Abstract
Chronic wasting disease (CWD) is a transmissible spongiform encephalopathy (TSE) of deer and elk, and little is known about its transmissibility to other species. An important factor controlling interspecies TSE susceptibility is prion protein (PrP) homology between the source and recipient species/genotypes. Furthermore, the efficiency with which the protease-resistant PrP (PrP-res) of one species induces the in vitro conversion of the normal PrP (PrP-sen) of another species to the protease-resistant state correlates with the cross-species transmissibility of TSE agents. Here we show that the CWD-associated PrP-res (PrPCWD) of cervids readily induces the conversion of recombinant cervid PrP-sen molecules to the protease-resistant state in accordance with the known transmissibility of CWD between cervids. In contrast, PrPCWD-induced conversions of human and bovine PrP-sen were much less efficient, and conversion of ovine PrP-sen was intermediate. These results demonstrate a barrier at the molecular level that should limit the susceptibility of these non-cervid species to CWD.
Keywords: cell-free conversion/chronic wasting disease (CWD)/prion protein (PrP)/scrapie/transmissible spongiform encephalopathy (TSE)
Introduction
The apparent transmission of bovine spongiform encephalopathy (BSE) to humans (Bruce et al., 1997; Hill et al., 1997) and other mammalian species emphasizes the importance of considering the potential for cross-species transmission of other animal transmissible spongiform encephalopathy (TSE) diseases. A high percentage (up to 15%) of free-ranging deer and elk in parts of north-eastern Colorado and south-eastern Wyoming are infected with chronic wasting disease (CWD) (Miller et al., 2000). CWD-infected cervids have also been identified in game farms in several other western US states. Although it appears that natural transmission of CWD between cervids is important in the maintenance of the CWD epidemic, the origin of CWD and the mode of transmission between wild animals are not understood. Furthermore, it is not clear whether the disease can be transmitted to humans who hunt and eat these animals or to domestic livestock whose range may overlap with infected cervids. The transmissibility of CWD to animals should be tested experimentally in vivo, but such tests will take years to complete because of the long incubation periods commonly encountered in interspecies TSE transmissions. Because of these problems, and the fact that CWD transmissibility cannot be tested directly in humans, we have sought alternative clues to the potential interspecies transmissibility of CWD.
In TSE diseases, the host’s protease-sensitive prion protein (PrP-sen or PrPC) is converted to a protease-resistant isoform (PrP-res). Transgenic studies have demonstrated the importance of PrP sequence homology between the sources and recipients of TSE infections (Prusiner et al., 1990). Such PrP homology is also important in PrP-res formation, as demonstrated in scrapie-infected neuroblastoma cells (Priola et al., 1994; Priola and Chesebro, 1995) and cell-free reactions (Kocisko et al., 1995; Bossers et al., 1997, 2000; Raymond et al., 1997; Horiuchi et al., 2000). In cell-free reactions, PrP-res directly induces the conversion of PrP-sen to a protease-resistant state that is biochemically indistinguishable from PrP-res (Kocisko et al., 1994). Although the products of this reaction have not yet been shown to be infectious, this apparent self-propagating activity of PrP-res correlates with infectivity in denaturation studies (Caughey et al., 1997) and shows striking strain and species specificities that reflect important biological parameters of these diseases (Bessen et al., 1995; Kocisko et al., 1995; Bossers et al., 1997, 2000; Raymond et al., 1997). For instance, the efficiency of conversion reactions between PrP-res and PrP-sen of different species correlates with the relative TSE transmissibility between those species in vivo (Kocisko et al., 1995; Bossers et al., 1997, 2000; Raymond et al., 1997; Chabry et al., 1999; Horiuchi et al., 2000). Collectively, the above studies provide evidence that an important control point in interspecies TSE infections is the molecular compatibility between incoming PrP-res and the endogenous PrP-sen. In the present study, we have compared the efficiency with which PrP-res from CWD-infected cervids (PrPCWD) induced conversion of PrP-sen molecules from other species. Although multiple factors control whether CWD actually transmits to other species, our analysis provides an initial assessment of the relative likelihoods that incoming PrPCWD, if delivered to an appropriate anatomical site, can initiate new, potentially pathogenic PrP-res formation.
