHE II score was 7.7. There were 2 deaths, 3 traumatic amputations, and 3 surgical amputations. Of the 2 deaths, 1 (patient 9 S, Fig. 1) died of cardiac arrest following cerebral herniation and concomitant Acinetobacter pneumonia; the second (patient 47 R sp, Fig. 1) suffered cardiac arrest following foot gangrene necessitating below-knee amputation, Clostridium difficile colitis requiring colectomy, multiorgan failure, and Acinetobacter stump infection. All 6 amputations occurred prior to NNMC admission, mostly while in Iraq, and none were due to Acinetobacter infection.
TABLE 1.
Clinical characteristics of 46 hospitalized service members and retirees during Acinetobacter outbreak at NNMC
| Characteristic | No. of patients (%) | Mean | Range |
|---|---|---|---|
| All 46 service members and retireesa | |||
| Male sex | 42 (91) | ||
| Female sex | 4 (9) | ||
| Age (yr) | 29.25 | 19-84 | |
| Injury severity score | 39 (85)b | 21 | 1-43 |
| APACHE II score | 40 (87)c | 7.7 | 0-37 |
| No. of days hospitalized at LRMC | 37 (80)d | 3.5 | 1-15 |
| No. of days in ICU at LRMC | 25 (54)d | 3.4 | 1-8 |
| No. of days hospitalized at NNMC | 46 (100) | 28 | 1-114 |
| No. of days in ICU at NNMC | 34 (74) | 9.6 | 1-38 |
| No. of days of readmission to hospital | 18 (39) | 1.5 | 1-4 |
| 38 wounded service members only | |||
| Injury type | |||
| Improvised explosive device | 24 (63) | ||
| Fragment/shrapnel | 8 (21) | ||
| Gunshot wound | 7 (18) | ||
| Blunt trauma | 2 (5) | ||
| Multiple injury types | 1 (3) | ||
| Wound location | |||
| Multiple | 27 (71) | ||
| Head/neck | 18 (47) | ||
| Face | 8 (21) | ||
| Chest | 12 (32) | ||
| Abdomen | 8 (21) | ||
| Extremity | 30 (79) |
Includes 38 combat wounded, 2 accident victims, and 6 retirees/spouses.
Includes 38 wounded and 2 accident victims. ISS not available for 1 patient.
Includes all 46 hospitalized patients; APACHE II scores not available for 6 patients.
Includes 38 wounded and 2 accident victims only. LRMC, Landstuhl Regional Medical Center, Germany.
Officer candidate school (OCS) trainees are routinely screened for methicillin-resistant Staphylococcus aureus (MRSA) with nasal cultures. Three Acinetobacter isolates were incidentally discovered in our lab from 2 patients' surveillance cultures; no clinical data were available as these patients were asymptomatic, and hence, they are not included in Table 1.
Species identification and strain typing.
Altogether, there were 65 isolates from the 46 patients and two trainees available for molecular and phenotypic characterization. Seven patients had multiple isolates with identical profiles in consecutive specimens, indicating colonization with one strain; these 10 replicates were removed from statistical analysis, leaving 55 isolates. Two isolates from one specimen (patient 13 S) had the same AFLP genotype but differed in antimicrobial susceptibility. We removed one of these isolates from statistical analysis as well, leaving 54 unique isolates. Three battlefield casualties (12 S, 14 S, and 27 S) and 1 trainee (7 Tr) had 2 strains identified from separate clinical sites as inferred from AFLP typing. Three patients (12 S, 14 S, and 4 S) had Acinetobacter isolated during hospitalization at LRMC in Germany (Fig. 1, black dots). All isolates belonged to A. baumannii, except for 2 (trainee 7 Tr and patient 35 S) that were identified to Acinetobacter genomic species 13TU and “between 1 and 3,” respectively (Fig. 1; Table 2 ).
TABLE 2.
