Abstract
Viruses with RNA genomes often capture and redirect host cell components to assist in mechanisms particular to RNA-dependent RNA synthesis. The nidoviruses are an order of positive-stranded RNA viruses, comprising coronaviruses and arteriviruses, that employ a unique strategy of discontinuous transcription, producing a series of subgenomic mRNAs linking a 5′ leader to distal portions of the genome. For the prototype coronavirus mouse hepatitis virus (MHV), heterogeneous nuclear ribonucleoprotein (hnRNP) A1 has been shown to be able to bind in vitro to the negative strand of the intergenic sequence, a cis-acting element found in the leader RNA and preceding each downstream ORF in the genome. hnRNP A1 thus has been proposed as a host factor in MHV transcription. To test this hypothesis genetically, we initially constructed MHV mutants with a very high-affinity hnRNP A1 binding site inserted in place of, or adjacent to, an intergenic sequence in the MHV genome. This inserted hnRNP A1 binding site was not able to functionally replace, or enhance transcription from, the intergenic sequence. This finding led us to test more directly the role of hnRNP A1 by analysis of MHV replication and RNA synthesis in a murine cell line that does not express this protein. The cellular absence of hnRNP A1 had no detectable effect on the production of infectious virus, the synthesis of genomic RNA, or the quantity or quality of subgenomic mRNAs. These results strongly suggest that hnRNP A1 is not a required host factor for MHV discontinuous transcription or genome replication.
RNA viruses constitute a major class of animal and plant pathogens. All members of this group (with the exception of retroviruses) encode RNA-dependent RNA polymerases (RdRPs) necessary for replication and, in some cases, transcription of their genomes. Despite this common requirement, the strategies that RNA viruses use to synthesize RNA are highly diverse and are intimately linked to how genes are organized within the viral genome, and whether the genome is positive-stranded (i.e., message-sense), negative-stranded, or double-stranded. In a number of cases, it has been demonstrated that host cellular proteins play critical roles in viral RNA-dependent RNA synthesis (1, 2). Strong evidence for host factor participation comes from biochemical studies of viral RdRPs (3) and genetic analyses of particular viruses and their hosts or surrogate hosts (4). In addition, putative host factors are often identified on the basis of their in vitro binding to cis-acting regulatory elements in the viral genome (1).
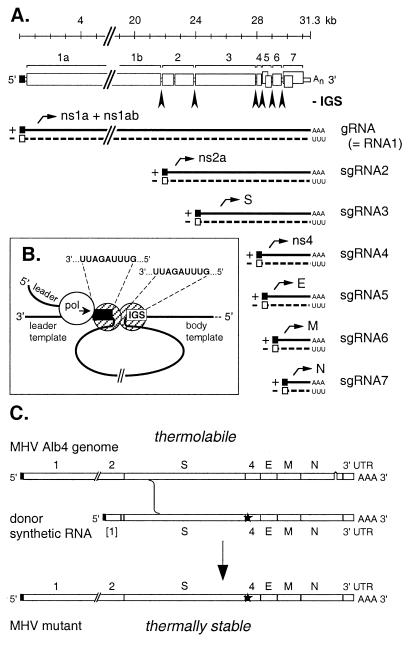
Coronaviruses and arteriviruses, the families that make up the order Nidovirales, are positive-stranded RNA viruses that use a unique scheme of discontinuous transcription as a means of gene expression (5). This process has been well studied for the coronavirus mouse hepatitis virus (MHV), but its underlying mechanism has not been resolved, in large part because of the enormous size of the MHV genome (31 kb) and the extreme difficulty of establishing a robust system for in vitro RNA synthesis. In vivo, initiation of MHV infection deposits the genome in the host cell cytoplasm. Here, it first acts as an mRNA, its 22-kb gene 1 being translated into the viral RdRP (the products of polyproteins ns1a and ns1ab) (Fig. 1A). The polymerase, which assembles on the cytoplasmic face of intracellular membranes, then copies the genome into a full-length negative strand that serves as the template for genome amplification. Expression of the downstream-encoded structural proteins depends on subsequent synthesis of a 3′-coterminal set of subgenomic (sg) RNAs, each consisting of the 70-nt leader sequence from the 5′ end of the genome fused, at an intergenic sequence (IGS), to the 3′ end of the genome. The IGSs contain a consensus nonanucleotide, 5′-AAUCUAAAC-3′ (in the positive sense) that is identical to the 3′ end of the leader sequence. Because the IGSs precede the principal ORFs of the genome, each sgRNA is a functionally monocistronic mRNA for the translation of the structural proteins S, E, M, or N or the nonstructural proteins ns2a or ns4 (Fig. 1A). Two points about nidovirus sgRNA formation are generally accepted. First, sgRNAs are created by some type of discontinuous synthesis, rather than by splicing of a genomic precursor (5). Second, at least one stage of their synthesis requires base-pairing interactions between the positive strand of the leader and the negative strand of the IGS (5–7). Potentially, leader–body fusion could be aided by a looping out of the transcription template mediated by proteins bound to the common regions of the leader and IGS (Fig. 1B). However, the exact nature of the discontinuous leader–body fusion step, whether it occurs during positive-strand or negative-strand RNA synthesis, and the significance of the negative-strand counterparts of the sgRNAs are all matters of controversy (8–10).