Results
Cloning, expression and labeling of cervid PrP-sen variants
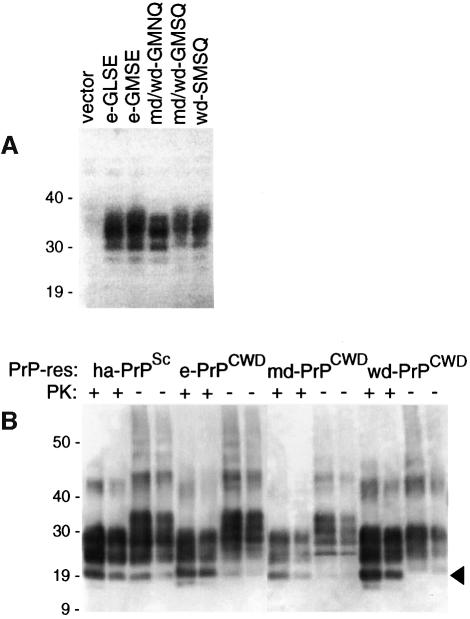
To test the ability of PrPCWD to induce the cell-free conversion of cervid PrP-sen molecules as a benchmark for other cross-species comparisons, we first needed to obtain suitable cervid PrP-sen and PrPCWD molecules. Cervid PrP has five known allelic variants, which are defined in Figure 1. Since PrP sequence variations strongly influence TSE susceptibility in non-cervid species, it was of interest to compare conversions of the various cervid PrP variants. Open reading frames encoding each of these variants were cloned and expressed in tissue culture cells. Analysis of the expressed PrP-sen molecules by immunoblotting showed that they had similar size ranges, glycoform ratios and expression levels (Figure 2A). For conversion reactions the [35S]PrP-sen molecules were labeled in the presence of a glycosylation inhibitor, tunicamycin, to simplify the banding pattern on SDS–polyacrylamide gels (Figure 3A); in previous studies, a lack of N-linked glycans had little or no influence on the species specificity of conversion reactions (Raymond et al., 1997).
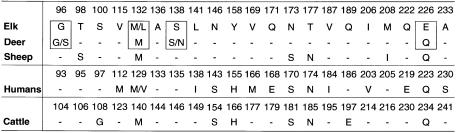
Fig. 1. PrP amino acid sequence variations at residues that differ amongst Rocky Mountain elk (Cervus elaphus nelsoni), mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), sheep (ov-AQ), humans (hu-M or hu-V at residue 129) and cattle (bo). All the known cervid, human and bovine polymorphisms are represented, but only one of 11 known sheep allelic forms is shown (see text). Variations amongst cervids occur at residues 96, 132, 138 and 226 (boxed). In elk, these residues are either G, M, S and E (e-GMSE) or G, L, S and E (e-GLSE), respectively (O’Rourke et al., 1999). Corresponding residues G, M, S and Q (md/wd-GMSQ) and G, M, N and Q (md/wd-GMNQ) are found in both mule deer and white-tailed deer, while to date the residues S, M, S and Q (wd-SMSQ) have been found only in white-tailed deer. Corresponding residue numbers for each species are provided above the single letter amino acid codes.
Fig. 2. (A) Expression of cervid PrP-sen variants in tissue culture cells. Immunoblot of lysates of cells of hamster origin transfected with the expression vector alone or vectors containing cervid PrP genes of the designated genotype (defined in the legend to Figure 1). The multiple bands are similar to those seen with other species’ PrP-sen and are due to variable Asn-linked glycosylation. (B) Immunoblot of PrP-res preparations (±PK treatment) from scrapie-infected hamsters (ha-PrPSc) (a positive control for quantitation) and CWD-infected elk (e-PrPCWD), mule deer (md-PrPCWD) and white-tailed deer (wd-PrPCWD). The arrowhead designates the ∼19 kDa unglycosylated band. The bands above the 40 kDa marker are likely to represent covalently linked or SDS-insoluble PrP-res multimers that are commonly seen in gels of PrP-res from various species. In each case, the lighter of the two lanes represents a 3-fold dilution of the darker lane, which in the case of ha-PrPSc contained 150 ng of PrP-res. The primary antiserum (R505, provided by J.Langeveld, ID-Lelystad, The Netherlands) was raised against a synthetic peptide corresponding to cervid residues 100–111, a sequence conserved amongst cervids, rodents and other species (van Keulen et al., 1995; Raymond et al., 1997). Positions of molecular mass markers are designated in kDa.