Summary of characters and origin of Acinetobacter isolates from 46 patientsa and two officer candidate school (OCS) trainees
| Group | AFLP type | Species | No. of individuals | Identification with (EU) clonesb | Specimen(s) from which isolates originated (no.) | Ward (no. of isolates) | Hospital days of first culture per patient, median (range) | Resistance: no. of antibiotics | OXA gene (no. of isolates) |
|---|---|---|---|---|---|---|---|---|---|
| Patients | 2 | Between 1 and 3 | 1 | NA | Wound (1) | Ward (1) | 1 | 1 | |
| 3 | A. baumannii | 1 | Unique | Respiratory tract (1) | ICU (1) | 1 | 0 | ||
| 4 | A. baumannii | 1 | Unique | Wound (1) | Ward (1) | 1 | 5 | ||
| 5 | A. baumannii | 3 | III | Wound (2); blood (1); respiratory tract (1) | Ward (2); ICU (2) | 1 (1-3) | 8 or 9 | ||
| 6 | A. baumannii | 1 | Unique | Wound (1) | Ward (1) | 1 | 0 | ||
| 7 | A. baumannii | 1 | Unique | Wound (1) | Ward (1) | 13 | 0 | ||
| 9 | A. baumannii | 4 | II | Wound (1); respiratory tract (2); skin (2) | ICU (5) | 3 (1-9) | 7-10 | ||
| 10 | A. baumannii | 1 | II | Urine (1) | Ward | 1 | 6 | ||
| 11 | A. baumannii | 6 | II | Blood (1); respiratory tract (2); skin (3) | Ward (2); ICU (4) | 6 (1-20) | 7 or 8 | ||
| 12 | A. baumannii | 1 | I | Skin (1) | ICU (1) | 1 | 9 | ||
| 13 | A. baumannii | 1 | I | Urine (1) | Ward (1) | 10 | 9c | OXA-23 (1) | |
| 15 | A. baumannii | 1 | Unique | Unknown (1) | Landstuhl (1) | NAg | 6d | OXA-58 (1) | |
| 16 | A. baumannii | 14 | Novel clone | Wound (9); blood (1); respiratory tract (6); skin (2); unknown (1) | ICU (11); ward (7); Landstuhl (1) | 3 (1-28) | 4-9e | OXA-58 (16) | |
| 17 | A. baumannii | 2 | Novel clone | Wound (2) | Ward (2) | 7 (2-12) | 7 | ||
| 18 | A. baumannii | 10 | Unique | Wound (4); respiratory tract (1); skin (7); urine (1); catheter tip (2); unknown (1) | ICU (11); ward (4); Landstuhl (1) | 2 (1-30) | 8 or 9f | OXA-23 (16) | |
| 19 | A. baumannii | 1 | Unique | Skin (1) | ICU (1) | 1 | 8 | ||
| Trainees | 1 | 13TU | 1 | NA | Nares (1) | NA | NA | 0 | |
| 8 | A. baumannii | 1 | Unique | Nares (1) | NA | NA | 3 | ||
| 14 | A. baumannii | 1 | Unique | Nares (1) | NA | NA | 1 |
Two distinct AFLP types in 4 individuals each.
Clones sensu Ørskov and Ørskov (26); NA, not applicable (non-A. baumannii); EU clones I to III (Dijkshoorn et al., 1996 [4], and van Dessel et al., 2004 [37]).
Imipenem and meropenem resistant.
Carbapenem susceptible.
Ten isolates imipenem resistant and 14 isolates meropemen resistant.
All 16 isolates imipenem and meropenem resistant.
NA, not applicable.
Nineteen AFLP profiles (genotypes) were distinguished at the strain cutoff level of 90% (Fig. 1; Table 2), and 6 of these profiles (genotypes 5, 9, 11, 16, 17, and 18) were observed across multiple patients (Fig. 1). The 13 remaining profiles were observed in 1 patient each. Genotypes 16 and 18 were most frequent, in 14 (1 retiree and 13 service members) and 10 (2 spouses and 8 service members) patients, respectively. The 3 isolates from patients admitted to LRMC belonged to AFLP profile 15 (patient 12 S), 16 (patient 14 S), and 18 (patient 4 S) (Fig. 1, black dots), suggesting that NNMC predominant strains 16 and 18 also occurred at LRMC.
Antimicrobial susceptibility and OXA gene determination.