Figure 1.
Summary of coronavirus RNA synthesis and genetics. (A) A schematic of the gene organization of the MHV genome is shown at the top. Intergenic sequences (IGSs) are marked by arrowheads. Beneath this are represented the positive-stranded genomic RNA (gRNA) and subgenomic RNA species (sgRNA2–7), as well as their negative-strand counterparts, all of which are synthesized in the cytoplasm during the course of infection. Leader and antileader sequences are indicated by ■ and □, respectively. (B) A model for MHV discontinuous transcription mediated by looping out of the template. In this scheme, fusion of the leader to the body of an sgRNA occurs via quasicontinuous synthesis across a junction created by dimerization of proteins bound to identical sequences found at the end of the leader template and at the IGS. (C) Construction of site-specific mutations in the MHV genome by targeted RNA recombination. Mutations (★) are transduced into the MHV genome via in vivo recombination between a synthetic donor RNA and the genome of a recipient virus, the N gene deletion mutant Alb4, which can be selected against on the basis of its thermolability. UTR, untranslated region.
Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 has recently been identified as a host cellular component that binds in vitro to probe RNAs containing the negative-sense strand of the MHV IGS (11, 12). This protein is one member of a large set of proteins that bind to cellular pre-mRNA shortly after transcription and function in mRNA maturation and transport. In particular, hnRNP A1 has been shown to participate in alternative splice site selection, mRNA export from the nucleus to the cytoplasm, and telomere biogenesis and maintenance (13–17). A number of properties of hnRNP A1 make it very attractive as a potential host factor integrating proposed steps in discontinuous transcription: its putative specificity as an RNA-binding protein, its RNA strand-annealing activity, and its propensity to dimerize. These functions would ideally contribute to the template looping-out and strand-switching events contemplated by one model of MHV transcription (Fig. 1B; refs. 1, 11, and 12). Two molecules of hnRNP A1, bound, respectively, to the negative strand of the leader template and to the negative strand of an IGS, could dimerize to bring about association of these two distant parts of the antigenome. Subsequently, one or both hnRNP A1 monomers could assist in annealing of the nascent leader RNA to the IGS template before its elongation into an sgRNA.
However, despite its congruity with some requirements of the model, the possible correlation between hnRNP A1 binding to the IGS and MHV transcription has not been extensively examined. As yet, two transcriptionally nonfunctional mutant IGS sequences have been shown to bind hnRNP A1 less well than the wild-type sequence (11, 12). More generally, it has been difficult to define the sequence specificity of hnRNP A1 binding to biologically relevant RNA substrates (18–20). In addition, the significance of hnRNP A1 binding to the negative-strand IGS is based on the presupposition that sgRNA leader–body fusion occurs during positive-strand RNA synthesis, whereas there is considerable evidence that the fusion event occurs during negative-strand RNA synthesis (5, 7, 9).
To examine the relationship of hnRNP A1 to MHV RNA synthesis in vivo, we took a genetic approach. From the standpoint of the virus, we tested whether an IGS preceding a nonessential gene could be functionally replaced by an engineered high-affinity hnRNP A1 binding site or whether insertion of this site adjacent to the same IGS would enhance transcription or skew the locus of leader–body fusion. Our results implied that an hnRNP A1 binding site alone is not sufficient to determine MHV sgRNA synthesis. More definitively, from the standpoint of the host cell, we compared MHV replication in a mutant cell line that does not express hnRNP A1 with that in the same cell line in which this candidate host factor was restored to wild-type levels. Our results strongly suggested that hnRNP A1 is not necessary for MHV replication, and its absence has no detectable quantitative or qualitative effect on MHV RNA synthesis.
Materials and Methods
Cells and Virus.
Wild-type MHV strain A59 and all mutant virus stocks were grown in mouse 17 clone 1 (17Cl1) cells. The mouse erythroleukemia CB3 and CB3A cell lines (17, 21), kindly provided by Benoit Chabot (Université de Sherbrooke, Québec, Canada) were carried in suspension in medium containing 400 μg/ml G418 (GIBCO/BRL). To measure single-step growth, CB3 and CB3A cells were harvested at 6–8 × 105 cells per ml and infected with wild-type MHV at a multiplicity of 1 plaque-forming unit (pfu) per cell. After incubation at 37°C for 2 h, cells were washed twice and resuspended in 50 ml of medium. At indicated times after infection, aliquots were taken, and titers of released virus were determined by plaque assay.