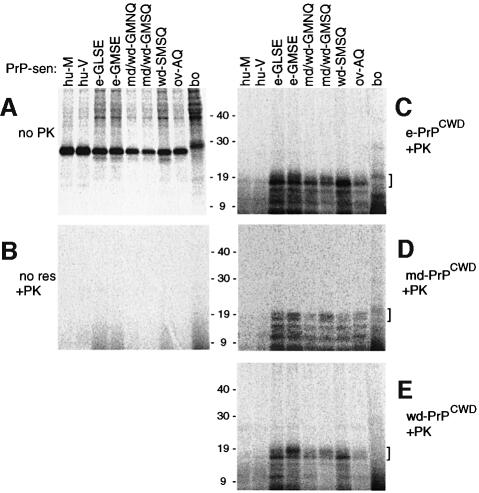
Fig. 3. Cell-free conversion reactions driven by 250 ng of PrPCWD from cervids. (A) Immunoprecipitates of [35S]PrP-sen of the designated species/polymorphism without PK digestion. (B) Negative control, PK-digested conversion reactions in which PrP-res was omitted. (C–E) PK-digested conversion products from reactions driven by e-PrPCWD, md-PrPCWD and wd-PrPCWD, respectively. Brackets on the right side of panels indicate the 17–20 kDa conversion products that were used in the quantitation shown in Figure 4. The lanes in (B)–(E) contain 10 times the reaction equivalents loaded in the lanes of (A).
PrPCWD-induced conversions of cervid PrP-sen variants
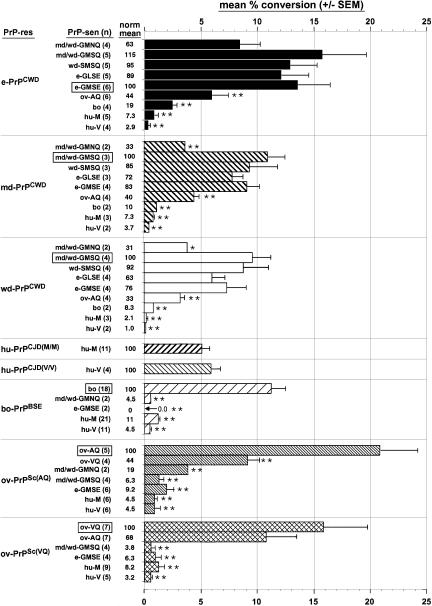
Unlabeled PrPCWD was purified from brain tissue pools of either CWD-affected mule deer, white-tailed deer or elk, and immunoblotting analysis showed proteinase K (PK)-resistant bands of the expected size (∼19–30 kDa) (Figure 2B). Conversion reactions between the various cervid [35S]PrP-sen and PrPCWD molecules generated 17–19 kDa, PK-resistant [35S]PrP products similar to those seen in conversion reactions with other PrP species (Kocisko et al., 1994; Raymond et al., 1997), i.e. 6–8 kDa lower in apparent molecular weight than the full-length [35S]PrP-sen substrate (marked with brackets in Figure 3C–E). These products were also the size expected for unglycosylated, PK-treated PrP-res derived from brain (arrowhead in Figure 2B). Smaller PK-resistant [35S]PrP conversion products were also observed but the formation of these products is not as species-dependent as the formation of the larger products (Kocisko et al., 1995; Chabry et al., 1999). The conversion efficiency (i.e. percentage of the input [35S]PrP-sen converted to the 17–19 kDa PK-resistant [35S]PrP bands) is shown in Figure 4. PrPCWD from elk induced the most efficient conversions of each of the cervid [35S]PrP-sen variants but deer PrPCWD preparations also gave strong conversions of these molecules. The conversions of the mule deer/white-tailed deer (md/wd)-GMNQ variant PrP-sen were least efficient amongst the inter-cervid conversion reactions even in the reactions induced by md-PrPCWD predominantly from md/wd-GMNQ/GMSQ heterozygotes (see Materials and methods). Otherwise, there was no apparent statistically significant advantage in having exact sequence homology between the PrPCWD and the cervid [35S]PrP-sen molecules. These results indicated that PrPCWD induces substantial conversion of all cervid PrP-sen variants to the protease-resistant form.
