Abstract
Increasing levels of plasmid vector-mediated activation of innate immune signaling pathways is an approach to improve DNA vaccine-induced adaptive immunity for infectious disease and cancer applications. Retinoic acid-inducible gene I (RIG-I) is a critical cytoplasmic double-stranded RNA (dsRNA) pattern receptor required for innate immune activation in response to viral infection. Activation of RIG-I leads to type I interferon (IFN) and inflammatory cytokine production through interferon promoter stimulator 1 (IPS-1)-mediated activation of interferon regulatory factor 3 (IRF3) and NF-κB signaling. DNA vaccines coexpressing antigen and an expressed RNA (eRNA) RIG-I agonist were made, and the effect of RIG-I activation on antigen-specific immune responses to the encoded antigen was determined. Plasmid vector backbones expressing various RIG-I ligands from RNA polymerase III promoters were screened in a cell culture assay for RIG-I agonist activity, and optimized, potent RIG-I ligands were developed. One of these, eRNA41H, combines (i) eRNA11a, an immunostimulatory dsRNA expressed by convergent transcription, with (ii) adenovirus VA RNAI. eRNA41H was integrated into the backbone of DNA vaccine vectors expressing H5N1 influenza virus hemagglutinin (HA). The resultant eRNA vectors potently induced type 1 IFN production in cell culture through RIG-I activation and combined high-level HA antigen expression with RNA-mediated type I IFN activation in a single plasmid vector. The eRNA vectors induced increased HA-specific serum antibody binding avidity after naked DNA intramuscular prime and boost delivery in mice. This demonstrates that DNA vaccine potency may be augmented by the incorporation of RIG-I-activating immunostimulatory RNA into the vector backbone.
Methods to increase DNA vaccine-induced innate immune responses to improve adaptive immunity are needed to enable the general application of DNA vaccination in large animals and humans. The innate immune system is present in essentially all cell types and can be directly triggered by virus- or bacterium-specific pathogen-associated molecular patterns (PAMPs). PAMPs trigger immediate antiviral or antibacterial responses, such as induction of RNA degradation, translation inhibition or cell death pathways, and secretion of stimulatory signals, such as interleukin-12 (IL-12), IL-4, and type I interferon (IFN), that activate and differentially regulate the adaptive immune response (23).
A number of RNA and DNA PAMPs activate innate immunity through Toll-like receptor (TLR) signaling, for example, double-stranded RNA (dsRNA; TLR3), single-stranded RNA (ssRNA; TLR7, TLR8), and unmethylated CpG DNA (TLR9). In addition to TLRs, cytoplasmically localized B-DNA can induce interferon regulatory factor 3 (IRF-3) through IFI16-STING-TANK binding kinase 1 (TBK-1) signaling (58) and the inflammasome through activation of the AIM2 (absent in melanoma 2) receptor (reviewed in references 15 and 20) (Fig. 1A). As well, cytoplasmic dsRNA signaling pathways, such as the recently identified retinoic acid-inducible gene I (RIG-I; ligand is a 5′-PPP-containing short, blunt dsRNA) and melanoma differentiation-associated gene 5 (MDA5; ligand is a long dsRNA [44]) RIG-I-like helicase (RLH) pathways that activate IRF3 through interferon-β promoter stimulator 1 (IPS-1; also known as MAVS, Cardif, or VISA) signaling are also critical determinants required for innate immune activation in response to viral infection (reviewed in references 21 and 23). Agonists that activate these signaling pathways have a potential application as new-generation adjuvants (26).
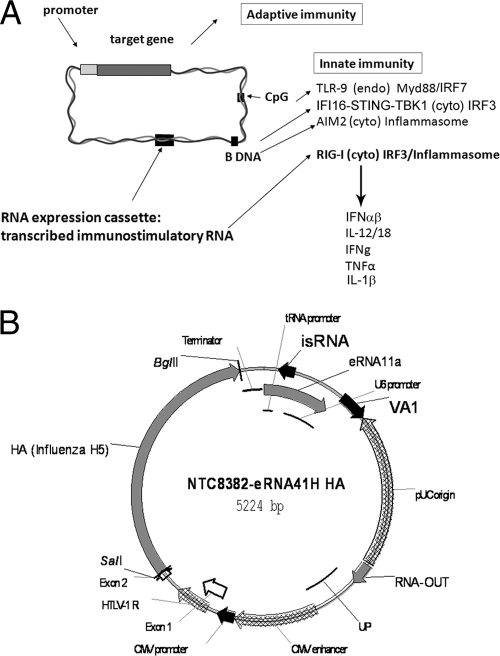
FIG. 1.
RIG-I-activating DNA vaccines. (A) Innate immune signaling in response to DNA and vector-encoded RIG-I PAMPs. Cytoplasmic (cyto) DNA may activate cytoplasmic receptors ZBP1 (DAI [not shown]), the IFI16 cytoplasmic receptor signaling through STING and TBK-1 (58), or the inflammasome through AIM2, while endosomal (endo) DNA activates TLR9 (15). Cytoplasmic RIG-I agonist RNA may activate IPS-1/IRF3 and the inflammasome through RIG-I and potentially also induce TLR3 (dsRNA) or TLR7 or -8 (ssRNA) activation in the transfected cell through endosome autophagy (not shown) or in bystander cells through uptake of RNA released by cell death. (B) Map of NTC8382 antibiotic-free (RNA-OUT) influenza virus H5 HA expression vector containing chimeric CMV-HTLV-1 R promoter and immunostimulatory RNA (isRNA) RIG-I agonist eRNA41H (eRNA11a and VA1).
The RLH pathway contributes to the adjuvant activity of poly(I:C), a dual ligand for TLR3 and MDA5 (24), demonstrating that RLH agonists may have adjuvant application. Consistent with this, the TBK-1-activating N-terminal caspase recruitment domain (N-CARD) of IPS-1 had adjuvant activity to improve humoral and cellular responses to protein vaccines (18). Interestingly, induction of adaptive immune responses to influenza virus or lymphocytic choriomeningitis virus infection required TLR not RLH signaling (reviewed in reference 21). This may reflect a difference between responses that control natural infections and responses to immunization.
DNA (e.g., unmethylated CpG oligonucleotide TLR9 agonist [reviewed in reference 22])- and RNA [e.g., synthetic poly(I:C) TLR3 and MDA5 agonist; ssRNA TLR7 and -8 agonists]-based adjuvants are made synthetically and are nonspecifically administered (reviewed in reference 4). For example, recently a bifunctional RIG-I-activating Bcl2-specific short interfering RNA (siRNA) was utilized to kill tumor cells after systemic administration (46). However, nonspecific administration of large doses of RNA and DNA may not be safe, and there is a need for molecules that can be codelivered with a DNA vaccine specifically to antigen-expressing cells.
DNA PAMPs present in the vector backbone mediate the immunogenicity of plasmid vectors. For example, if a plasmid vector is delivered to the endosome (e.g., naked or liposomal plasmid delivery), unmethylated CpG motifs in the backbone stimulate innate immune signals through TLR9, resulting in improved adaptive immune responses against the transgene product. Alternatively, if plasmid is cytoplasmically delivered, B-DNA can induce innate pattern receptors through IFI16 (TBK-1 effector) or the inflammasome through AIM2 signaling. For electroporation delivery, wherein plasmid is delivered directly to the cytoplasm, TBK-1 has been implicated as a key component required for induction of adaptive humoral and cellular responses to DNA vaccines (16; reviewed in reference 15) (Fig. 1A).
Increasing plasmid vector-directed innate immune activation through inclusion of novel backbone-encoded innate immunity agonists or signaling molecules improves DNA vaccine-induced adaptive immunity (17). For example, overexpression of IRF3 or IRF7 by expression plasmid coadministration increased humoral and cellular responses after naked intramuscular (i.m.) delivery (8, 51). For use in a licensed vaccine, innate immune agonists would preferably not be an endogenous protein that could potentially induce autoimmune responses. Ideally, these agonists would also not themselves induce an adaptive immune response since this would limit repeat usage as well as generate variable results in a population due to attenuated responses in individuals with prior exposure (i.e., preexisting immunity). An RNA-based innate immune agonist would meet these criteria.
RIG-I recognizes RNA from positive-sense ssRNA flavivirus and negative-sense RNA virus and paramyxoviruses (37, 56) and siRNA (35). The RIG-I ligand is a 5′-PPP-containing short, blunt dsRNA (14, 43, 52). 5′-PPPs are generally removed from, or masked, on self-RNA species; this is a structural basis for the distinction between self- and non-self-RNA (reviewed in reference 52). For example, the 5′-PPP is removed from RNA polymerase II-produced mRNA during capping prior to nuclear export. RNA polymerase I promoters will likewise not produce unmodified cytoplasmic RNA, since rRNA is heavily processed and modified in the nucleolus prior to nuclear export. However, RNA polymerase III (RNA Pol III) promoters are small, express to levels up to 10-fold higher than Pol II promoters, do not interfere with Pol II-directed antigen expression (J. A. Williams, world patent application WO2006078979), and, critically, have the potential to retain 5′-PPP.
Since plasmid vectors are already TLR9 and cytoplasmic DNA receptor agonists, incorporation of RLH ligands into the vector backbone of an antigen-expressing DNA vaccine may have targeted immunostimulation application by amplification of type I IFN and cytokine production in transfected cells (Fig. 1A). We report herein that RNA Pol III directed expression of immunostimulatory RIG-I agonist RNA from the vector backbone-activated innate immunity and improved adaptive immune responses.
MATERIALS AND METHODS
Strains and plasmids.