Plasmid Constructs.
Plasmid templates for donor RNAs for targeted recombination were derived from pCFS8 (22), which encodes a 7.9-kb T7 RNA polymerase transcript containing all MHV ORFs from the S gene (gene 3) through the 3′ end of the genome (Fig. 1). A 31-bp region of pCFS8 encompassing IGS4 was replaced with a linker eliminating the IGS and flanked by BspEI and RsrII sites. The BspEI–RsrII segment was then replaced with oligonucleotide cassettes to construct plasmids pXS13 and pXS17, which correspond to the win and igs-win mutants, respectively. For pXS13, the cassette replacement (in the positive sense) was 5′-CCGGCACCCUAAGUCCCUAUCAUA-3′, and for pXS17, it was 5′-CCGGACAGAAaaucuaaacAAUUUACACCCUAAGUCCCUAUCAUA-3′; the complement of the winner sequence is underlined, and the consensus IGS motif is indicated in lowercase. The sequences of all constructs were confirmed by automated DNA sequencing.
A plasmid, pGA, was constructed for in vitro synthesis of mouse hnRNP A1 mRNA. RNA from 17Cl1 cells was reverse transcribed with a negative-sense primer, 5′-TCCTCTCCTGCTAAGCTT-3′, and this primer and positive-sense primer 5′-CGGGGATCCACCGTCATGTCTAAG-3′ were used for PCR amplification. The 1,016-bp cDNA product was digested with BamHI and HindIII and inserted into pGEM3ZF(+) (Promega) downstream of the T7 promoter. The sequence of the insert, containing the 963-bp hnRNP A1 ORF and flanking regions, was found to be identical to that reported previously (GenBank accession no. M99167; ref. 21). Capped RNA transcripts were synthesized by using a T7 RNA polymerase kit (Ambion, Austin, TX).
Site-Directed Mutagenesis of MHV.
Targeted RNA recombination was carried out as described (23, 24). Briefly, mouse L2 spinner cells were infected with the temperature-sensitive MHV mutant Alb4 at 33°C for 2 h. Infected cells were then transfected with in vitro-synthesized donor RNA by electroporation and plated onto monolayers of 17Cl1 cells, which were incubated at 33°C for a further 44 h. Candidate recombinants were identified as viruses able to form wild-type-sized plaques at the nonpermissive temperature (39°C; ref. 24). Recombinants were screened for the presence of mutations in the IGS4 region and the absence of the Alb4 N gene deletion by reverse transcription–PCR (RT-PCR) of RNA isolated from infected 17Cl1 cells. Final verification of site-directed mutations was obtained by direct RNA sequencing of MHV genomic RNA extracted from virions purified by two cycles of equilibrium centrifugation on potassium tartrate gradients as described (25).
Leader–body junctions of wild-type and mutant sgRNA4 were determined by RT-PCR of RNA isolated from infected cells by using primers specific for gene 4 and the leader (22). PCR products were cloned into the vector pCRII-TOPO (Invitrogen), and the inserts of multiple independent clones were sequenced.
Immunoprecipitation and Western Blotting.
Mouse L2 spinner, CB3, and CB3A cells were harvested at 6–8 × 105 cells per ml and lysed in buffer containing 10 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.65% Nonidet P-40, 1.25 mM PMSF, 0.01% sodium azide, 1 μM pepstatin A, 1 μM leupeptin, and 0.25 μM aprotinin. Cell lysates were used directly or were immunoprecipitated with mAb 9H10 (kindly provided by Gideon Dreyfuss, University of Pennsylvania, Philadelphia; ref. 19) and protein A Sepharose (Amersham Pharmacia) at 4°C for 1 h, and analyzed by 12.5% SDS/PAGE. Proteins were transferred to polyvinylidine difluoride membranes (Millipore) and probed with the same mAb, and bound antibody was detected by enhanced chemiluminescence (ECL kit, Amersham Pharmacia). A standard for hnRNP A1 was produced by in vitro translation of synthetic hnRNP A1 RNA in a rabbit reticulocyte lysate (Amersham Pharmacia) labeled with 900 μCi/ml [35S]methionine (Amersham Pharmacia; 1 Ci = 37 GBq). Radiolabeled hnRNP A1 was detected by fluorography after decay of the chemiluminescent signal.
Metabolic Labeling of RNA.