Fig. 4. Conversion efficiencies of [35S]PrP-sen proteins in cell-free reactions with equivalent amounts of different PrP-res molecules. The results show the mean percentage of the input 24–27 kDa [35S]PrP-sen converted to the 17–20 kDa PK-resistant [35S]PrP bands (an apparent doublet) as described in the text. The number of separate determinations for each combination (n) is shown in parentheses. The results include determinations obtained with two PrPCWD preparations from each pool of cervid brain tissue, which gave similar results. PrPCJD-M/M and PrPCJD-V/V designate human PrP-res derived from the brains of CJD patients homozygous for hu-M or hu-V PrP, respectively. Within the groups of values for each type of PrP-res, the statistical significance of the difference of the means relative to the maximum mean percentage conversion of PrP-sen of the same species (boxed) was assessed with a one way ANOVA by Dunnett’s multiple comparison test: *P <0.05; **P <0.01. The ‘norm mean’ column shows means within each PrP-res group normalized to the homologous conversion (boxed).
PrPCWD-induced conversions of human PrP-sen variants
Considering that hunters and other humans are potentially exposed to CWD infectivity, we tested the efficiency of conversion reactions between PrPCWD and human PrP-sen molecules. Wild-type human PrP has two common allelic forms that encode either methionine (hu-M) or valine (hu-V) at codon 129. This human codon (No. 129) corresponds to a polymorphic codon (No. 132) of elk (Figure 1). Although discrete ∼18 kDa, PK-resistant [35S]PrP products were sometimes visible in reactions of [35S]hu-M PrP-sen or [35S]hu-V PrP-sen with all of the three cervid PrPCWD preparations, the efficiencies of these conversions were >14-fold lower than the inter-cervid conversion reactions (Figures 3C–E and 4) and >5-fold weaker than the homologous conversion reactions induced by human PrP-res from the brains of Creutzfeldt–Jakob disease (CJD) patients homozygous for hu-M or hu-V PrP (Figure 4). These results provide evidence for very weak compatibility between PrPCWD and human PrP-sen molecules in conversion reactions. Although the mean conversion efficiency of hu-M was higher than hu-V for each type of PrPCWD, the difference between these means was not statistically significant by the Mann–Whitney U test.
PrPCWD-induced conversions of bovine PrP-sen
Since cattle can share range land with deer and elk in CWD endemic areas, we tested the efficiency of PrPCWD-induced conversions of bovine (bo) [35S]PrP-sen. An ∼20 kDa band was observed occasionally as a PK-resistant conversion product, but the efficiency of these conversions was an average of at least 5- to 12-fold weaker than the corresponding inter-cervid conversion reactions and the homologous conversion of bovine [35S]PrP-sen induced by BSE-associated PrP-res (PrPBSE) (Figures 3C–E and 4).
PrPCWD-induced conversions of ovine PrP-sen
Both domestic and bighorn sheep (Ovis canadensis) can also share habitats with CWD-infected cervids, so we tested PrPCWD-induced conversions of ovine PrP-sen. Ovine PrP exists in at least 11 allelic forms, several of which have been tested in previous cell-free conversion studies using ov-PrPSc and PrPBSE (Bossers et al., 1997, 2000; Raymond et al., 1997). In this study, we used ovine-AQ [35S]PrP-sen (Figure 1). This allele is associated with scrapie susceptibility in domestic sheep (Hunter et al., 1994) and is identical to the sequence found in bighorn sheep (K.O’Rourke, unpublished data). The PrPCWD-induced conversions of [35S]ov-PrP-AQ were less than half as efficient as the corresponding homologous cervid and ovine PrP reactions but several fold more efficient than the corresponding PrPCWD-induced conversions of bovine and human [35S]PrP-sen molecules (Figures 3C–E and 4).