Acinetobacter strains were resistant (including intermediate susceptibility) to 0 to 10 antibiotics per isolate (median of 8); all isolates were susceptible to colistin. The Acinetobacter 13TU, “between 1 and 3,” and 4 A. baumannii isolates were all resistant to 0 to 1 antibiotic. Multidrug resistance (≥3 agents) was observed in 13 of the 19 genotypes (Table 2). Resistance or intermediate susceptibility to imipenem and/or meropenem was observed in genotype 13 (one patient), genotype 16 (12 patients, 14 isolates), and genotype 18 (10 patients, 16 isolates). PCR for ISAba1 was negative in 5 strains (genotypes 2, 3, 6, 7, and 8; resistance to 0 to 3 antibiotics noted) (Table 2). All A. baumannii strains were OXA-51 PCR positive, and Acinetobacter 13TU and “between 1 and 3” strains were negative. OXA-23 was detected in genotypes 13 and 18, imparting imipenem and meropenem resistance throughout. OXA-58 was observed in genotype 15 strains (imipenem- and meropenem-susceptible phenotype) and in 16 of 19 isolates of genotype 16 (13 of them with imipenem and/or meropenem resistance). All genotypes found in more than 1 patient were MDR, emphasizing the correlation between multidrug resistance and epidemicity of strains. Of note, the number of antibiotic resistances in replicate isolates from 4/7 patients varied.
Comparison of strains to those of the EU clones and other strains.
Isolated strains compared to our AFLP database revealed that genotypes 12 and 13 correspond to EU clone I, clustering at ∼80% similarity (Fig. 1B). Likewise, genotypes 9 to 11 corresponded to EU clone II and genotype 5 corresponded to EU clone III (Fig. 1B; Table 2). Furthermore, genotypes 16 and 17 corresponded to a tentative novel international clone (clustering level, ≥80%) comprising 14 strains from different cities and countries obtained over 3 decades (data not shown) and containing susceptible and MDR strains requiring future detailed characterization. Two strains of this clone (Fig. 1) were recently characterized by multilocus sequence typing (MLST) to belong to clonal complex CC32 (3). Other genotypes, including the predominant genotype 18, did not correspond to any known strains or clones in our database. Further comparison showed that MDR strains 5, 9, 10 to 13, and 17 possess ≥90% similarity to strains from various European countries and beyond. Interestingly, genotype 11, found in 6 service members, was by AFLP >95% similar to the “T-strain” found in Iraq casualties in the United Kingdom, LRMC, and the United States (31) and subsequently identified as “ST-11” by another isolate identification technique (multilocus PCR) at Walter Reed Army Medical Center (WRAMC) (10).
Genotypes in association with the origin in time and space of strains.
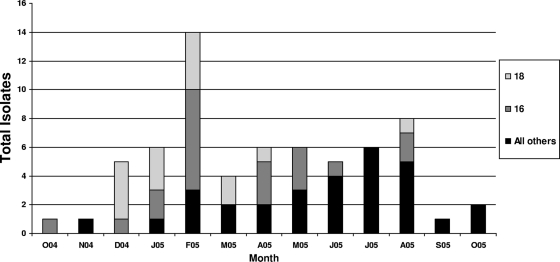
Acinetobacter was most prevalent in February 2004 at NNMC with genotypes 16 and 18 predominating. Genotype 16 was present from October 2004 until August 2005. Genotype 18 predominated from December 2004 to April 2005, with others being sporadic throughout the year (Fig. 2).
FIG. 2.
AFLP type by month October 2004 to 2005. Three isolates were from Landstuhl: AFLP type 18 (December 2004), AFLP 15 (February 2005), and AFLP type 16 (April 2005). O04 to O05, October 2004 to October 2005, respectively.
Before admission to NNMC, patients had, on average, been admitted to 2 hospitals, including field hospitals in Iraq. From Iraq they were transferred to LRMC; 32 then directly transferred to NNMC and 6 came via other hospitals. Two (17 S and 35 S) were readmissions at 3 and 12 months after initial NNMC hospitalization. Seventeen were Acinetobacter culture positive <1 day after admission to NNMC. Six of 13 with profile 16 had overlap with 1 or 2 others in time or space, either in LRMC or NNMC or in Iraq (2 patients). Overlap in time or space with 1 or 2 others was also observed in 6/8 with profile 18, 3/4 with profile 9, and 3/6 with profile 11 (data not shown). Three of six retirees (or family members) admitted solely to NNMC had cultures positive for 1 of the epidemic strains (genotype 5, 1 case; genotype 18, 2 cases).
Clinical impact.