Viral RNA was labeled metabolically essentially as described (26). Briefly, 17Cl1 cells infected at a multiplicity of 5 pfu per cell were labeled with 100 μCi/ml [33P]orthophosphate (ICN) in the presence of 20 μg/ml actinomycin D, which inhibits cellular RNA synthesis, from 6 through 8 h after infection. For CB3 and CB3A cells, as well as L2 cells in suspension, cells infected at a multiplicity of 1 pfu per cell were labeled with 100 μCi/ml [5,6-3H]uridine (Amersham Pharmacia) in the presence of 20 μg/ml actinomycin D from 6 through 10 h after infection. Isolated RNA was analyzed by electrophoresis in 1.0% agarose containing formaldehyde and visualized by fluorography; 33P-labeled RNA was quantitated with a PhosphorImager (Molecular Dynamics).
Results
Generation and Characterization of MHV win and igs4-win Mutants.
To test the in vivo functionality of hnRNP A1 during MHV transcription, we sought to examine whether directed binding of this protein to a transcriptionally active region within the MHV genome could substitute for the presence of an IGS or confer an advantage to sgRNA synthesis from an adjacent IGS. Although it is not resolved how hnRNP A1 discriminates among biological substrates with different sequences, one highly specific ligand named “winner” (19) has been identified by a randomized RNA selection and amplification technique (27). Even though it corresponds to no known cellular RNA sequence, the winner 20-mer, 5′-UAUGAUAGGGACUUAGGGUG-3′, has a 10-fold greater affinity for hnRNP A1 than any other RNA tested (19, 20). Moreover, it has been shown that winner is able to replace functionally the exonic splicing silencer in exon 2 of the tat gene of HIV-1 (15). Winner can also capture hnRNP A1 for cooperation with HIV-1 Rev protein in nuclear export of unspliced mRNA (16). These demonstrations provide precedents, then, for the use of winner to recruit hnRNP A1 for bona fide biological roles.
We incorporated the winner sequence into the MHV genome by targeted RNA recombination (28). Site-directed mutagenesis of positive-stranded RNA viruses is usually carried out by manipulation of genomic cDNA, from which infectious RNA can be transcribed (29, 30). It has not yet been possible to assemble a full-length cDNA clone for MHV, which has the largest known RNA viral genome; however, a full-length clone of another coronavirus has recently been reported (31). Full-length clones have also been used to study arteriviruses, which have genomes on the order of 13 kb. For MHV, we have used an alternative method that exploits the high rate of coronavirus RNA–RNA recombination to engineer specific mutations throughout the entire structural gene region of the 31-kb genome (23, 24, 28). In this technique, new mutants are constructed from a conditional lethal mutant, Alb4, which has a deletion within the N gene, encoding the viral nucleocapsid protein. Donor synthetic RNA containing the mutations of interest, as well as restoring the deleted portion of the N gene, is transfected into Alb4-infected cells, and recombinants are then isolated by counter-selecting the thermolabile Alb4 parent (Fig. 1C).
Two mutants were generated in this manner (Fig. 2): one, designated win, in which winner completely replaced IGS4, and a second, igs4-win, with winner immediately downstream of IGS4. In each case, the complement of the winner 20-mer (above) was incorporated into the positive-strand genome (Fig. 2) such that winner would be present in the negative strand on genome replication, in accord with the reported binding of hnRNP A1 to the negative strand of the IGS in vitro (11, 12). The IGS4 region was chosen for these constructs because gene 4 is absolutely nonessential for MHV growth (22), and neither mutant had any aberration in growth phenotype detectable on the basis of plaque size or infectious titer.
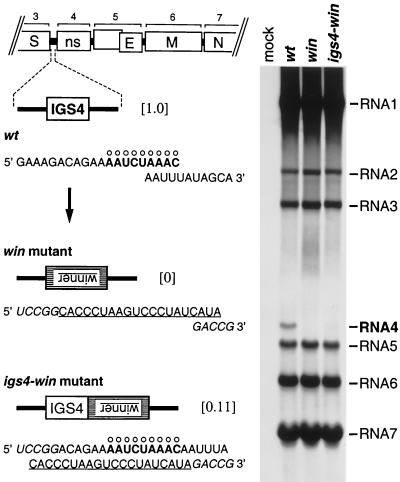
Figure 2.
Insertion of a high-affinity hnRNP A1 binding site in place of IGS4 or adjacent to IGS4. Left shows the sequences, in the positive sense, of the IGS4 regions of wild-type MHV (wt), the win mutant, and the igs4-win mutant. The consensus IGS motif is marked with ○, and the complement of the winner sequence is underlined. Residues derived from the BspEI and RsrII sites, used to construct the donor RNA transcription vectors for the mutants, are indicated in italics. The numbers in brackets are the relative levels of RNA4 synthesized by each virus. Right shows viral RNA metabolically labeled with [33P]orthophosphate, in the presence of actinomycin D, in 17Cl1 cells that were mock-infected or infected with wild-type MHV, the win mutant, or the igs4-win mutant. RNA was separated by agarose gel electrophoresis and visualized by fluorography.