Cross-species conversions induced by ov-PrPSc(AQ), ov-PrPSc(VQ), hu-PrPCJD(M/M), hu-PrPCJD(V/V) and PrPBSE
For comparison with the PrPCWD-induced conversions, selected cross-species conversions induced by ovine, human and bovine PrP-res were performed (Figure 4). Of the ovine and bovine PrP-res types, ov-PrPSc(AQ) induced the strongest conversion of cervid PrP-sen molecules, but even these conversions were at least 5-fold less efficient than the homologous conversion of ov-AQ [35S]PrP-sen. Conversions of human [35S]PrP-sen molecules (hu-M and hu-V) by PrPBSE and ov-PrPSc(VQ) were reported in a previous study (Raymond et al., 1997). More replicates of the previous conversions as well as conversions of hu-M and hu-V PrP-sen induced by ov-PrPSc(AQ) are shown in Figure 4. These conversions of human PrP-sen by bovine and ovine PrP-res are all >10-fold less efficient compared with the corresponding homologous conversions and are close to the limit of detection for conversion product.
Discussion
Comparing cell-free conversion efficiencies and transmissibilities
The cell-free PrP conversion reaction is a test of the molecular compatibility between PrP-res and PrP-sen of different sequences. While this compatibility seems necessary for TSE disease transmission in vivo, it is clearly not sufficient. Other factors such as dose, strain and route of infection, the stability of the infectivity inside and outside the host and the efficiency of its delivery to the nervous system are also important in determining actual transmission rates between species. In assessing the correlation between cell-free conversion efficiency and interspecies transmissibility, the biggest challenge is the quantitative comparison of transmissibility. We anticipate that the cell-free conversion efficiency would correlate best quantitatively with relative intracerebral transmission titers between the relevant species. Use of intracerebral inoculations would bypass most of the other potentially important factors mentioned above that might be more pronounced in peripheral infections. Relative infectivity titer is the most significant practical parameter in comparing infectious doses between species. Unfortunately, there are few bioassay titration data available for interspecies transmissions.
Nonetheless, in side by side tests performed with multiple types of PrP-res, many fold (5 to >50) stronger conversion efficiencies have been observed with PrP-sen molecules from highly susceptible animals than with those from clearly resistant species/genotypes (Kocisko et al., 1995; Bossers et al., 1997, 2000; Raymond et al., 1997; Chabry et al., 1999). Intermediate efficiencies (2- to 4-fold weaker than homologous) have been observed with PrP-sen from animals that are susceptible but apparently less so than the original host species. For instance, quantitative comparisons are available for relative transmissibilities of BSE to cattle and mice. In this case, BSE infectivity titers per gram are 103 higher when inoculated into cattle compared with mice (Bradley, 1999), while in cell-free reactions PrPBSE induces the conversion of bovine PrP-sen ∼3- to 4-fold more efficiently than murine PrP-sen (Raymond et al., 1997). The ratio of intracerebral infectivity titers of hamster 263K scrapie in Syrian hamsters versus RML mice is at least 107, while the 263K PrPSc-induced conversion of hamster PrP-sen is at least 7-fold more efficient than mouse PrP-sen under conditions similar to those used in the present study (Kocisko et al., 1995; Chabry et al., 1999). The ratio of intracerebral infectivity titers of Chandler mouse scrapie in RML mice versus Syrian hamsters is ∼105 while the mouse PrP-res-induced conversion of hamster PrP-sen is ≥5-fold less efficient than mouse PrP-sen (Kocisko et al., 1995; Chabry et al., 1999). Based on the available information, it seems that the log of the relative intracerebral transmission titer might be roughly proportional to the relative cell-free conversion efficiency on a linear scale. However, far more quantitative transmission data between various species will be required to establish the fit between these parameters.