Kanamycin-resistant (Kanr) and ampicillin-resistant (Ampr) plasmids were grown in Escherichia coli strain DH5α [λ− φ80dlacZΔM15 Δ(lacZYA -argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1; Invitrogen, Carlsbad, CA], while antibiotic-free (AF) plasmids were grown in NTC4862 (DH5α attλ::P5/6 6/6-RNA-IN-SacB, Cmr).
Plasmid NTC8382 (Fig. 1B) is an AF selection pUC origin cytomegalovirus (CMV)-human T-cell leukemia virus 1 (HTLV-1) promoter expression vector that contains a 140-bp DraIII-KpnI RNA-based sucrose-selectable AF marker (RNA-OUT) (32). NTC7382 is a Kanr version of this vector. gWIZ-HTLV-I R AF is a CMV-HTLV-1 R promoter RNA-OUT AF derivative of the Kanr CMV promoter pUC origin gWIZ DNA vaccine plasmid (Genlantis, San Diego, CA). In these vectors, influenza virus A/Vietnam/1203/2004 serotype H5N1 hemagglutinin (HA), secreted alkaline phosphatase (SEAP), or enhanced green fluorescent protein (EGFP) transgenes are expressed from the CMV-HTLV-1 R promoter (32). The vectors also optionally encode immunostimulatory-expressed RNA (eRNA).
pDNAVACCUltra-EGFP U6 was made by cloning the murine U6 promoter into the NotI site of pDNAVACCUltra-EGFP vector (67). Inserts for U6 expression were cloned into an HpaI/NheI-cleaved vector. The first transcribed base is the first base after the blunt vector-encoded HpaI site. pDNAVACCUltra-HA had the H5 HA transgene substituted for EGFP. Constitutive RIG-I (cRIG-I) was a pDNAVACCUltra vector expressing the N-terminal CARD of murine RIG-I (56). This fragment was PCR amplified from pUNO-mRIG-I (InvivoGen, San Diego, CA) with primers (5′-ccttcaGTCGACatgacagcggagcagcggcagaatctg-3′ and 5′-ccttctAGATCTtcatttcaatgggctgtgtaaattattagaagac-3′), digested with SalI and BglII (uppercase), and cloned downstream of the CMV promoter in pDNAVACCUltra.
Molecular biology.
Vectors were made by standard restriction digestion-mediated transfer of fragments between vectors as described previously (68). The insertions, deletions, and substitutions in vectors were made by inverse PCR, using amplification primers containing terminal AarI type IIS restriction enzyme sites for seamless site-directed mutagenesis (J. A. Williams, world patent application WO2008153733). For cell culture and immunization applications, low-endotoxin (<100 endotoxin units [EU]/mg) plasmid DNA was purified using Nucleobond AX 2000 or AX 10000 columns (Macherey Nagel, Düren, Germany).
RIG-I ligands.
DNA vaccine vectors containing eRNA hairpins were made by cloning annealed primer inserts that generate HpaI (5′ blunt end, first base is +1 from the transcription start)- and NheI-compatible fragments into the HpaI/NheI-cleaved pDNAVACCUltra-EGFP U6 vector. The final lowercase “g” of the insert sequences below is the first base of the vector-encoded NheI site. All hairpin sequences contained a microRNA loop (tgtgctgtc) for exportin 5 cytoplasmic export.
In Bgal 728 (5′-ACTACACAAATCAGCGATTTGtgtgctgtcCAAATCGCTGATTTGTGTAGTtttttg-3′), the 21-bp stems of Bgal 728 are uppercase, and the microRNA loop is lowercase. This eRNA was designed to have a 3-bp 3′ overhang after RNA Pol III termination after TTTT.
In dsRNA43 (5′-AAGAAATTATTCTACACAAATCAGCGATTTACTTTGTTCTTTGtgtgctgtcCAAAGAACAAAGTAAATCGCTGATTTGTGTAGAATAATTTCTTttttg-3′), the 43-bp stems are uppercase, and the microRNA loop is lowercase. The eRNA was designed to have a 2-bp 3′ overhang after RNA Pol III termination after TTTT.
In dsRNA45 (blunt) (5′-AAAAGAAATTATTCTACACAAATCAGCGATTTACTTTGTTCTTTGtgtgctgtcCAAAGAACAAAGTAAATCGCTGATTTGTGTAGAATAATTTCTTTTttg-3′), the two additional 5′ bases to convert dsRNA43 to a blunt 45-bp stem dsRNA after RNA Pol III termination after TTTT are underlined. The 45-bp stems are uppercase, and the microRNA loop is lowercase.
C26 is a pDNAVACCUltra-EGFP-mU6 clone that expressed the protein kinase R (PKR) inhibitor C26 (9, 11)/GAAA (72) (5′-gggagagagGTCACTGAcTAAGTTGGcgaaagTTGATTTAgTCAGTGACaagaaggaaggttttttg-3′). The bases that form two dsRNA stems (separated by a cgaaag loop) are uppercase. The 9-bp single-stranded leader (lowercase) and 11-bp tail (lowercase, underlined) are shown. To adapt for RNA Pol III expression, a terminator (tttttt) was added after the 11-bp tail. Termination after TTTT would extend the single-stranded tail to 15 bp.
pDNAVACCUltra5 EGFP vectors that contained the adenovirus serotype 5 (Ad5) VA RNAI (VA1) gene, or both the VA1 and VA RNAII (VA2) genes, in either orientation, were constructed. The VA RNA genes, including the RNA Pol III terminator and upstream sequences, were PCR amplified from Ad5 genomic DNA and cloned into the blunt StuI site of the parent vector. This is a permissive site between the prokaryotic replication origin and the eukaryotic Pol II terminator (Fig. 1B). The resultant constructs with single or double inserts were sequence verified. VA1 was transcribed clockwise relative to the pDNAVACCUltra vector (Fig. 1B) and comprised sequences 10597 to 10790 of the Ad5 genome (GenBank accession number X02996). The VA1 and VA2 clone comprised sequences 10589 to 11049 of the Ad5 genome. VA2-only constructs were made by PCR-mediated site-directed deletion of the VA1 gene from VA1 and -2.
The pDNAVACCUltra5 EGFP eRNA11, -11a (114-bp dsRNA), and -11b constructs contained a short dsRNA based on the first 100-bp fragment of gene segment 3 of influenza virus A/PR/8/34 (H1N1) (PR8) (27). These vectors express dsRNA by convergent transcription from the human tRNA(Val) and murine U6 promoters. The human tRNA(Val) and linker sequence was as described previously (25). The insert sequences and predicted RNA Pol III start and stop sites are shown in Fig. 2C.
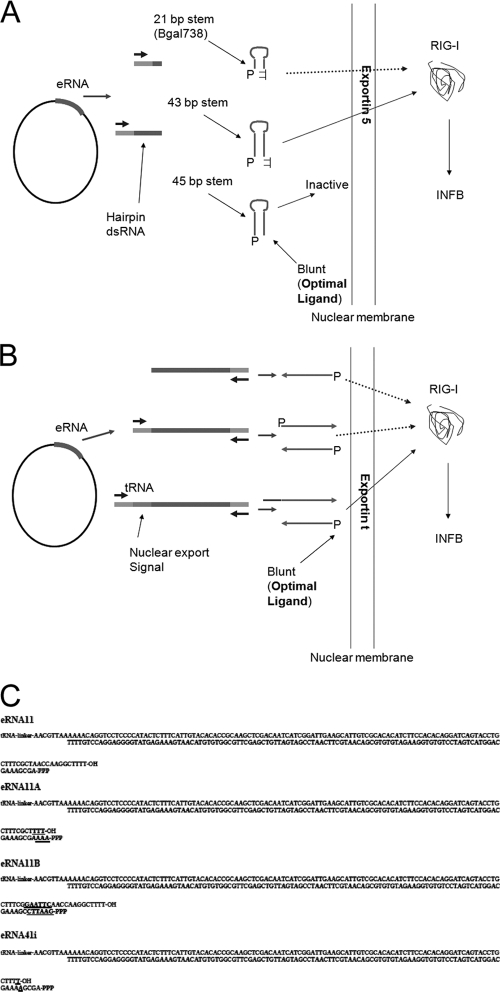
FIG. 2.
Assay for RIG-I activation by plasmid-borne eRNA generated by hairpin (A) and convergent transcription (B). Transfected plasmid vectors enter the nucleus, and RNA Pol III transcribes the encoded eRNA sequence, retaining the 5′-PPP (P). The optimal configuration for RIG-I activation (blunt dsRNA with 5′-PPP) is not permissive for exportin 5-mediated hairpin RNA export (A). (C) DNA sequences for convergently transcribed eRNAs 11, 11a, 11b, and 41i with predicted tRNA-expressed RNA stop site (top strands, TTTT-OH) and predicted convergent U6-expressed RNA start (bottom strands, 5′-PPP) and stop sites (bottom strands, TTTT). The U6 promoter 5′-PPP base was designed to have a recessed 5′-PPP-A (eRNA11) or 5′-PPP-G (eRNA11b) or a blunt 5′-PPP-A (eRNA11a) or 5′-protruding 5′-PPP-A (eRNA41i). The 3-bp insert in eRNA11 used to create eRNA11a is bolded and underlined. The 6-bp insert modification from eRNA11 used to create eRNA11b is bolded and underlined. The 1-bp T insert in eRNA11 to create eRNA41i is bolded and underlined (eRNA41i plasmid also encodes VA1). The eRNA11f construct has the AACG sequence immediately after the tRNA-linker in eRNA11a mutated to TTTT (creating a TTTTTT RNA Pol III terminator).