The predominant source was wound cultures (37%), followed by skin surveillance samples (30%) and respiratory specimens (19%) (Table 3). AFLP type 16 comprised predominantly wound isolates (40%) and respiratory isolates (33%). Urine isolates showed no predominant genotype. Genotype 18 comprised predominantly wound (45%) and skin surveillance (36%) isolates, most of which were isolated <2 days from initial hospitalization. Surveillance isolates caused subsequent infections in 1 patient, 4 S (respiratory and catheter tip infections, genotype 18). Patient 14 S had genotype 16 isolated from skin surveillance at both Landstuhl and NNMC. Most respiratory and surveillance cultures were from ICU patients; most wound cultures were from patients on surgical wards. AFLP type was not significantly related to culture source for any of the 4 major types (16, 18, 11, and 9) by Fisher's exact test.
TABLE 3.
Sources of 54 nonreplicate cultures by predominant AFLP type with antimicrobial susceptibilities
| AFLP type (n) | Culture sourcea |
No. susceptible to drugb/no. of total isolates |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wound | Respiratory | Skin surveillance | Bloodstream | Urine | Unknown | CTZ | OFX | TET | AM-S | IMP | TMS | MER | AMK | TOB | PIP | GEN | NET | |
| Inpatient | ||||||||||||||||||
| 16 (15) | 6 | 5 | 2 | 1 | 0 | 1 | 10/15 | 0/15 | 2/15 | 15/15 | 7/15 | 0/15 | 4/15 | 15/15 | 4/15 | 0/15 | 4/15 | 15/15 |
| 18 (11) | 5 | 0 | 4 | 0 | 1 | 1 | 8/11 | 0/11 | 0/11 | 11/11 | 0/11 | 0/11 | 0/11 | 11/11 | 0/11 | 0/11 | 0/11 | 11/11 |
| 11 (6) | 0 | 2 | 3 | 1 | 0 | 0 | 0/6 | 0/6 | 0/6 | 0/6 | 6/6 | 0/6 | 6/6 | 5/6 | 6/6 | 0/6 | 0/6 | 6/6 |
| 9 (4) | 1 | 1 | 2 | 0 | 0 | 0 | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 | 4/4 | 1/4 | 1/4 | 0/4 | 0/4 | 1/4 |
| All others (15) | 8 | 2 | 2 | 1 | 2 | 0 | 6/15 | 5/15 | 4/15 | 10/15 | 14/15 | 6/15 | 14/15 | 10/15 | 7/15 | 3/15 | 4/15 | 13/15 |
| Outpatient | ||||||||||||||||||
| All others (3) | 0 | 0 | 3 | 0 | 0 | 0 | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 1/3 | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 |
| Total | 20 | 10 | 16 | 3 | 3 | 2 | ||||||||||||
No significant association between any source and AFLP type (Fisher's exact test, P values not shown).
CTZ, ceftazidime; OFX, ofloxacin; TET, tetracycline; AM-S, ampicillin-sulbactam; IMP, imipenem; TMS, trimethroprim-sulfamethoxazole; MER, meropenem; AMK, amikacin; TOB, tobramycin; PIP, piperacillin; GEN, gentamicin; NET, netilmicin.