Metabolic labeling of wild-type- and mutant-infected cells (Fig. 2) revealed the standard pattern of RNA synthesis for wild-type MHV (23, 26): sgRNA4, the least abundant MHV transcript, was present at a 22-fold lower molar level than sgRNA7, the most abundant transcript. Cells infected with the win mutant produced absolutely no sgRNA4 (Fig. 2), a result confirmed by RT-PCR. Cells infected with the igs4-win mutant exhibited a 9-fold decrease in sgRNA4 synthesis compared with wild-type MHV (Fig. 2), and sequencing of eight cloned RT-PCR products from this transcript population showed that all had the canonical 5′-AAUCUAAAC-3′ leader–body fusion junction. Thus, in the win mutant, the inserted hnRNP A1 binding site did not function as an IGS, nor did it perturb the transcription pattern at other IGSs relative to the wild type. In the igs4-win mutant, the inserted winner sequence did not enhance transcription of sgRNA4, nor did it displace the site of leader–body fusion. These results, although negative, prompted us to more stringently test the proposed role of hnRNP A1 in MHV RNA synthesis.
MHV Replication and RNA Synthesis in a Cell Line That Does Not Express hnRNP A1.
To determine more conclusively whether the availability of hnRNP A1 affects RNA synthesis by MHV, we obtained two cell lines, CB3 and CB3A, that differ only in their ability to express this protein. CB3 is a murine BALB/c erythroleukemia cell line that has a retroviral insertion near the Hnrpa1 gene, encoding hnRNP A1, and it has lost the second Hnrpa1 allele (21). As a result of these alterations, CB3 cells produce no detectable hnRNP A1 protein and have a 200- to 500-fold reduction in hnRNP A1 mRNA (13, 17). CB3 cells have been stably transformed with a retroviral vector encoding neomycin resistance. CB3A cells have been transformed with the same vector containing the murine hnRNP A1 coding sequence (17). The lack of hnRNP A1 in CB3 cells has been documented to result in defects in alternative splice site selection (13, 32) and maintenance of telomere length (17). Whereas the loss of hnRNP A1 is not lethal (21), it is disadvantageous; in our hands, CB3 cells exhibit somewhat slower growth kinetics (a doubling time of 17.5 h) compared with that of CB3A cells (14 h).
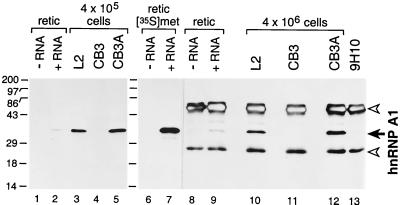
Initially, we confirmed the phenotype of CB3 cells by Western blotting, either with or without prior immunoprecipitation, by using a mAb specific for hnRNP A1 (Fig. 3). No hnRNP A1 protein was detected in as many as 4 × 106 CB3 cells, whereas CB3A cells were seen to have restored hnRNP A1 expression to levels equivalent to those of mouse L2 cells. Additionally, the identity of the 34-kDa protein recognized by the mAb was verified with parallel in vitro translations of synthetic hnRNP A1 mRNA (Fig. 3). It should be noted that the mAb we used can also react with A1B, a 38-kDa minor splice variant of hnRNP A1, expression of which is seen in some cell lines but not others (13, 17). We observed a faint band at 38–39 kDa in the Western blots of all immunoprecipitated samples, but because this also appeared in the reticulocyte control lanes and the control lane of the mAb alone, we do not believe it represents A1B protein.
Figure 3.
CB3 cells produce no detectable hnRNP A1. hnRNP A1 (solid arrow) was detected in cell lysates by Western blotting (lanes 1–5) or by immunoprecipitation followed by Western blotting (lanes 8–13) with mAb 9H10. Blots were visualized by detection of chemiluminescence. Open arrowheads denote IgG heavy and light chains. As a control, rabbit reticulocyte lysate was programmed with synthetic RNA encoding hnRNP A1 (lanes 2, 7, and 9) or with H2O (lanes 1, 6, and 8) and labeled with [35S]methionine; lanes 6 and 7 are a fluorogram of lanes 8 and 9. Molecular masses (kDa) of marker proteins are indicated on the left.