eRNA11d is an eRNA11 vector derivative modified by the deletion of the downstream U6 promoter to produce an ssRNA from the tRNA(Val) promoter only. eRNA18 is an eRNA11 derivative in which the tRNA(Val) gene was deleted to produce an ssRNA from the U6 promoter only. H1-mU6 IFN dsRNA is an eRNA11 derivative with the human H1 RNA Pol III substituted for the tRNA(Val) promoter. eRNA11f is an eRNA11a derivative with a TTTTTT RNA Pol III terminator inserted downstream from the tRNA(Val) promoter (Fig. 2C).
eRNA11c is a derivative in which the influenza virus sequence in eRNA11B (EcoRI GAATTC-AclI AACGTT fragment) was replaced with a 78-bp fragment of the highly structured HCV 3′ nontranslated region (NTR) sequence (56) from a synthetic gene (IDT Technologies, Coralville, IA). The cloned sequence (eRNA11B flanks underlined [Fig. 2C]) is 87 bp long: 5′-AACGTTAAAAAAGTCACGGCTAGCTGTGAAAGGTCCGTGAGCCGCTTGACTGCAGAGAGTGCTGATACTGGCCTCTCTGCAGATCAAGTGAATTC-3′.
eRNA41H is the combination of pDNAVACCUltra VA1 and eRNA11a in the configuration shown in Fig. 1B. eRNA41 is the same as eRNA41H, except that it contains eRNA11. eRNA41i contains a 1-bp T insert in the eRNA11 component to create eRNA41i (Fig. 2C). Derivatives with internal deletions of the 114-bp dsRNA influenza virus eRNA11a sequences to result in dsRNA lengths of 34 and 54 bp were made, as well as an internal insertion of 50 bp to increase dsRNA length to 164 bp. eRNA41H-MV has the measles virus (MV) leader RIG-I agonist (45) (with A-to-T substitutions to remove TTTT terminators) substituted for eRNA11a (between the AAAA and the TTTT [underlined below]) (see Fig. 2C), resulting in the following 88-bp transcribed dsRNA insert: 5′-AAAAGCAAATTGTTAATCATTATTTGCAATATATTAAAGATAACTTTGATAATACGAAGTTTCTATTAATAGCTTTGTCTTTGCTTTT-3′.
The terminal 9 bp from each end are complementary and are predicted by Mfold analysis (76) to form a snapback blunt hairpin stem (ssRNA from the U6 promoter). A derivative predicted by Mfold analysis to form an 18-bp blunt hairpin stem (single-stranded RNA from the U6 strand) (underlined bases below) was made by the insertion of the bolded base pairs into eRNA41H-MV, resulting in a 97-bp dsRNA insert: 5′-AAAAGCAAAGACAAAGCTTTGTTAATCATTATTTGCAATATATTAAAGATAACTTTGATAATACGAAGTTTCTATTAATAGCTTTGTCTTTGCTTTT-3′.
eRNA41H-PKR1 is pDNAVACCUltra-EGFP eRNA41H with the VA1 component engineered to contain an internal deletion from after the promoter B box to the beginning of the terminal minihelix within the VA1 gene (Ad5 sequence base pairs 10696 to 10740 deleted) that was predicted by Mfold to form a smaller stable minihelix, with the central domain deleted. In eRNA41H-PKR2, the VA1 gene was modified to express a 10-bp hairpin structure replacing the central domain with the sequence shown below. Modified VA1 (see Fig. 4B) bases are underlined, and the hairpin stems are uppercase: 5′-GGTTACCGCGAATTCAACGgaaaCGTTGAATTCGCGGTAACC-3′.
FIG. 3.
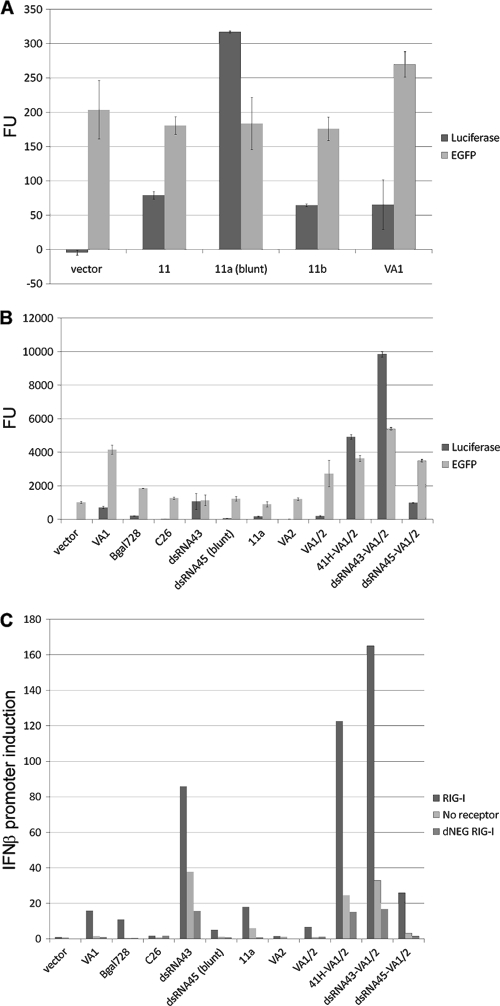
eRNA optimization. (A) RIG-I-dependent activation of IFN-β promoter-luciferase reporter (Luciferase) and vector-encoded EGFP expression (EGFP). Increased immunostimulation was observed with blunt-ended eRNA (11a) compared to that of 3′-recessed eRNA (11 or 11b). Luciferase and EGFP fluorescence units (FU) are shown. Each well was transfected with 100 ng RIG-I plasmid, 100 ng pI25luc, and 200 ng pDNAVACCUltra EGFP eRNA test vector and assayed for RIG-I activation 29 h after transfection. (B) Synergistic activation of IFN-β promoter by combination of either convergent eRNA11a or hairpin dsRNA43 with VA1. VA1 also increased transgene expression. Luciferase and EGFP fluorescence units are shown. Each well was transfected with 200 ng RIG-I plasmid, 200 ng pI25luc, 400 ng pDNAVACCUltra-EGFP, and 200 ng pDNAVACCUltra EGFP eRNA test vector and assayed for RIG-I activation 29 h after transfection. (C) VA1-mediated synergistic activation of IFN-β promoter using eRNA11a ligand does not require RIG-I receptor overexpression (RIG-I); synergy was observed in cells without RIG-I receptor overexpression (No receptor) and is partially inhibited by transfection of dominant negative RIG-I (dNEG RIG-I). Fold induction of the IFN-β promoter-luciferase reporter (luciferase/EGFP ratio of pDNAVACCUltra eRNA test plasmid divided by luciferase/EGFP ratio of pDNAVACCUltra parent plasmid) is shown. VA2 was not a RIG-I ligand or expression enhancer in this assay. Each well was transfected with 200 ng receptor plasmid (hRIG-I, dNEG RIG-I, or pDNAVACCUltra-HA for no receptor), 200 ng pI25luc, 400 ng pDNAVACCUltra-EGFP, and 200 ng eRNA test vector and was assayed for RIG-I activation 29 h after transfection.
FIG. 4.
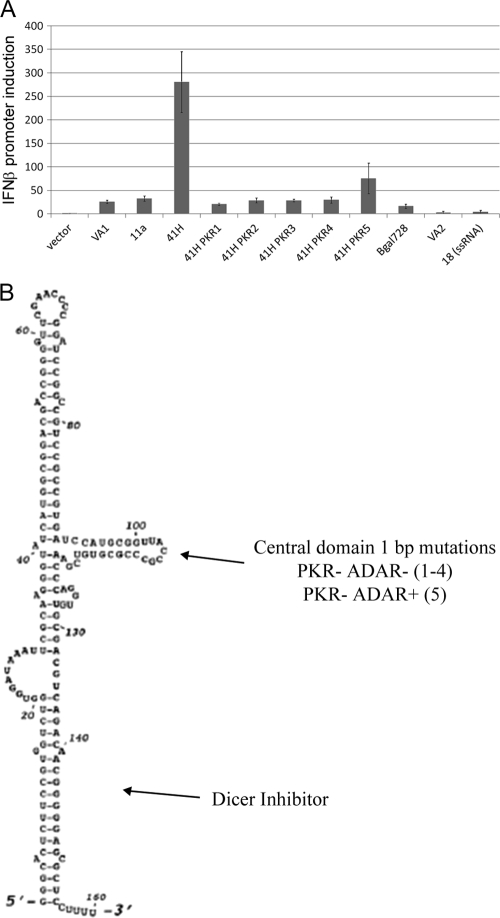
Adenoviral VA1 improved eRNA11a-mediated RIG-I activation through ADAR inhibition. (A) RIG-I activation of IFN-β promoter-luciferase reporter (luciferase/EGFP ratio of pDNAVACCUltra eRNA test plasmid divided by luciferase/EGFP ratio of pDNAVACCUltra parent plasmid). Synergistic RIG-I activation by plasmid-borne VA1 and eRNA11a combination (41H) and improved RIG-I activation with eRNA41H-PKR5 (41H PKR5) that retains partial VAI-mediated ADAR inhibition was observed. Each well was transfected with 200 ng hRIG-I plasmid, 200 ng pI25luc, 400 ng pDNAVACCUltra-EGFP, and 200 ng eRNA test vector. RIG-I activation was assayed 29 h after transfection. (B) VA1 structure with PKR, ADAR, and Dicer/exportin 5 inhibitor domains indicated.
eRNA41H-PKR3 (L3) and eRNA41H-PKR4 (L4) are a pDNAVACCUltra-EGFP eRNA41H derivative with the VA1 component modified in the central domain. These L3 and R1 mutations are 2-bp changes in the central region that are inactive for PKR inhibition yet are transcribed at high levels in cell culture and maintain a VA1-like RNA secondary structure (34).
eRNA41H-PKR5 (pm91) is a pDNAVACCUltra-EGFP eRNA41H derivative with the VA1 component modified in the central domain to incorporate the 1-bp pm91 mutation (49).