The most common genotypes, 18 and 16, appeared related to illness severity, as expressed by the APACHE II score and elevated ISS trauma score. Genotype 18 (RR, 5.4 [95% CI, 1.3 to 23.4]), genotype 16 (RR, 2.9 [95% CI, 0.6 to 14.4]), and infection/colonization with ≥2 genotypes (RR, 6.3 [95% CI, 1.4 to 29.4]) were all associated with a higher risk of having an APACHE II score of >10, compared to the other genotypes. The same was true for patients with an ISS of >25 for genotypes 18 and 16 and >2 types (RR, 2.7 [95% CI, 0.9 to 8.0], 2.9 [95% CI, 1.0 to 7.8], and 4.8 [95% CI, 2.0 to 11.3], respectively). Genotype 18 or 16 did not appear related to length of hospital or ICU stays, however, as RRs were only mildly elevated (data not shown). Carbapenem therapy was defined as receipt of imipenem and/or meropenem for at least 3 doses or 24 h during the period between time of injury and time of culture at NNMC. Out of 14 patients with type 16, 9 (64%) had previous carbapenem therapy, compared to 4 out of 27 (15%) in the patients with types other than 16 and 18 (two patients had two types and were counted twice in this analysis) (RR, 4.3 [95% CI, 1.6 to 11.6]). Similar results were found for patients with type 18: 7 out of 10 subjects (70%) with this type had previous carbapenems (RR, 4.7 [95% CI, 1.8 to 12.7]). Furthermore, the mean number of antibiotics that was used in the time between injury and positive culture at NNMC in patients with type 16 was higher than that in those with types other than 16 and 18, i.e., 2.3 versus 0.7 (mean difference, 1.6 [95% CI, 0.8 to 2.5]). Similarly in type 18 the mean number of antibiotics was 2.8 versus 0.7 (mean difference, 2.1 [95% CI, 1.4 to 2.9]). The 2 deaths had the predominant (drug-resistant) genotypes 16 and 18. Of the 3 traumatic amputations, 1 had both genotype 16 and genotype 18, 1 had genotype 18, and 1 had genotype 17. Of the 3 surgical amputations, 2 had genotype 18 and 1 had genotype 16. Ten patients met the CDC definition (15) for hospital-associated pneumonia, of which 6 cases were caused by Acinetobacter.
DISCUSSION
Diversity.
Nineteen strains of Acinetobacter baumannii were found in NNMC during 2004 to 2005. The two predominant strains, AFLP profiles (genotypes) 16 and 18, have not been described before and were genotypically distinct from the European clones I to III (Fig. 1). Genotype 16, however, grouped with a tentative novel clone in our AFLP database. Both genotype 16 and genotype 18 expressed OXA-23- or -58-mediated carbapenem resistance and appeared epidemic. Genotype 16 predominated among wound and respiratory isolates; genotype 18 predominated among skin surveillance isolates. A recent Army hospital study with similar sample size but an earlier study period (2003 to 2005) is an interesting contrast to our findings (16). We found higher rates of carbapenem resistance (50% versus 24%) and higher rates of OXA-23 (25% versus 11%), and OXA-58 (31% versus 12%) carbapenemases present. This might be due to differences in strains available for study or due to increasing empirical utilization of carbapenems since 2003 and, consequently, the emergence of carbapenem-resistant strains. As a comparator group and to determine if Acinetobacter originates from healthy people's normal microbial flora, we also analyzed incidentally found Acinetobacter isolates from healthy trainees undergoing MRSA screening (Table 2) for their genotypic relatedness. These individuals were not included in the outcome analyses of wounded service members, however. In contrast to the predominant hospital isolates, the 3 isolates found in 2 trainees entering the military were 13TU or A. baumannii with resistance to only 1 to 3 antibiotics and genetically distinct. This is consistent with previous studies (13) showing that MDR A. baumannii strains are probably acquired nosocomially, not before injury.
Comparison with EU clones and other strains.
Clones have been previously defined by Ørskov and Ørskov as “cultures from different origin in time and space, but so similar in genotype and phenotype that a common origin is likely” (26). Using AFLP analysis correlated with our database of international strains and clones, 6 genotypes were identified to belong to European clones I to III, while genotype 17 and the predominant genotype 16 belonged to a tentative novel international clone (Table 2; Fig. 1). Some genotypes of the present study (e.g., genotypes 12 [clone I], 5 [clone II], and 11 [clone III]) were highly similar (∼90%) to strains found elsewhere (Fig. 1) including those in Iraq casualties (“T-strain”) (31). It is of note that genotypes at WRAMC (February 2004 to 2005) (16) and in a study of 2005-2006 isolates from NNMC (38) showed predominantly EU clones II and III. The emergence of strains of these clones, mostly in critically ill patients, emphasizes that these organisms must have particular attributes favoring success in this population.
Therefore, unambiguous identification (e.g., sequence-based methods like multilocus variable-number tandem repeat analysis [MLVA]) based on a standard protocol will be important for future studies on these organisms. Resources for Acinetobacter MLST-based clone identification can be found at www.pasteur.fr/mlst and http://pubmlst.org/.
Origin in time and space.