Interpretation of PrPCWD-induced conversion reactions
The present data show that there is little effect of inter-cervid variations in PrP sequence on the efficiency of the PrPCWD-induced conversion reactions other than a somewhat reduced conversion efficiency of md/wd-GMNQ PrP-sen. In contrast, lower efficiencies were observed with PrPCWD-induced conversions of the PrP-sen of non-cervids. Since the rank order efficiency of all the PrPCWD-induced conversions of various aglycosyl PrP-sen molecules is e-GLSE, e-GMSE, md/wd-GMSQ, wd-SMSQ > md/wd-GMNQ, ov-AQ > bo > hu-M, hu-V, we expect that the same rank order would also apply to the relative susceptibilities of species expressing those PrP-sen variants to equivalent exposures to CWD infectivity. Thus, these data suggest that CWD transmission amongst mule deer, white-tailed deer and elk will not be limited by PrP sequence differences between cervids. The moderate efficiency of conversion of ov-AQ PrP-sen suggests that CWD transmission to domestic or bighorn sheep of this genotype would be reduced, but not eliminated by incompatibility with PrPCWD. However, it should be noted that no transmissions have been documented yet, and that bighorn sheep held with CWD-infected cervids have not developed TSE disease (M.W.Miller, unpublished data). For cattle and humans, it is likely that susceptibility to CWD would be severely, but perhaps not completely, limited by the lack of conversion compatibility of the respective PrP isoforms. It is uncertain how these low levels of conversion compare with the conversion of PrP-sen from a truly resistant host species because no such species has been documented for CWD. Nonetheless, discrete conversion products were sometimes observed, albeit very weakly, in human PrP-sen reactions with ovine PrPSc, PrPBSE or PrPCWD. This suggests that after exposure of humans to scrapie, BSE or CWD, potentially pathogenic conversion to human PrP-res might follow, if only rarely or inefficiently.
Clearly, it is premature to draw firm conclusions about CWD passing naturally into humans, cattle and sheep, but the present results suggest that CWD transmissions to humans would be as limited by PrP incompatibility as transmissions of BSE or sheep scrapie to humans. Although there is no evidence that sheep scrapie has affected humans, it is likely that BSE has caused variant CJD in ∼74 people (definite and probable variant CJD cases to date according to the UK CJD Surveillance Unit). Given the presumably large number of people exposed to BSE infectivity, the susceptibility of humans may still be very low compared with cattle, which would be consistent with the relatively inefficient conversion of human PrP-sen by PrPBSE. Nonetheless, since humans have apparently been infected by BSE, it would seem prudent to take reasonable measures to limit exposure of humans (as well as sheep and cattle) to CWD infectivity as has been recommended for other animal TSEs.
Materials and methods
Cloning and expression of cervid PrP genes
Six cervid PrP open reading frames [two elk, three deer (white-tailed deer/mule deer); Figure 1] were cloned and expressed as follows. Open reading frames were PCR amplified from genomic DNA using either primer set 1 (5′-CAGGTTAACACCCTCTTTATTTTGCAG-3′ and 5′-ACCTCTAGAAGATAATGAAAACAGGAAG-3′) or the degenerated primer set 2 [5′-CAGGCCGGCCACCATGGTGAAAAGCCA (C/T)-3′ and 5′-CAGGCCGGCCACCATGGCGAACCTT(A/G)G-3′ as forward primers, and 5′-ACCTCTAGACCTATCCTACTATGAGAAAAATGAGG-3′ as the reverse primer]. Oligonucleotides of primer set 1 hybridize 11–30 nucleotides pre- or 8–29 nucleotides post-open reading frame respectively, while oligonucleotides of primer set 2 hybridize at the first five or the last eight codons of the PrP open reading frames respectively. Primer set 2 was only used to obtain PrP PCR product of the white-tailed deer 138N variant. Open reading frames were subsequently cloned (HpaI–XbaI) in cloning vector pTZnot, a derivative of pTZ18R (VecBase accession No. VB0071). The sequences of both strands were confirmed using an ABI-373A sequencer.
PrP allelic variants of elk (e-GLSE and e-GMSE; DDBJ/EMBL/GenBank accession Nos AF156182 and AF156183, respectively) and deer (wd-SMSQ, md/wd-GMSQ and md/wd-GMNQ; accession Nos AF156184–6, respectively) were subcloned (NotI–SalI) into expression vector pECV7 (NotI–XhoI) as described previously (Bossers et al., 1997; Raymond et al., 1997). Since the md/wd-GMNQ allelic variant (obtained from white-tailed deer in this case) contained an incomplete N-terminal signal sequence (missing first 11 amino acids), its signal sequence was replaced by the signal sequence of the md/wd-GMSQ variant using the SmaI restriction site at approximately codon 42 of the PrP open reading frame. SalI-linearized expression vectors were electroporated into hamster cells (provided by N.L.Hubrechts), which express little or no endogenous PrP (vector only lane in Figure 2A), and stably transfected single-cell clones were selected for hygromycin-B and high levels of PrP expression by immunoblotting using the R505 anti-peptide antibody (described in the legend to Figure 2).