Cell culture.
Adherent HEK293 (human embryonic kidney), A549 (human lung carcinoma), NIH 3T3 (murine embryonic fibroblast), and L929 (murine areolar fibroblast) cell lines were obtained from the American Type Culture Collection (Manassas, VA). Cell lines were propagated in Dulbecco's modified Eagle's medium (DMEM)-F-12 medium containing 10% fetal bovine serum (FBS) and split (0.25% trypsin-EDTA) using Invitrogen (Carlsbad, CA) reagents and conventional methodologies.
The vectors were tested for RIG-I activation using an in vitro cell culture assay. EGFP target gene expression was also monitored in the same experiments as a surrogate assay for PKR activation (PKR inhibits translation, leading to loss of protein expression). All assays were performed using 24-well cell culture plates. For each well, unless otherwise noted, 0.1 μg pI25luc (a reporter plasmid, with the luciferase gene driven by the IFN-β promoter), 0.1 μg of the cytoplasmic RNA receptor (either human RIG-I [hRIG-I], murine RIG-I [mRIG-I], human MDA5, or dominant negative RIG-I control [DeNy] [pUNO-hRIG-I, pUNO-mRIG-I, pUNO1-hMDA5, or pDeNy-hRIG-I, respectively]; Invivogen, San Diego, CA), 0.16 μg pDNAVACCUltra-EGFP (EGFP reporter plasmid), and 0.04 μg (40 ng) of the test plasmid were mixed in 50 μl DMEM (no serum) and cotransformed into the cell line using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or Superfect (activated dendrimer; Qiagen, Hilden, Germany) as described by the manufacturers. Cells were grown in DMEM-F-12 plus 10% FBS at 37°C with 5% CO2. The transfection medium was removed after 4 h of incubation. Expression (EGFP) and IFN-β activation (luciferase) were determined 24 to 35 h posttransfection using the FLX800 microplate fluorescence reader, directly for EGFP in black plates, and after activation with the Promega (Madison, WI) luciferase assay system in opaque plates for luciferase. To determine IFN-α promoter activation, the pNIFTY2-IFA-SEAP (a reporter plasmid, with the SEAP gene driven by the mouse IFN-α promoter; Invivogen) was substituted for pI25luc in the assay, and SEAP expression was visualized in the wells using HEK-BLUE SEAP detection buffer (Invivogen).
Western blot analysis for influenza virus HA expression was performed on protein extracts from transfected HEK293 cells by using rabbit anti-HA H5N1 IgG (eEnzyme, Gaithersburg, MD) primary antibody and the amplified alkaline phosphatase immunoblot assay kit and AP conjugate substrate kit (Bio-Rad, Hercules, CA).
RT-PCR.
Cytoplasmic RNA was isolated from HEK293 cells using the protein and RNA isolation system (PARIS kit; Ambion, Austin, TX) and quantified by A260. Samples were DNase treated (DNA-free DNase, Ambion, Austin, TX) prior to reverse transcriptase PCR (RT-PCR) using the Agpath-ID one-step RT-PCR kit (Ambion, Austin, TX) with either TaqMan eRNA11 or VA1-targeted minor groove binder (MGB) probes with flanking primers in a TaqMan gene expression assay using the StepOne real-time PCR system (Applied Biosystems, Foster City, CA). TaqMan eRNA11 assay was with 6-carboxyfluoroscein (6FAM)-CACCGCAAGCTCGA-MGBNFQ probe and flanking primers (5′-GACAATGCTTCAATCCGATGA-3′ and 5′-TCCTCCCCATACTCTTTCATTGTAC-3′), and TaqMan VA1 assay was with 6FAM-CCGCGTGTCGAAC-MGBNFQ probe and flanking primers (5′-CCGTGATCCATGCGGTTAC-3′ and 5′-CGTTGTCTGACGTCGCACAC-3′). Strand-specific cDNA was made by heat denaturing DNase-treated cytoplasmic RNA and a single primer for 2 min at 95°C, cooling to 48°C, adding AgPath RT-PCR mix and incubating 30 min to make single-strand cDNA, and a 10-min step at 95°C to inactivate reverse transcriptase. The resultant single-strand-specific cDNAs were then quantified in a standard TaqMan gene expression assay (no reverse transcriptase) using Applied Biosystems (Foster City, CA) TaqMan reagents. DNase treatment conditions were confirmed by the lack of detectable amplification in control RT-PCRs run without the initial 48°C reverse transcriptase step using 25× PCR buffer that was heat treated to inactivate the heat-labile reverse transcriptase (but not Taq DNA polymerase). The pDNAVACCUltra-eRNA41H vector was used for the eRNA11 and VA1 RT-PCR standard curves.
Murine studies. (i) HA plasmid naked DNA immunization.
Groups of five mice were immunized with plasmid DNA in an IACUC-approved study (JW-NX3-08) performed at the OLAW-certified Aldevron animal facility, Fargo, ND. Ten micrograms of HA plasmid in 100 μl of phosphate-buffered saline (PBS) was injected bilaterally intramuscularly (i.m.) into the tibialis cranialis muscles of female BALB/c mice (6 to 8 weeks old) in a prime-boost dose (days 0 and 21). Serum samples were taken on day 49, and anti-HA-specific total IgG, IgG2a, and IgG1 titers were determined by standard enzyme-linked immunosorbent assay (ELISA) using plates coated with the E. coli-produced recombinant A/Vietnam/1203/2004 HA HA2 domain as described previously (32). Antibody avidity was determined using thiocyanate elution as described previously (48).
(ii) HA plasmid naked DNA plus EP immunization.
Groups of eight mice were immunized with plasmid DNA in an IACUC-approved study (GEN 08-02) performed at the OLAW-certified Explora BioLabs animal facility, San Diego, CA. Three micrograms of HA plasmid (or 2 μg HA combined with 1 μg cRIG-I plasmid) in 20 μl of saline was injected intramuscularly into the quadriceps muscles of female BALB/c mice (6 to 8 weeks old) in a prime-boost dose (days 0 and 21) prior to electroporation (EP). Electroporation was performed using the Elgen 1000 EP system using a 2-needle array (27 G), 4 mm apart, two 60-ms pulses at 50 mA. Serum and spleens were harvested on day 35. Serum was tested as described above for anti-HA-specific total IgG, IgG2a, and IgG1 titers and antibody avidity. Epitope-specific CD8 T-cell frequencies were determined using a mouse IFN-γ enzyme-linked immunospot assay (ELISPOT) kit (R&D Systems, Minneapolis, MN) and splenocytes stimulated with 1 μg/ml of the immunodominant conserved influenza virus HA major histocompatibility complex (MHC) class I molecule H-2Kd peptide HA518 (IYSTVASSL) or a negative-control peptide. SEAP levels in murine serum on days 4, 7, and 14 (three mice per group) after day 0 electroporation delivery of SEAP transgene vectors (Aldevron study JW-NX1-07) were determined using the Phospha-Light SEAP reporter gene assay system from Applied Biosystems (Foster City, CA) according to the manufacturer's instructions.
(iii) gag plasmid naked DNA immunization.
eRNA41H was cloned into the pITR p55gag (pITR p43-LAMP/gag wt [wild type]) vector to create pITR p43-LAMP/gag wt-eRNA41H. pITR is an Ampr CMV promoter vector with the eukaryotic region flanked by adeno-associated virus (AAV) long terminal repeats (2). Murine evaluation was performed at the OLAW-certified Johns Hopkins University animal facility. Mice were immunized subcutaneously with 50 μg of plasmid in PBS on days 0 and 28, spleens were harvested on day 38, and epitope-specific CD8 and CD4 IFN-γ, tumor necrosis factor alpha (TNF-α), IL-4, and IL-2 and IL-10-secreting T cell frequencies were determined by ELISPOT assay as described previously (2).
RESULTS
The murine U6 RNA Pol III promoter was used to express 5′-PPP RNAs. While U6 small nuclear RNA contains a gamma-monomethyl phosphate cap, capping of this RNA is dependent on downstream RNA sequences. U6 promoter-expressed heterologous RNAs are not capped and retain the 5′-PPP (54).
Hairpin exportin 5 eRNA DNA vaccines.
To activate RIG-I, an RNA Pol III-transcribed RNA must be exported from the nucleus to the cytoplasm (RIG-I is cytoplasmic) while presenting a 5′-PPP in a dsRNA configuration. While blunt 5′-PPP is the optimal RIG-I ligand, 3′ overhangs of 2, 3, or 10 bp retain one-third, one-fourth, or 1/10 the activity of the blunt ligand, respectively (52). Export of RNA Pol III transcripts can be mediated by exportin 5, which transfers >16-bp hairpin double-stranded (hdsRNA) minihelix sequences with a 3′ overhang to the cytoplasm (reviewed in reference 19). This may be a 2-bp 3′ overhang, such as generated by Drosha processing, or longer overhangs such as the CAAAA five-base overhang in VA1. The presence of a 5′ overhang is inhibitory for export (71) and RIG-I activation (52).
DNA vaccine vectors containing U6 promoter-expressed hdsRNA were made. The pDNAVACCUltra-EGFP vaccine vector was modified to encode the murine U6 RNA Pol III promoter (J. A. Williams, world patent application WO2006078979). Target hdsRNAs (Bgal728, dsRNA43) were cloned into the vector as well as a PKR aptamer (C26). Bgal728 and dsRNA43 hdsRNAs encode 21- or 43-bp stems, respectively, with 2- to 3-bp 3′ overhangs for exportin 5 recognition. A control derivative of dsRNA43 (dsRNA45) was made by the addition of two bases to the 5′ end to encode a predicted blunt 45-bp stem hdsRNA. While blunt ligands are optimal for RIG-I activation (35, 52), it is unknown if blunt minihelixes will be exported by exportin 5 (42).