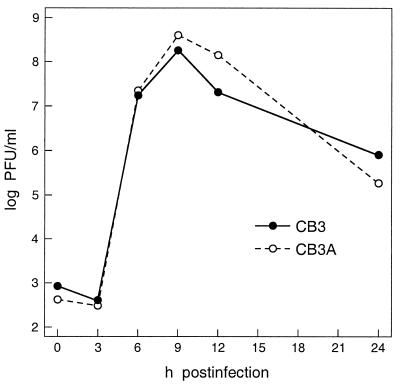
We next examined the ability of MHV to replicate in the two cell lines by a single-step growth assay. The resulting growth curves revealed virtually identical kinetics of viral replication in CB3 and CB3A cells (Fig. 4). The small difference that was observed between the two we attribute to the slower growth rate of CB3 cells. In both cell lines MHV reached maximal titers exceeding 108 pfu/ml at 9 h after infection. It should be noted that these titers are comparable to those obtained with the optimal host cell lines used for tissue culture of this virus. Furthermore, both cell lines showed the same clear signs of the cytopathic effect typical for cytolytic infection by MHV. At earlier times after infection, we observed clustering of cells indicative of syncytia formation in suspension culture. Later in infection, cells swelled to roughly twice their normal size and lysed thereafter. These data argue strongly that hnRNP A1 is not required for MHV growth in vivo.
Figure 4.
Growth of wild-type MHV in CB3 (hnRNP A1-deficient) and CB3A (hnRNP A1-restored) cells. Cells were infected at a multiplicity of 1 pfu per cell for 2 h. After removal of inoculum (t = 0 h), cells were washed and resuspended in medium, and aliquots of supernatant, taken at the indicated times, were titered for infectivity by plaque assay on mouse L2 cells.
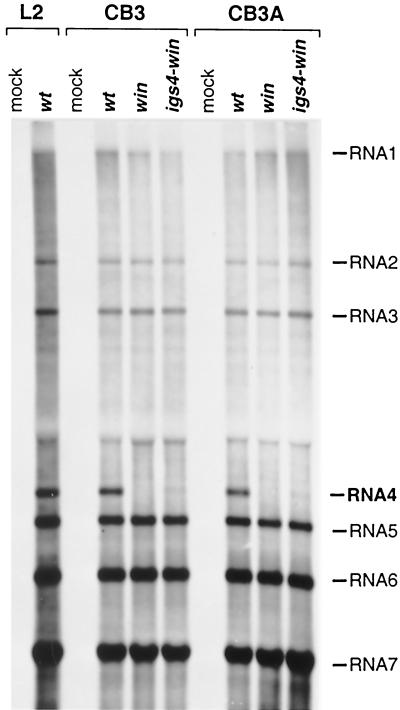
Clearly, MHV would not be able to produce infectious progeny virus in CB3 cells if it were unable to carry out sgRNA synthesis. However, if hnRNP A1 played an auxiliary, rather than essential, role in leader–body fusion, we would expect that its absence would differentially affect the multiple IGSs, which appear in different contexts in the MHV genome and function with different efficiencies. To directly examine MHV RNA synthesis in CB3 and CB3A cells, we metabolically labeled viral RNA in cells infected with wild-type MHV, or with the win or igs4-win mutants. This experiment revealed that there were no detectable differences among the abilities of the cell lines to support wild-type or mutant viral RNA synthesis (Fig. 5). CB3 and CB3A cells both produced the same amounts of sgRNAs and in the same relative ratios. Moreover, their pattern of MHV RNA synthesis was identical to that of L2 cells (Fig. 5 and data not shown) and 17Cl1 cells (Fig. 2). In particular, CB3 cells infected with the igs4-win mutant displayed the same reduced level of sgRNA4 as did cells expressing hnRNP A1. Therefore, this reduction of sgRNA4 was not due to physical hindrance of the RdRP by hnRNP A1 tightly bound to the winner sequence in the antigenome, but, rather, was likely an effect of altering the sequence context of IGS4 by the adjacent insertion of winner. Collectively, the data obtained with the CB3 and CB3A cell lines strongly support the conclusion that hnRNP A1 is not necessary for MHV discontinuous transcription or genome replication in vivo.
Figure 5.
MHV RNA synthesis in CB3 (hnRNP A1-deficient) and CB3A (hnRNP A1-restored) cells. Viral RNA was metabolically labeled with [3H]uridine, in the presence of actinomycin D, in CB3 and CB3A cells that were mock-infected or infected with wild-type MHV (wt), the win mutant, or the igs4-win mutant; mock-infected and wild-type MHV-infected mouse L2 cells were included for comparison. RNA was separated by agarose gel electrophoresis and visualized by fluorography.