We found no correlation between geographic location of traumatic injury and genotype (data not shown), suggesting that the environment at time of wounding is an unlikely source of infection. Our findings do not support one facility as the source but reaffirm findings (30) of epidemic genotypes occurring across many facilities with the majority of isolates comprising multiple genotypes. Among 23 patients with contemporary hospitalization at any facility and similar genotypes, all were hospitalized for at least 1 day at LRMC. Eight patients had contemporary hospitalizations at LRMC. We found AFLP types 16 and 18 present during hospitalization at LRMC in 2 patients; both had contemporary stays at both LRMC and in Baghdad, suggesting acquisition at one of those hospitals. Furthermore, almost half of all combat injuries had a positive culture within 24 h of admission. Hence, acquisition of several strains at or before LRMC is probable. However, because isolates antecedent to LRMC admission were not available, a precise source is elusive. A recent study of surveillance specimens (18) examined longitudinal transmission dynamics in 54 patients hospitalized via the same process as that in our study. No A. baumannii strains were isolated in Iraq, 1 was found at LRMC, and 3 positives were followed by 7 infections in a Texas hospital. The authors explained the lack of positive isolates from Iraq and Germany on the basis of culture methods lacking the sensitivity to detect low levels of Acinetobacter colonization when it first occurs. Clearly a better method of detecting A. baumannii both in the hospital environment and among normal host flora is needed. One such method could be enrichment cultivation on acetate-mineral water (7).
We noted a large increase in AFLP genotypes 16 and 18 in the months of December 2004 to February 2005 (Fig. 2), coinciding with a casualty peak following the Second Battle of Fallujah. Outbreaks of nosocomial MDR Gram-negative infections following a mass influx of casualties have been described in earthquakes (25) and tsunamis (17, 33). This phenomenon is hypothetically due to decreased infection control adherence and increased empirical antibiotic use. Our study is unique in that it describes an outbreak of MDR and carbapenem-resistant Acinetobacter in such a setting. It is a concern that trauma patients transferred across multiple hospitals become potential vectors for regional or global spread of carbapenem-resistant Acinetobacter.
Eight patients had contemporary stays of 3 to 28 days at NNMC with a patient infected with a similar genotype representing probable acquisition at NNMC. Noncombat casualties are presumed to have acquired the organism nosocomially. Of these, patients 39 sp, 40 R, and 47 R sp had predominant genotypes 16 and 18, and patients 16 R sp, 37 R, and 44 R had genotypes 4, 10, and 13, respectively. Eight more patients had contemporary hospitalization with a patient with similar genotype of only 1 day prior to developing a positive culture. Whether or not this is sufficient exposure for nosocomial transmission is unclear, but a health care worker recently caught pneumonia after merely suctioning an infected patient briefly, suggesting that minor exposures might be sufficient (38).
Clinical impact.
We found a significant association with more severe injury and colonization/infection with MDR A. baumannii (here also carbapenem resistant) but less association with length of hospital or ICU stay. However, these relations should be interpreted as simple associations, as any causal or temporal inference is difficult in the current design. These patients also received more carbapenem antibiotics prior to culture, a previously described risk (28). While this might be a cause-and-effect phenomenon (sicker patients being more likely to get the most powerful antibiotics at the first fever), recent analysis of combat casualties' immune response may also explain this phenomenon. Casualties with surgical wound dehiscence had significantly higher APACHE II scores and ISSs (14) and persistent significantly higher levels of proinflammatory cytokines interleukin-6 (IL-6), IL-8, and macrophage inflammatory protein 1α (MIP-1α). The severe blast injury so prevalent in this conflict overwhelms the immune system, a sustained hyperinflammatory state ensues, the host cannot physiologically self-regulate, and wound healing is impaired. It is possible that this phenomenon also predisposes young healthy patients to A. baumannii infection heretofore seen only in immunocompromised patients and should be studied further. Additionally, our mean blood transfusion requirement was 4 units (range, 0 to 18) among casualties; this predisposes to infection (8, 9) probably via an exchange-transfusion-like effect on immunity.