The EGFP plasmids containing these U6 promoter-driven hdsRNAs were tested in vitro in a human cell culture (HEK293) transfection assay for RIG-I-dependent activation of a cotransfected IFN-β promoter-luciferase reporter (Fig. 2A). The C26 PKR aptamer was inactive, while the Bgal728 ligand was very weakly active, requiring RIG-I overexpression (Fig. 3B and C). However, dsRNA43 was much more potent, activating the IFN-β promoter with or without RIG-I overexpression (Fig. 3B and C). Inhibition of dsRNA43-mediated signaling by transfection of dominant negative RIG-I demonstrated that IFN-β promoter activation was mediated by RIG-I (Fig. 3C). In contrast, dsRNA45 was a weak RIG-I agonist (Fig. 3B and C). We hypothesize that either this blunt dsRNA was not exported from the nucleus or that it was processed by Drosha to create an exportable 2-bp 3′ overhang RNA that lacks RIG-I agonist activity.
Convergent exportin-t eRNA DNA vaccines.
Export of RNA Pol III tRNA transcripts is mediated by exportin-t (reviewed in reference 19). tRNA promoters, such as tRNA(Val), will generally produce cytoplasmic RNAs lacking 5′-PPP, since normal processing of tRNAs in the nucleus removes the 5′-PPP. However, fusions to the C terminus of tRNA(Val) that maintain tRNA folding without 5′-end removal have been utilized to export tRNA(Val)-ribozyme fusions to the cytoplasm using a defined linker (25). While unprocessed tRNA has limited secondary structure and is unlikely to be an RIG-I ligand, an exported 3′ cis-linked RNA may be used to create a downstream acceptor strand for a complementary 5′-PPP RNA.
A convergently transcribed 114-bp dsRNA containing the first 100-bp fragment of gene segment 3 (27) of influenza virus A/PR/8/34 (H1N1) (PR8), mutated as necessary to remove potential RNA Pol III termination sequences, was selected for evaluation (eRNA11). We hypothesized that this RNA encoded an RIG-I ligand; it may normally be made with a 5′-PPP since it is at the 5′ end of the genome and RIG-I activates IFN-β production in influenza virus A-infected cells (39). As well, this genomic region has potent in vivo stimulatory activity (11).
A pDNAVACCUltra vector expressing this RNA (eRNA11) convergently from tRNA(Val) and U6 RNA Pol III promoters activated RIG-I (Fig. 3A) in a human cell culture transfection assay (Fig. 2B). eRNA11 has a recessed 5′-PPP-A. Mutagenized versions with recessed 5′-PPP-G and blunt 5′-PPP-A ends were screened for improved activity. The convergently transcribed blunt 5′-PPP-A RIG-I agonist eRNA11a was optimal (Fig. 3A). RIG-I activation by eRNA11a does not require overexpression of the RIG-I receptor. As well, inhibition of eRNA11a-mediated signaling by transfection of dominant negative RIG-I demonstrated that IFN-β promoter activation was mediated by RIG-I (Fig. 3C).
Variation of the dsRNA length from 114 bp to 34, 54, and 164 bp by internal modification slightly reduced RIG-I agonist activity (data not shown). Transcription of both strands was needed for RIG-I activation since vector modifications that interfere with either U6 or tRNA(Val) transcription abolish the activity (Table 1). Substitution of the tRNA(Val) promoter with the H1 promoter dramatically reduced cytoplasmic levels of the corresponding RNA and RIG-I activation (Table 1). This was suggestive that tRNA-mediated export of the cis-linked RNA was critical for RIG-I ligand formation. However, cytoplasmic levels of RNA produced from the U6 promoter were not increased by transcription of the tRNA(Val) strand. As well, RIG-I activation by eRNA11a does not correlate with the absolute levels of cytoplasmic tRNA or U6-transcribed RNA (Table 1). We hypothesize that RIG-I was activated by a subset of the cytoplasmic U6 and tRNA-transcribed complementary RNAs that have “annealed” into dsRNA with a blunt U6 strand-encoded 5′-PPP-A.
TABLE 1.
Cytoplasmic RNA exporta
| eRNA | eRNA structureb | Fold IFN-β inductionc | Mean ± SD pg cytoplasmic RNA/150 ng total RNA (RT-PCR)d |
||
|---|---|---|---|---|---|
| tRNA/H1 strand | U6 strand | Total RNA | |||
| 11a | tRNA------U6 | 18.4 | 1.48 ± 0.01 | 0.57 ± 0.02 | 1.16 ± 0.02 |
| 11d | tRNA------ | 2.6 | 2.36 ± 0.13 | 0.19 ± 0.03 | 4.34 ± 0.32 |
| 18 | ------U6 | 2.7 | 0.09 ± 0.00 | 1.01 ± 0.07 | 0.72 ± 0.03 |
| 11f | tRNA-T------U6 | 0 | 0.16 ± 0.00 | 0.90 ± 0.28 | 0.38 ± 0.02 |
| dsU6-H1 | H1------U6 | 2.9 | 0.20 ± 0.01 | 1.15 ± 0.06 | 1.02 ± 0.02 |
| 41H (VA1 + 11a) | tRNA------U6 + hairpin RNA | 112.9 | 0.62 ± 0.03 | 0.33 ± 0.09 | 1.17 ± 0.01 |
| VA1 | Hairpin RNA | 11.9 | ND | ND | 0.02 ± 0.00 |
Transfection of HEK293 cells with eRNA plasmid and hRIG-I receptor plasmid was as described in Materials and Methods. Cytoplasmic RNA was isolated 27 h posttransfection.
Convergent RNA Pol III promoters are shown flanking expressed RNA (------). T, TTTTTT RNA Pol III terminator.
Standardized luciferase value (luciferase/EGFP ratio) divided by the vector control standardized luciferase value.
ND, not determined.
The activity of the influenza virus gene segment 3-based eRNA11a construct was compared to two other known RIG-I ligands expressed by convergent tRNA-U6 transcription. Similar activity to eRNA11a was obtained with eRNA11a-MV in which the influenza virus RNA was substituted with the AT-rich measles virus RNA leader, a known RIG-I ligand (45). A modified MV insert, with an 18-bp blunt terminal stem formed by snapback hybridization of the ssRNA expressed from the U6 promoter, also had similar activity (data not shown). Interestingly, substitution of a structured RNA (the HCV 3′ NTR) for the influenza virus 100-bp RNA (eRNA11C) resulted in lower levels of RIG-I activation (data not shown). In contrast, strong RIG-I activation with in vitro-transcribed HCV 3′ NTR RNA has been reported (56). Reduced activity in our vector expression system may be due to inhibition of complementary strand annealing-mediated RIG-I ligand formation by the strong snapback single-stranded RNA secondary structure. These results demonstrated that RIG-I ligands identified in the literature do not necessarily function as such when expressed from plasmid backbones since the alterations necessary for nuclear export (e.g., convergent tRNA transcription) may abolish activity. As expected, since MDA5 is activated by long dsRNA (44), none of these ligands had detectable MDA5-mediated IFN-β activation (data not shown).
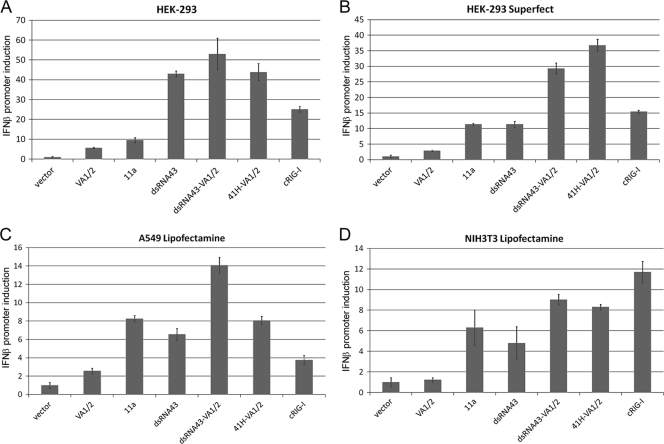
RIG-I activation by eRNA11a and dsRNA43 was observed with lipofection and dendrimer delivery, and in human (HEK293 and A549 [see Fig. 5]) and murine (NIH 3T3 [see Fig. 5] and L929 [data not shown]) cell lines. This demonstrates that RIG-I activation with these ligands is not delivery or cell line specific.
FIG. 5.
eRNA11a activated RIG-I in human and murine cell lines with different transfection reagents. Fold induction of the IFN-β promoter (as in FIG. 3) for eRNA11a and dsRNA43, with and without VA1/2, is shown. Each well was transfected with 200 ng hRIG-I plasmid, 200 ng pI25luc, 400 ng pDNAVACCUltra-EGFP, and 200 ng eRNA test vector (or cRIG-I), and RIG-I activation was assayed 30 h after transfection. (A) HEK293 with Lipofectamine 2000 (lipofection); (B) HEK293 with Superfect; (C) A549 (human) with Lipofectamine 2000; (D) NIH 3T3 (murine) with Lipofectamine 2000. RIG-I and control plasmid were also transfected into Huh7 (RIG-I-responsive cell line) and Huh7.5 (RIG-I mutant) human hepatoma cell lines and fold induction of IFN-β mRNA versus GAPDH mRNA control determined by RT-PCR. However, signaling sensitivity in this cell system was not amenable to this analysis since transfected DNA gave high background signaling in the assay (data not shown).
VA1 synergistically increased the activity of eRNA DNA vaccines.