Discussion
The preponderance of eukaryotic viruses have RNA genomes, and the majority of these replicate purely in the cytoplasm without having any direct transactions with the nucleus of the host cell, its DNA genome, or its transcriptional apparatus. The most stringent requirement imposed by this unique arrangement is the need to encode an RdRP. Nevertheless, the relatively limited capacity of RNA virus genomes argues that cellular host factors may also be appropriated to serve in replication and transcription. There is a steadily increasing list of host proteins proposed to be involved in viral RNA synthesis (1). Confirmation of the roles these proteins play will depend on corroborating evidence from biochemical, molecular biological, and genetic experiments (2–4).
For MHV, hnRNP A1 has been put forward as a candidate factor mediating the leader–body fusion event that is the hallmark of nidovirus sgRNA synthesis (1, 12). Evidence supporting this supposition was originally derived from in vitro studies. hnRNP A1 was found to bind to synthetic RNA molecules containing either the negative-strand IGS or the negative strand of the leader sequence, and it bound with reduced affinity to two mutated, transcriptionally nonfunctional IGS sequences (11, 12). Subsequent studies led to the proposed involvement of hnRNP A1 in a ribonucleoprotein containing the anti-IGS, the antileader, and the viral nucleocapsid protein (33, 34), although a requirement for the nucleocapsid protein in coronavirus RNA synthesis also has not been conclusively demonstrated; for arteriviruses, it is now clear that the nucleocapsid protein is not required for RNA transcription or replication (35). Very recently, it was shown that a modest overexpression of hnRNP A1 resulted in a marked increase in the kinetics of MHV RNA synthesis, leading to an expansion of the proposed role of this factor to include participation in genome replication as well as transcription (36). However, it was not established whether this resulted from a direct effect on viral RNA synthesis or, indirectly, from an enhancement of translation of viral protein. Alternatively, hnRNP A1 overexpression may have acted via effects on cellular gene expression through modification of alternative splicing, nucleocytoplasmic mRNA transport, or translation. Additionally, expression of a truncated form of hnRNP A1 resulted in its localization to the cytoplasm and inhibited the replication of MHV (36). It was not demonstrated whether this inhibition also could occur with an unrelated virus, and it thus may have been a nonspecific effect of unregulated overexpression of a general RNA binding protein in the cytoplasm.
In vitro, the measurement of equilibrium binding of hnRNP A1 to the negative-strand IGS (or mutants thereof) remains to be determined, so it is not yet known where the MHV IGS falls in the panoply of hnRNP A1 binding site affinities (19, 20). In vivo, it is questionable whether hnRNP A1 would have access to the negative-strand IGS, because intracellular negative-strand MHV RNA is entirely duplexed with its positive-strand counterpart (37), which is at least 10-fold more abundant (8). Additionally, elegant work with an infectious clone of the arterivirus equine arteritis virus has shown that if the IGS consensus sequence, in this case the five nucleotides 5′-UCAAC-3′, was completely changed at a given IGS to 5′-AGUUG-3′, then synthesis of the corresponding sgRNA was abolished. However, transcription at this IGS was restored (and simultaneously abolished at every other IGS) when the leader homology region was also changed to 5′-AGUUG-3′ (7). This finding appears to preclude sequence-specific recognition of the IGS by an RNA-binding protein.
In light of these considerations, we searched for ways of testing the participation of hnRNP A1 in MHV transcription by using mutants of both the virus and the host cell. In preliminary experiments, the highest affinity hnRNP A1 binding site known (“winner”; ref. 19) was engineered into the MHV genome in place of, or adjacent to, a particular IGS. In other systems, winner has been shown to be able to functionally replace natural hnRNP A1 binding sites involved in splice site selection or nuclear export of mRNA (15, 16). However, our data showed that winner could not functionally substitute for an IGS, enhance transcription from an adjacent IGS, or competitively inhibit other IGSs in the genome. These findings may be taken to indicate that hnRNP A1 is not sufficient to determine leader–body fusion in MHV transcription, but these negative results may also be attributable to other causes. Although there are examples of aberrant MHV transcription where leader–body fusion occurs in the absence of a recognizable IGS (22), it nevertheless remains possible that alterations in primary sequence in the win mutant abrogated any possible leader–body base-pairing interactions that would have allowed discontinuous transcription to proceed. Similarly, a possible required RNA secondary structure in the proximity of the IGS may have been altered by the insertion of winner in the igs4-win mutant.
To more definitively assess the importance of hnRNP A1 in MHV RNA synthesis, we analyzed MHV growth in the CB3 cell line, which does not express hnRNP A1. This deficiency resulted in no detectable restriction of MHV replication. Most strikingly, neither the quantity nor the quality of MHV sgRNA transcription was affected by the absence of hnRNP A1. We cannot rule out the possibility that the absence of hnRNP A1 in CB3 cells results in the induction of a different protein that duplicates its activity. However, such a hypothetical redundant factor would have the remarkable property that it could exactly and completely replace hnRNP A1 in MHV transcription but only partially replace hnRNP A1 in normal cellular roles such as alternative splice site selection and telomere maintenance, which are markedly affected in CB3 cells (13, 17, 32). Further, it would have to possess the same relative affinities as hnRNP A1 for each of the individual IGSs in the MHV genome to maintain the same relative ratios of all of the sgRNAs.