Labeling and purification of [35S]PrP-sen
[35S]PrP-sen was immunoprecipitated from [35S]methionine-labeled tissue culture cells expressing specific PrP-sen molecules (Kaneko et al., 1997; Raymond et al., 1997). md/wd-GMNQ, md/wd-GMSQ, wd-SMSQ, e-GLSE and e-GMSE [35S]PrP-sen molecules were immunoprecipitated using R521 or R522, polyclonal antisera against sheep peptide 94–105 (van Keulen et al., 1995; provided by J.Langeveld, ID-Lelystad, The Netherlands). [35S]PrP-sen, isolated from tissue culture cells expressing ov-AQ, bo, hu-M and hu-V, was immunoprecipitated as previously described by Raymond et al. (1997).
Cell-free conversion reaction
The protocol used was similar to that described previously (Raymond et al., 1997). Briefly, for each reaction the selected immunopurified [35S]PrP-sen (20 000–50 000 c.p.m.) was incubated with 250 ng of the appropriate unlabeled purified PrP-res (see below). The PrP-res was pretreated with 2.0–2.5 M guanidine hydrochloride (Gdn–HCl) at 37°C for 1–3 h, prior to adding to the [35S]PrP-sen and diluting to 1 M Gdn–HCl in 50 mM sodium citrate pH 6.0, 5 mM cetylpyridinium chloride and 1.25% sarkosyl. The final volume for each reaction was 22 µl and the incubations were at 37°C for 3 days. Post-incubation, 2 µl were removed for the minus PK control and the remaining 20 µl were diluted to 50 µl in 0.05 M Tris–HCl pH 8.0 and 0.13 M NaCl, then treated with 20 µg/ml PK at 37°C for 1 h to determine whether [35S]PrP-sen was converted to PK-resistant forms. Proteins in both the PK-treated and untreated portions were methanol precipitated and run on 16% SDS–polyacrylamide gels (NOVEX) with subsequent autoradiography. Analyses were carried out using a PhosphorImager (Molecular Dynamics).
Purification of PrP-res
PrP-res was isolated from the brains of clinically affected animals as described previously for other types of PrP-res (Raymond et al., 1997; Caughey et al., 1999). In the case of the PrPCWD, brain tissue pools were used. Of the 28 animals contributing to the mule deer brain pool, 24 were GMSQ/GMNQ heterozygotes, three were GMSQ/GMSQ homozygotes and one was of unknown genotype. Of the seven total white-tailed deer brain samples, five were GMSQ/GMSQ homozygotes and two were GMNQ/GMSQ heterozygotes. Of nine total elk brain samples, seven were GMSE/GMSE homozygotes and two unknown. The yields of PrPCWD from the mule deer, white-tailed deer or elk brain pools were ∼3, 6 and 6 µg/g brain equivalent, respectively, as estimated by semi-quantitative immunoblotting. Two independent preparations from each cervid species gave consistent results in conversion reactions.
Acknowledgments
Acknowledgements
We gratefully acknowledge the following: (i) technical assistance of Ruth de Vries and Robert Hayter; (ii) the kind provision of PrPCJD from Drs James Hope and Shu Chen; (iii) critical reading of the manuscript by Drs Bruce Chesebro, Suzette Priola and Kim Hasenkrug; and (iv) graphics assistance by Gary Hettrick, Bob Evans and Anita Golden.