Cytoplasmic dsRNA can directly induce dsRNA receptors adenosine deaminase acting on RNA (ADAR), PKR, 2′-5′ oligoadenylate synthetase (OAS; leads to RNase L activation), RIG-I, and MDA5. Activation of RIG-I and MDA5 leads to interferon and inflammatory cytokine (e.g., TNF-α) production, while PKR and RNase L activation inhibits protein synthesis through eukaryotic initiation factor 2-α phosphorylation and RNA cleavage, respectively, thus reducing antigen production. ADAR activation leads to dsRNA A-to-I editing (reviewed in reference 62). To be compatible with robust DNA vaccine antigen expression, a RIG-I ligand must therefore not activate PKR or OAS.
PKR is activated by long (>30-bp) dsRNAs that can interact with both dsRNA binding domains. eRNA11a and dsRNA43 are therefore probable PKR agonists. PKR activation is inhibited by a variety of structured viral RNAs, such as adenoviral virus-associated RNAI (VA1) (33). VA1 has a tRNA-like RNA-internal A and B box promoter and is transcribed by RNA Pol III into a structured RNA with a 5-bp 3′ overhang (Fig. 4B) that is efficiently transferred to the cytoplasm by exportin 5 (13). Most VA1 retains the 5′-PPP (60). When expressed from a plasmid backbone, VA1 enhanced transgene expression and activated RIG-I (Fig. 3A and B). However, VA1, like Bgal728, was a weak ligand, requiring RIG-I receptor overexpression for RIG-I activation (Fig. 3C). The related adenoviral RNA VA2 has the same terminal stem and 5-bp 3′ overhang as VA1 but is not a PKR inhibitor due to central domain sequence differences (33). VA2 does not increase transgene expression or activate RIG-I (Fig. 3B). This demonstrated that inhibition of RNA interference or competitive binding to La protein (i.e., shielding RNA from RIG-I [7]) does not mediate these effects, since these functions are conserved between VA1 and VA2 (1, 31, 36).
Strikingly, synergistic RIG-I activation (and VA1-mediated improved transgene expression) was obtained with eRNA41H, a combination of VA1 and eRNA11a, in cells with or without RIG-I receptor overexpression (Fig. 3B and C; Table 1). In contrast, no effect on RIG-I activation was observed when the eRNA11a or VA1 plasmids were combined with Bgal728 or C26 eRNA plasmids in RIG-I receptor-overexpressing cells (data not shown). Combination of VA1 and dsRNA43 or dsRNA45 increased RIG-I activation only in cells in which the RIG-I receptor was overexpressed (Fig. 3C).
Enhanced RIG-I activation was also obtained with vectors with different orientations of the VA1 and eRNA11a components or cotransfection of separate VA1 and eRNA11a plasmids (data not shown). This demonstrated that the VA1/eRNA11a interaction was trans-acting. RIG-I activation was observed over a broad range of eRNA41H plasmid transfection amounts, including 40, 20, or 10 ng/well (data not shown). The eRNA41H vector also induced the IFN-α promoter in the RIG-I assay (data not shown). VA1/eRNA11a-enhanced RIG-I activation was not delivery specific and was observed using dendrimer or lipofection transfection (Fig. 5). In human cells, VA1/eRNA11a induced the IFN-β promoter more effectively than a constitutive RIG-I (cRIG-I) comparator plasmid (Fig. 5). The eRNA11a blunt ligand was optimal in combination with VA1, since eRNA41H had stronger activity than eRNA41 (VA1 plus eRNA11 recessed 5′-PPP) and eRNA41i (VA1 plus eRNA11 protruding 5′-PPP) (data not shown). VA1/eRNA11a-enhanced RIG-I activation was weaker but detectable in murine cell lines NIH 3T3 (Fig. 5) and L929 (data not shown). The weaker effect of VA1 in murine cells may reflect evolutionary divergence from the human cognate of the target protein for Ad5 VA1 regulation. eRNA41H also activated murine RIG-I (data not shown). This further demonstrated that eRNA-mediated RIG-I activation was not delivery or cell line specific, and eRNA ligand recognition by RIG-I was conserved between humans and mice.
RT-PCR demonstrated that similar levels of cytoplasmic VA1 were present in cells transfected with pDNAVACCUltra-VA1 and eRNA41H and other vectors with alternative orientations of eRNA11a and VA1 (data not shown). A derivative with two copies of VA1 had twice the level of cytoplasmic RNA (data not shown). This demonstrated that the VA1 promoter was insensitive to the presence of the U6 promoter or other alterations in flanking sequences and that the synergistic enhancement of RIG-I activation was not due to increased VA1 transcription. The synergistic RIG-I activation was also not the result of increased eRNA11a cytoplasmic RNA (total RNA or either strand) (Table 1).
The RNA interference pathway may compete with RIG-I for available dsRNA ligands. Once exported to the cytoplasm, 2-bp 3′ overhang hairpins are substrates for Dicer-mediated loop cleavage, RNA unwinding, and ssRNA presentation to Argonaute proteins of the RNA-induced silencing complex (RISC) that mediates RNA interference (71). When VA1 is expressed to high levels, the dsRNA terminal stem weakly activated OAS (10) and competitively inhibited exportin 5 and Dicer, effecting nonspecific suppression of RNA interference (1, 31) (Fig. 4A). VA1, through the structured central domain, is also a potent specific inhibitor of PKR (33) and ADAR (28). VA1 mutagenesis was utilized to determine which of these activities mediates the VA1/eRNA11a synergistic enhancement of RIG-I activation.
Internal alterations of the VA1 central domain (eRNA41H-PKR1 and -2 [Fig. 4A]), or substitution of VA2 for VA1 (data not shown), eliminated the VA1/eRNA11a synergistic RIG-I activation effect. This suggested that PKR or ADAR inhibition was critical, since VA2 and the modified VA1 RNAs maintained dsRNA stem structures and should retain similar RNA interference and OAS activation to the VA1 molecule. eRNA41H-PKR3 and -4 incorporated central domain mutations that inactivated VA1-mediated PKR inhibition. These RNAs were transcribed and exported to the cytoplasm in cell culture (34). Cytoplasmic export was verified for eRNA41H-PKR3 using RT-PCR (data not shown). However, these modified VA1 RNAs abolished the VA1-eRNA11a RIG-I activation enhancement (eRNA41H-PKR3 and -4 were equivalent to the eRNA11a component alone [Fig. 4A]). Most VA1 central domain mutations inactivate both PKR and ADAR inhibition. However, eRNA41H-PKR5 incorporated the unusual pm91 (49) 1-bp change in the central region that is inactive for PKR inhibition but retains partial ADAR inhibitory activity (28). This mutation also retained partial eRNA11a-dependent increased RIG-I activation (Fig. 4A), indicating that inhibition of ADAR, not PKR, was critical.
eRNA HA DNA vaccine improves HA-specific antibody binding avidity after naked DNA delivery.
The RIG-I agonist eRNA41H was integrated into the backbone of various DNA vaccine vectors (32) (J. A. Williams, world patent applications WO2006078979 and WO2008153733) expressing detoxified influenza virus H5N1 A/Vietnam/1203/2004 hemagglutinin (HA) (32). These vectors potently induced the IFN-β promoter in the RIG-I cell culture assay and expressed high levels of HA protein in HEK293 cells by Western blot analysis (data not shown).
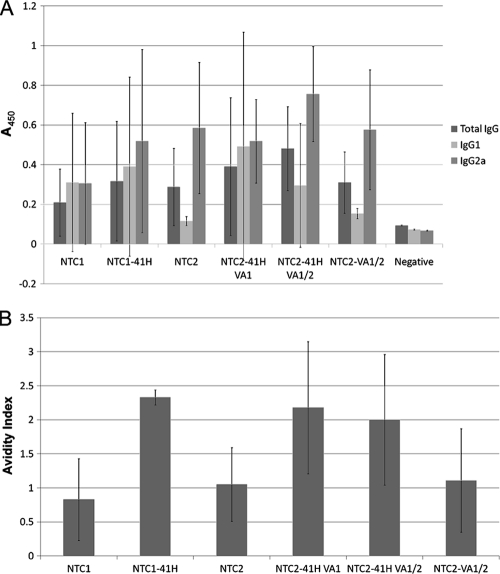
Antigen-specific humoral immune responses were evaluated in BALB/c mice after intramuscular (i.m.) delivery of 10 μg naked plasmid DNA in prime (day 0) and boost (day 21) injections. The eRNA41H HA vectors had similar HA-specific serum antibody titers (Fig. 6A). However, due to the low DNA dosage, the antibody titers were highly variable; thus, it is possible that eRNA41H had subtle effects on HA antibody titers that were not detectable in this study. In contrast, HA-specific antibody binding avidity was improved, compared to that of HA or VA1/2 HA vectors, on both day 21 (prime only [data not shown]) and day 49 (prime-boost [Fig. 6B]) with two different vector backbones. The improved avidity of eRNA41H HA compared to that of VA1/2 HA demonstrated that the effect is not mediated by VA1-mediated enhancement of HA expression.
FIG. 6.
RIG-I-activating eRNA41H improved humoral immune response to DNA vaccine-encoded antigen after prime (day 0) and boost (day 21) i.m. immunizations (no electroporation). (A) Antibody titers in seroconverted mice (day 49). ELISA well absorbances (average ± standard deviation [SD]) for IgG (1/12,500), IgG1 (1/500), and IgG2a (1/500) are shown. The high variability was attributed to the low dose (10 μg) used in the study. (B) Antibody avidity in seroconverted mice (day 49). The concentration (molarity) of sodium thiocyanate that elutes 50% of prebound antibody (avidity index) is shown. NTC1 is the AF NTC8382 CMV-HTLV-I R promoter vector (FIG 1B). NTC2 is the gWIZ HTLV-I R AF vector.