Taken together, our results strongly suggest that hnRNP A1 is not a required host factor for MHV discontinuous transcription or genome replication and that it does not play any significant role in MHV RNA synthesis in general. In part, our work has depended on the fortuitous availability of a mutant host cell line defective in the component under consideration. More universal procedures for the validation of other proposed host factors will likely require the establishment of both a murine (or surrogate host) genetic system, where the relevance of a candidate host factor can be tested directly, and an efficient in vitro RNA replication and transcription system, in which reconstitution of MHV RNA synthesis can be achieved from isolated components and precursors.
Acknowledgments
We are grateful to Dr. Benoit Chabot for providing the CB3 and CB3A cell lines and to Dr. Gideon Dreyfuss for the 9H10 mAb. We thank the Molecular Genetics Core of the Wadsworth Center for oligonucleotide synthesis and DNA sequencing. This work was supported in part by Public Health Service Grant AI 39544 from the National Institutes of Health.
Abbreviations
- RdRP
RNA-dependent RNA polymerase
- MHV
mouse hepatitis virus
- sg
subgenomic
- IGS
intergenic sequence
- hnRNP
heterogeneous nuclear ribonucleoprotein
- pfu
plaque-forming unit
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lai M M C. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 2.Strauss J H, Strauss E G. Science. 1999;283:802–804. doi: 10.1126/science.283.5403.802. [DOI] [PubMed] [Google Scholar]
- 3.Das T, Mathur M, Gupta A K, Janssen G M C, Banerjee A K. Proc Natl Acad Sci USA. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa M, D'ez J, Restrepo-Hartwig M, Ahlquist P. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Most R G, Spaan W J M. In: The Coronaviridae. Siddell S G, editor. New York: Plenum; 1995. pp. 11–31. [Google Scholar]
- 6.van der Most R G, de Groot R J, Spaan W J M. J Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J M, Snijder E J. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethna P B, Hung S-L, Brian D A. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawicki S G, Sawicki D L. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong Y S, Makino S. J Virol. 1992;66:3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Lai M M C. J Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Bani M-R, Lu S-J, Rowan S, Ben-David Y, Chabot B. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchette M, Chabot B. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caputi M, Mayeda A, Krainer A R, Zahler A M. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Najera I, Krieg M, Karn J. J Mol Biol. 1999;285:1951–1964. doi: 10.1006/jmbi.1998.2473. [DOI] [PubMed] [Google Scholar]
- 17.LaBranche H, Dupuis S, Ben-David Y, Bani M-R, Wellinger R J, Chabot B. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 18.Nadler S G, Merrill B M, Roberts W J, Keating K M, Lisbin M J, Barnett S F, Wilson S H, Williams K R. Biochemistry. 1991;30:2968–2976. doi: 10.1021/bi00225a034. [DOI] [PubMed] [Google Scholar]
- 19.Burd C G, Dreyfuss G. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdul-Manan N, Williams K R. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-David Y, Bani M R, Chabot B, De Koven A, Bernstein A. Mol Cell Biol. 1992;12:4449–4455. doi: 10.1128/mcb.12.10.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer F, Stegen C F, Koetzner C A, Masters P S. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masters P S, Koetzner C A, Kerr C A, Heo Y. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer F, Peng D, Hingley S T, Weiss S R, Masters P S. J Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsue B, Masters P S. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Masters P S. Adv Virus Res. 1999;53:245–264. doi: 10.1016/S0065-3527(08)60351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racaniello V R, Baltimore D. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 30.Boyer J-C, Haenni A-L. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 31.Almazán F, González J M, Pénzes Z, Izeta A, Calvo E, Plana-Durán J, Enjuanes L. Proc Natl Acad Sci USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai Y, Lee D, Yu T, Chasin L A. Nucleic Acids Res. 1999;27:1126–1134. doi: 10.1093/nar/27.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Li H-P, Xue W, Lai M M C. Virology. 1999;264:115–124. doi: 10.1006/viro.1999.9970. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Wang Y. Virology. 1999;265:96–109. doi: 10.1006/viro.1999.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molenkamp R, van Tol H, Rozier B C D, van der Meer Y, Spaan W J M, Snijder E J. J Gen Virol. 2000;81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- 36.Shi S T, Huang P, Li H-P, Lai M M C. EMBO J. 2000;19:4701–4711. doi: 10.1093/emboj/19.17.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y-J, Liao C-L, Lai M M C. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]