References
- Bessen R.A., Kocisko,D.A., Raymond,G.J., Nandan,S., Lansbury,P.T.,Jr and Caughey,B. (1995) Nongenetic propagation of strain-specific phenotypes of scrapie prion protein. Nature, 375, 698–700. [DOI] [PubMed] [Google Scholar]
- Bossers A., Belt,P.B.G.M., Raymond,G.J., Caughey,B., de Vries,R. and Smits,M.A. (1997) Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl Acad. Sci. USA, 94, 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossers A., de Vries,R. and Smits,M.A. (2000) Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol., 74, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R. (1999) BSE transmission studies with particular reference to blood. Dev. Biol. Stand., 99, 35–40. [PubMed] [Google Scholar]
- Bruce M.E. et al. (1997) Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature, 389, 498–501. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J., Kocisko,D.A. and Lansbury,P.T.,Jr (1997) Scrapie infectivity correlates with converting activity, protease resistance and aggregation of scrapie-associated prion protein in guanidine denaturation studies. J. Virol., 71, 4107–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J., Priola,S.A., Kocisko,D.A., Race,R.E., Bessen,R.A., Lansbury,P.T.,Jr and Chesebro,B. (1999) Methods for studying prion protein (PrP) metabolism and the formation of protease-resistant PrP in cell culture and cell-free systems. Mol. Biotechnol., 13, 45–55. [DOI] [PubMed] [Google Scholar]
- Chabry J., Priola,S.A., Wehrly,K., Nishio,J., Hope,J. and Chesebro,B. (1999) Species-independent inhibition of abnormal prion protein (PrP) formation by a peptide containing a conserved PrP sequence. J. Virol., 73, 6245–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.F., Desbruslais,M., Joiner,S., Sidle,K.C.L., Gowland,I., Collinge,J., Doey,L.J. and Lantos,P. (1997) The same prion strain causes vCJD and BSE. Nature, 389, 448–450. [DOI] [PubMed] [Google Scholar]
- Horiuchi M., Priola,S.A., Chabry,J. and Caughey,B. (2000) Interactions between heterologous forms of prion protein: binding, inhibition of conversion and species barriers. Proc. Natl Acad. Sci. USA, 97, 5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Goldmann,W., Smith,G. and Hope,J. (1994) The association of a codon 136 PrP gene variant with the occurrence of natural scrapie. Arch. Virol., 137, 171–177. [DOI] [PubMed] [Google Scholar]
- Kaneko K., Wille,H., Mehlhorn,I., Zhang,H., Ball,H., Cohen,F.E., Baldwin,M.A. and Prusiner,S.B. (1997) Molecular properties of complexes formed between the prion protein and synthetic peptides. J. Mol. Biol., 270, 574–586. [DOI] [PubMed] [Google Scholar]
- Kocisko D.A., Come,J.H., Priola,S.A., Chesebro,B., Raymond,G.J., Lansbury,P.T. and Caughey,B. (1994) Cell-free formation of protease-resistant prion protein. Nature, 370, 471–474. [DOI] [PubMed] [Google Scholar]
- Kocisko D.A., Priola,S.A., Raymond,G.J., Chesebro,B., Lansbury,P.T.,Jr and Caughey,B. (1995) Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc. Natl Acad. Sci. USA, 92, 3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.W., Williams,E.S., McCarty,C.W., Spraker,T.R., Kreeger,T.J., Larsen,C.T. and Thorne,E.T. (2000) Epidemiology of chronic wasting disease in free-ranging cervids. J. Wildl. Dis., 36, 676–690. [DOI] [PubMed] [Google Scholar]
- O’Rourke K.I., Besser,T.E., Miller,M.W., Cline,T.F., Spraker,T.R., Jenny,A.L., Wild,M.A., Zebarth,G.L. and Williams,E.S. (1999) PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J. Gen. Virol., 80, 2765–2769. [DOI] [PubMed] [Google Scholar]
- Priola S.A. and Chesebro,B. (1995) A single hamster amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J. Virol. 69, 7754–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priola S.A., Caughey,B., Race,R.E. and Chesebro,B. (1994) Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol., 68, 4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. et al. (1990) Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell, 63, 673–686. [DOI] [PubMed] [Google Scholar]
- Raymond G.J. et al. (1997) Molecular assessment of the transmissibilities of BSE and scrapie to humans. Nature, 388, 285–288. [DOI] [PubMed] [Google Scholar]
- van Keulen L.J.M., Schreuder,B.E.C., Meloen,R.H., Poelen-Van Den Berg,M., Mooij-Harkes,G., Vromans,M.E.W. and Langeveld,J.P.M. (1995) Immunohistochemical detection and localization of prion protein in brain tissue of sheep with natural scrapie. Vet. Pathol., 32, 299–308. [DOI] [PubMed] [Google Scholar]