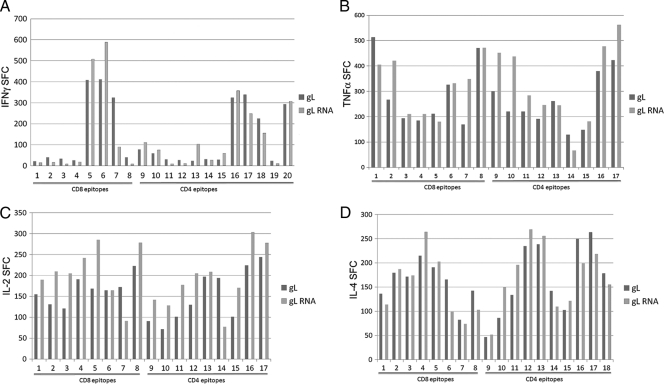
The effect of eRNA41H on antigen-specific cellular immune responses after naked plasmid immunization was then investigated. CD4 and CD8 T-cell responses to pITR p43-LAMP/gag wt vector, with or without eRNA41H, after subcutaneous naked DNA delivery were determined by ELISPOT. While many epitopes showed increased T-cell activation with eRNA41H, others did not (Fig. 7). There was no significant difference between CD8- or CD4-specific activation and no major differences with different cytokines with the exception that most CD8 epitopes with the IL-2 ELISPOT were increased with the eRNA41H vector. RIG-I activation in cell culture was confirmed with the pITR p43-LAMP/gag wt eRNA41H vector; as expected, the pITR p43-LAMP/gag wt parent vector was inactive.
FIG. 7.
eRNA41H effects on cellular immune response after prime (day 0) and boost (day 28) subcutaneous immunization (no electroporation). The number of spot-forming cells (SFC)/106 splenocytes (average ± SD) determined on day 38 by IFN-γ (A), TNF-α (B), IL-2 (C), or IL-4 (D) ELISPOT, using various CD4 or CD8 epitopes, are shown for pITR p43-LAMP/gag wt (gL) and pITR p43-LAMP/gag wt-eRNA41H (gL RNA). Similar results were obtained with IL-10 ELISPOT (data not shown).
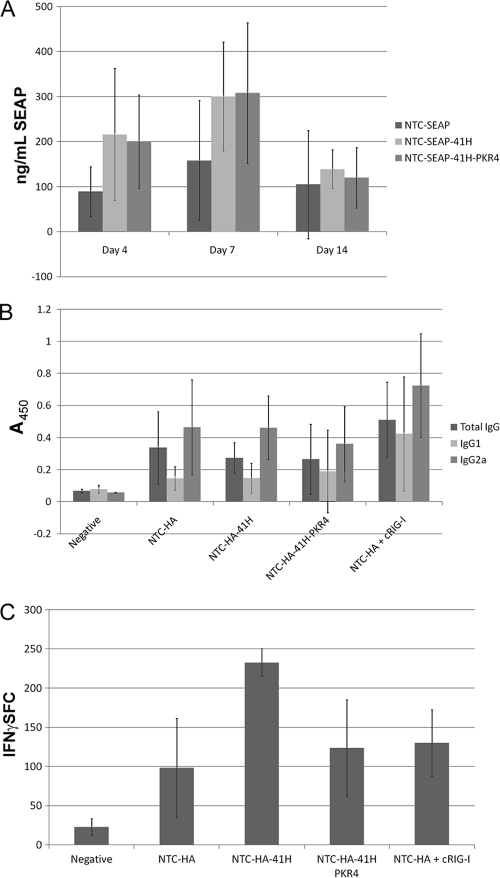
The effects of electroporation-enhanced DNA delivery on antigen-specific humoral and cellular immune responses to eRNA41H HA vector, compared to that of eRNA41H-PKR4 HA and HA vector controls, were also determined. SEAP versions of these vectors demonstrated that eRNA41H improved expression of SEAP in vivo after electroporation-mediated delivery to mice (Fig. 8A). Expression improvement required eRNA11a, since expression was also enhanced with the eRNA41H-PKR4 vector. Interestingly, eRNA41H had no effect on antibody titers (Fig. 8B) or avidity (data not shown) after electroporation-enhanced delivery. However, T-cell responses were induced 2-fold compared to that of the HA or eRNA41H-PKR4 HA vector groups (Fig. 8C). In contrast, immunization with cRIG-I improved antibody titers (Fig. 8B) and avidity (data not shown) but not T-cell response (Fig. 8C). The cRIG-I immunizations had the same total DNA quantity but less HA plasmid (2 μg HA plasmid and 1 μg cRIG-I plasmid; HA groups received 3 μg HA plasmid).
FIG. 8.
eRNA41H increased cellular immunity after electroporation delivery. (A) Expression of SEAP (average ± SD) in serum on days 0 (not shown; baseline) 4, 7, and 14 after a single i.m. dose (3 mice per group) of 3 μg NTC7382-SEAP (NTC-SEAP) or NTC7382-SEAP encoding eRNA41H (NTC-SEAP-41H) or eRNA41H-PKR4 (NTC-SEAP-41H-PKR4). (B) Groups of 8 mice were i.m. injected with 3 μg NTC7382-SEAP (Negative), NTC7382-HA vector (NTC-HA), NTC7382-HA vector encoding eRNA41H (NTC-HA-41H), NTC7382-HA vector encoding eRNA41H-PKR4 (NTC-HA-41H), or NTC-HA plus constitutive RIG-I (cRIG-I; 2 μg NTC7382-HA and 1 μg cRIG-I plasmid). ELISA well absorbances (average ± SD) for IgG (1/25,000), IgG1 (1/1000), and IgG2a (1/1000) are shown. (C) Numbers of spot forming cells (SFC)/106 splenocytes (average ± SD) as determined by IFN-γ ELISPOT to HA518 immunodominant peptide for the groups from panel B.
DISCUSSION
We report herein the creation and analysis of DNA vaccine vectors that coexpressed with antigen an immunostimulatory RIG-I-activating expressed RNA (eRNA). Plasmid-encoded RNA Pol III-expressed RIG-I agonists (hairpin dsRNA43 and convergently transcribed dsRNA eRNA11a) were identified using a cell culture transfection assay, and ligand conservation between murine and human RIG-I was demonstrated. A combinational RIG-I agonist eRNA41H (convergent eRNA11a and VA1) activated an IFN-β reporter in human and murine cell lines. eRNA41H DNA vaccine vectors combined high-level antigen expression with RNA-mediated type I IFN activation. RIG-I-activating DNA vaccines therefore activate polyvalent (RNA and DNA agonist) innate immune responses. This may be desirable, since highly effective viral vector vaccines, such as yellow fever vaccine 17D, and adenoviral vectors activate multiple innate immune pathways. Consistent with this, antibody avidity after naked DNA immunization was increased with eRNA41H influenza virus HA DNA vaccines. This is a significant improvement, since antibodies that bind antigen tightly often are more effective viral or toxin neutralizers.
Plasmid-mediated RIG-I activation is limited to transfected antigen-expressing cells. This targeted immunostimulation is a far safer approach than nonspecific administration of large doses of RNA or DNA adjuvants and avoids the potential cytokine storm that is possible with broadly delivered high-dose adjuvants that may alter serum titers of immune effector molecules.
RIG-I-activating DNA vaccines could also be expected to induce RIG-I effector-mediated antiviral defenses in transfected cells and may therefore have utility for tissue-targeted antiviral therapy, such as nasal sprays. This could be assessed in vitro or in vivo, for example, by administrating the plasmid vaccine intranasally or intratracheally prior to inhalation virus infection to determine if the vaccine affects a decrease in virus titer.
TLRs are differentially expressed in human cells compared to in mice. It is likely that the differences in TLR expression patterns, responsiveness, and immunology between mice and humans dramatically affect vaccine performance in humans (38). Such issues are not anticipated with ubiquitously expressed RIG-I (69).
The nonrepetitive eRNA41H vectors are preferable for manufacture compared to long hairpin-containing vectors since hairpins reduce plasmid copy number due to replication inhibition. For example, eRNA41H DNA vaccine vectors had high-yield plasmid manufacture up to 2,200 mg/liter (68), while reduced yields of 343 mg/liter were obtained with a vector containing the 43-bp dsRNA43 hairpin (66). However, hairpin vectors could be utilized for therapeutic strategies requiring a smaller manufacturing scale, such as cancer vaccines.
Production of RIG-I-activating RNAs from a plasmid vector presented some special considerations. The viral RNAs that RIG-I detects are generally produced in the cytoplasm using viral RNA polymerases that do not remove the 5′-PPP. As well, proteins bound to the 5′-PPP may mask the RNA from RIG-I detection, such as with endogenous 5S rRNA, which is ribosome associated after nucleosome assembly. Careful design is needed to obtain 5′-PPP cytoplasmic RNAs using the endogenous nuclear transcription and nuclear export capacities of a cell needed for expression from a plasmid. First, to obtain a 5′-PPP on a nuclear-produced RNA, an appropriate promoter must be utilized such that the 5′-PPP is not removed by capping (mRNA) or processing (e.g., tRNA, rRNA). The RNA must then be exported from the nucleus to the cytoplasm since plasmid transcription will occur in the nucleus while the RIG-I receptor is in the cytoplasm. The 5′-PPP-containing cytoplasmic RNA must then act as a ligand and activator of RIG-I. Plumet et al. failed to activate RIG-I using a plasmid vector in which the RNA Pol III H1 promoter was used to express a known RIG-I activator, measles viral leader (45). RIG-I activation was obtained using plasmid-driven expression of 10 copies of a native Epstein-Barr virus small RNA (EBER1), another known RIG-I ligand (50). However, plasmids containing 10 copies of a repetitive sequence are unstable in E. coli (65). An alternative approach, generation of RIG-I ligands by cytoplasmic RNA Pol III transcription of polyAT repeats (9), did not result in RIG-I activation when annealed DNA fragments polyAT40 (40 AT repeats), or AAAAAA-polyAT40TTTTTT (RNA Pol III terminators flanking polyAT40), were incorporated into pDNAVACCUltra-EGFP and pDNAVACCUltra-VA1-EGFP vector backbones (J. Williams, unpublished observations).