We noted more wound (37% versus 11%) and respiratory (30% versus 6%) infections but fewer bloodstream (6% versus 55%) infections than previously reported (16), and yet we find no significant association with AFLP type and source of culture. Genotype 18, a carbapenem-resistant strain, was highly prevalent in skin surveillance and wounds and yet not a cause of invasive disease (Table 3). Hence, although genotypes 16 and 18 were both MDR, carbapenem resistant, and epidemic, their virulence seemed to differ. However, our findings might be due to selection bias, as all combat casualties are screened for MDR organisms. Furthermore, our overall infection numbers were low, and this was not a case-control study. The lack of invasiveness does not diminish the importance of type 18 in the hospital or the need to aggressively screen at-risk populations for colonization, especially in light of the limited therapies available for this genotype. We noted spread of this clone (type 18) to the wound of 1 patient who subsequently died. All observed deaths had multiple comorbidities, so determining if Acinetobacter was causative, contributive, or merely commensal to death is difficult. Carbapenem resistance effect on mortality is difficult to analyze; a previous meta-analysis failed (11), but recently A baumannii carbapenem resistance did not increase mortality from infection with OXA-23-expressing organisms except for bacteremic patients or ICU infections (20). Trauma patients actually had improved outcomes with Acinetobacter infection compared to nontrauma patients. A prospective trial of OXA-23 impact on outcomes would greatly advance understanding of Acinetobacter pathophysiology.
In summary, we found multiple A. baumannii strains at our facility among combat wounded, including two predominant MDR and carbapenem-resistant strains. Several strains, including one predominant genotype, belonged to four international clones. Many questions remain regarding the dynamics and pathogenicity of this organism.
Acknowledgments
All authors do not have a commercial or other association that might pose a conflict of interest. Sources of financial support for all authors were none.
We thank Diana Temple for her assistance in preparing the manuscript.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Baker, S. P., B. O'Neill, W. Haddon, Jr., and W. B. Long. 1974. The injury severity score: a method for describing patients with multiple injuries and evaluating emergency care. J. Trauma 14:187-196. [PubMed] [Google Scholar]
- 2.Da Silva, G. J., et al. 2004. Long-term dissemination of an OXA-40 carbapenemase-producing Acinetobacter baumannii clone in the Iberian Peninsula. J. Antimicrob. Chemother. 54:255-258. [DOI] [PubMed] [Google Scholar]
- 3.Diancourt, L., V. Passet, A. Nemec, L. Dijkshoorn, and S. Brisse. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dijkshoorn, L., et al. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkshoorn, L., and A. Nemec. 2008. The diversity of the genus Acinetobacter, p. 1-34. In U. Gerischer (ed.), Acinetobacter molecular microbiology, 1st ed. Caister Academic Press, Norwich, United Kingdom.
- 6.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 7.Dijkshoorn, L., W. Van Vianen, J. E. Degener, and M. F. Michel. 1987. Typing of Acinetobacter calcoaceticus strains isolated from hospital patients by cell envelope protein profiles. Epidemiol. Infect. 99:659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunne, J. R., et al. 2009. Perioperative blood transfusion in combat casualties: a pilot study. J. Trauma 66:S150-S156. [DOI] [PubMed] [Google Scholar]
- 9.Dunne, J. R., M. S. Riddle, J. Danko, R. Hayden, and K. Petersen. 2006. Blood transfusion is associated with infection and increased resource utilization in combat casualties. Am. Surg. 72:619-625. [PubMed] [Google Scholar]
- 10.Ecker, J. A., et al. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44:2921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falagas, M. E., I. A. Bliziotis, and I. I. Siempos. 2006. Attributable mortality of Acinetobacter baumannii infections in critically ill patients: a systematic review of matched cohort and case-control studies. Crit. Care 10:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692-699. [DOI] [PubMed] [Google Scholar]
- 13.Griffith, M. E., et al. 2006. Acinetobacter skin colonization of US Army soldiers. Infect. Control Hosp. Epidemiol. 27:659-661. [DOI] [PubMed] [Google Scholar]
- 14.Hawksworth, J. S., et al. 2009. Inflammatory biomarkers in combat wound healing. Ann. Surg. 250:1002-1007. [DOI] [PubMed] [Google Scholar]
- 15.Horan, T. C., M. Andrus, and M. A. Dudeck. 2008. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control 36:309-332. [DOI] [PubMed] [Google Scholar]
- 16.Hujer, K. M., et al. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallman, O., C. Lundberg, B. Wretlind, and A. Ortqvist. 2006. Gram-negative bacteria from patients seeking medical advice in Stockholm after the tsunami catastrophe. Scand. J. Infect. Dis. 38:448-450. [DOI] [PubMed] [Google Scholar]
- 18.Kaspar, R. L., et al. 2009. Association of bacterial colonization at the time of presentation to a combat support hospital in a combat zone with subsequent 30-day colonization or infection. Mil. Med. 174:899-903. [DOI] [PubMed] [Google Scholar]
- 19.Knaus, W. A., E. A. Draper, D. P. Wagner, and J. E. Zimmerman. 1985. APACHE II: a severity of disease classification system. Crit. Care Med. 13:818-829. [PubMed] [Google Scholar]
- 20.Livermore, D. M., et al. 2010. Antimicrobial treatment and clinical outcome for infections with carbapenem- and multiply-resistant Acinetobacter baumannii around London. Int. J. Antimicrob. Agents 35:19-24. [DOI] [PubMed] [Google Scholar]
- 21.Nemec, A., et al. 2001. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 51:1891-1899. [DOI] [PubMed] [Google Scholar]
- 22.Nemec, A., L. Dijkshoorn, and T. J. van der Reijden. 2004. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J. Med. Microbiol. 53:147-153. [DOI] [PubMed] [Google Scholar]
- 23.Nemec, A., et al. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484-489. [DOI] [PubMed] [Google Scholar]
- 24.Nemec, A., M. Maixnerova, T. J. van der Reijden, P. J. van den Broek, and L. Dijkshoorn. 2007. Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J. Antimicrob. Chemother. 60:483-489. [DOI] [PubMed] [Google Scholar]
- 25.Oncul, O., et al. 2002. Hospital-acquired infections following the 1999 Marmara earthquake. J. Hosp. Infect. 51:47-51. [DOI] [PubMed] [Google Scholar]
- 26.Ørskov, F., and I. Ørskov. 1983. From the National Institutes of Health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the Enterobacteriaceae and other bacteria. J. Infect. Dis. 148:346-357. [DOI] [PubMed] [Google Scholar]
- 27.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez, F., et al. 2010. Antibiotic resistance determinants in Acinetobacter spp and clinical outcomes in patients from a major military treatment facility. Am. J. Infect. Control 38:63-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen, K., et al. 2007. Trauma-related infections in battlefield casualties from Iraq. Ann. Surg. 245:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott, P., et al. 2007. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin. Infect. Dis. 44:1577-1584. [DOI] [PubMed] [Google Scholar]
- 31.Turton, J. F., et al. 2006. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J. Clin. Microbiol. 44:2630-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turton, J. F., et al. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 33.Uckay, I., H. Sax, S. Harbarth, L. Bernard, and D. Pittet. 2008. Multi-resistant infections in repatriated patients after natural disasters: lessons learned from the 2004 tsunami for hospital infection control. J. Hosp. Infect. 68:1-8. [DOI] [PubMed] [Google Scholar]
- 34.van Belkum, A., et al. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1-46. [DOI] [PubMed] [Google Scholar]
- 35.van den Broek, P. J., et al. 2006. Epidemiology of multiple Acinetobacter outbreaks in The Netherlands during the period 1999-2001. Clin. Microbiol. Infect. 12:837-843. [DOI] [PubMed] [Google Scholar]
- 36.van den Broek, P. J., A. T. Bernards, T. J. van der Reijden, B. van Strijen, and L. Dijkshoorn. 2009. Can Escherichia coli be used as an indicator organism for transmission events in hospitals? Eur. J. Clin. Microbiol. Infect. Dis. 28:169-173. [DOI] [PubMed] [Google Scholar]
- 37.van Dessel, H., et al. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 38.Whitman, T. J., et al. 2008. Occupational transmission of Acinetobacter baumannii from a United States serviceman wounded in Iraq to a health care worker. Clin. Infect. Dis. 47:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wikler, M. A., and Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing: 15th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 40.Woodford, N., et al. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]
- 41.Yun, H. C., et al. 2006. Bacteria recovered from patients admitted to a deployed U.S. military hospital in Baghdad, Iraq. Mil. Med. 171:821-825. [DOI] [PubMed] [Google Scholar]
- 42.Zapor, M. J., and K. A. Moran. 2005. Infectious diseases during wartime. Curr. Opin. Infect. Dis. 18:395-399. [DOI] [PubMed] [Google Scholar]