The combination of VA1 and eRNA11a synergistically increased RIG-I activation. Perhaps eRNA11a binding to PKR, ADAR, Dicer, or other cellular receptors that limit ligand availability for RIG-I binding is competed by VA1. Alternatively, VA1 may activate an alternative receptor (e.g., OAS, MDA5, PKR) that synergizes with the eRNA11a RIG-I ligand to amplify RIG-I activation. Mutation analysis indicated that the VA1-mediated increase in eRNA11a potency correlated with ADAR inhibition. Different forms of ADAR modify (A-to-I edit) dsRNA in both the nucleus and the cytoplasm. Mammalian genomes contain many natural sense-antisense transcripts that have the potential to produce dsRNA. Long nuclear dsRNA is a substrate for ADAR. Export of the resultant extensively adenosine-to-inosine-edited RNA from the nucleus is inhibited (e.g., by binding to p54nrb [reviewed in reference 62]). Thus, long dsRNA (>30 bp) expressed in the nucleus will be edited by ADAR, and the modified versions are not exported to the cytoplasm. Consistent with this, a plasmid vector containing a long dsRNA (250 bp) expressed by convergent transcription of U6 and H1 promoters did not activate PKR or the interferon response after transfection and expression of the dsRNA (55). However, in our studies, the levels of cytoplasmic eRNA11a were not increased in the presence of VA1. This implies that ADAR does not prevent eRNA11a export and instead that A-to-I ADAR-edited cytoplasmic dsRNA may have reduced RIG-I agonist activity. Consistent with this hypothesis, ADAR-edited dsRNA suppresses poly(IC)-mediated IFN induction and inhibits IRF3 activation (61). This may be a natural feedback regulation mechanism that downregulates RIG-I signaling.
HA-specific antibody binding avidity after naked DNA intramuscular delivery was improved with eRNA HA vectors designed to activate IPS-1-mediated IRF3 signaling. The effect was not an indirect effect of VA1-mediated increased expression, since HA and HA-VA1 vectors had similar avidity.
The mechanisms controlling vaccination-induced antibody affinity maturation are complex. For example, for unknown reasons avidity and protective antibody response were improved with heterologous DNA prime/protein boost over homologous DNA or protein immunization (59). However, the improvement in antibody avidity with eRNA vectors was consistent with the reported loss of antibody affinity and reduced antibody-mediated protection after West Nile virus vector immunization of IPS-1 knockout mice compared to that of wild-type mice (57). Thus, IPS-1 or downstream IRF3 activation may be important for antibody maturation.
Plasmid delivered by naked injection will enter the cell through endosomal uptake, and unmethylated CpG residues bind endosomal TLR9, activating IRF7 through myeloid differentiation primary response gene 88 (MyD88) signaling (23). The combination of eRNA-mediated IRF3 activation with unmethylated CpG-mediated IRF7 activation may improve adaptive immunity. Alternatively, RIG-I activation of the inflammasome resulting in interleukin 1β production (47) may mediate the observed humoral response improvements. This model predicts that eRNA vectors will improve immune responses when combined with endocytosis-mediated plasmid delivery vehicles such as liposomal and nanoparticle formulations that internalize plasmid in endosomes.
Electroporation delivery transfers plasmid directly to the cytoplasm, where it activates IRF3 through TBK-1 signaling (53). This existing IRF3 activation may explain why antibody avidity was not improved with EP-delivered eRNA. Alternatively, electroporation itself has adjuvant effects, stimulating antigen-presenting cell recruitment; this effect eliminated the benefit of plasmid chemokine adjuvants MIP-1α/Flt3L that also recruit APCs (30) and may also be redundant with eRNA. The improved antibody avidity observed with cRIG-I overexpression may indicate that this protein adjuvant induces higher levels of IPS-1/IRF3 activation in vivo than is attainable with eRNA.
The TBK-1-activating N-CARD of IPS-1 improved humoral and cellular responses to protein vaccines (18). The eRNA ligand developed here also signals through IPS-1 but is superior for immunostimulation since, unlike protein ligands, it will not limit repeat usage of the DNA vaccine vector. As well, significant autoantibody safety concerns exist with a hybrid human protein like N-CARD. A targeted RNA ligand expressed in minute amounts only in transfected cells will present less regulatory concerns.
The effects of eRNA41H on cellular immunity were complex. A 2-fold increase in T-cell response to influenza virus HA was observed after EP delivery. However, with the exception of a less-than-2-fold eRNA-mediated improvement for IL-2 with most CD8 epitopes, no effect on CD8 and CD4 cellular immune response to HIV gag was observed after naked subcutaneous immunization. The high dosage of 50 μg used in the naked DNA study may have obscured the detection of weak adjuvant effects. As well, cellular responses to eRNA may be tissue specific. MDA5 activation was essential for the establishment of CD8 T-cell memory after vaccination with poly(I:C) adjuvant (63); an evaluation of RIG-I-activating eRNA to determine if it similarly enhances generation of cellular memory needs to be performed. Indeed, eRNA41H treatment increased the IL-2 level in CD8 epitopes; since it is known that IL-2 stimulates growth, differentiation, and survival of cytotoxic T cells, eRNA41H treatment may lead to an increase in mature CD8 T-cell number and enhanced generation of cellular memory.
Previous studies using HIV env and nef DNA vaccines coexpressing an RNA Pol II-directed 40-bp hairpin demonstrated that the hairpin increased T-cell response 2-fold to both antigens but had no effect on humoral response (29). This hairpin would not activate RIG-I since RNA Pol II transcripts are capped. Therefore, enhancement of cellular immunity with dsRNA in these studies may be due to activation of an alternative receptor, such as MDA5, TLR3, PKR, or ADAR, or activation of the inflammasome or apoptosis (see below). Consistent with this, cRIG-I did not improve T-cell response after electroporation (Fig. 8C). The relative contribution of different signaling pathways could be determined by vaccination of homozygous knockout mice. However, these results suggest that the hairpin dsRNA43 RIG-I ligand may be more potent than convergent eRNA11a for induction of cellular responses.
Incorporation of RLH ligands into the vector backbone of an antigen-expressing Ad or adeno-associated virus (AAV) vector may have targeted immunostimulation application. Adaptive immune responses to AAV vectors, which do not activate cytoplasmic DNA receptor signaling pathways, are TLR9 dependent (74). Ad vector DNA is also an agonist of TLR9, as well as cytoplasmic DNA receptors and the inflammasome (40, 41, 73) but not RIG-I/MDA5 (12). Poly(I:C), a dual ligand for TLR3 and MDA5, was an effective adjuvant for Ad vectors; the MDA5 pathway was critical for this adjuvant activity (24). As well, incorporation of a TLR3 agonist into an Ad vector backbone improved antibody responses to coexpressed influenza virus HA antigen after oral administration (S. N. Tucker, world patent application WO2007100908). However, in many tissues, RIG-I, not TLR3, is the dominant pathway for induction of cytokine responses to dsRNA. Thus, eRNA-modified Ad and AAV vectors may be superior for vaccination.
The RIG-I pathway is conserved in vertebrates (75) and is involved in innate immune signaling in fish (6) and ducks (although chickens do not have RIG-I [3]). Thus, RIG-I-activating DNA vaccines may have an application for improving DNA vaccination for various agricultural applications. RIG-I agonists, through an alternative IPS-1-dependent signaling pathway, activate proapoptotic proteins (70); this results in apoptosis in Bcl-XL-deficient tumor cells, but not Bcl-XL-protected primary cells, to overcome cancer cell resistance to apoptosis (5). As well, combinations of dsRNA and CpG have been demonstrated to synergistically enhance innate immune responses for tumor vaccination (64). Thus, RIG-I-activating DNA vaccines may have therapeutic potential for cancer treatment.
Acknowledgments
We thank Kim Hanson for purifying the plasmid DNA used in this study, Marni England-Hill for oversight of the murine study at Aldevron, and Rune Kjeken for help designing the murine study performed at Explora Laboratories.
This paper described work supported by NIH grants R44GM072141, R43 GM080768-01, and R43A1071660.
J.M.L., C.P.H., and J.A.W. have an equity interest in Nature Technology Corporation.
Footnotes
Published ahead of print on 24 November 2010.
REFERENCES
- 1.Andersson, M. G., et al. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 79:9556-9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda, L. B., et al. 2006. Dendritic cell-lysosomal-associated membrane protein (LAMP) and LAMP-1-HIV-1 gag chimeras have distinct cellular trafficking pathways and prime T and B cell responses to a diverse repertoire of epitopes. J. Immunol. 177:2265-2275. [DOI] [PubMed] [Google Scholar]
- 3.Barber, M. R., J. R. Aldridge, R. G. Webster, and K. E. Magor. 2010. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. U. S. A. 107:5913-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barchet, W., V. Wimmenauer, M. Schlee, and G. Hartmann. 2008. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr. Opin. Immunol. 20:389-395. [DOI] [PubMed] [Google Scholar]
- 5.Besch, R., et al. 2009. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 119:2399-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biacchesi, S., et al. 2009. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J. Virol. 83:7815-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitro, V., A. Musiyenko, M. A. Bayfield, R. J. Maraia, and S. Barik. 2008. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J. Virol. 82:7977-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramson, J. L., et al. 2003. Super-activated interferon-regulatory factors can enhance plasmid immunization. Vaccine 21:1363-1370. [DOI] [PubMed] [Google Scholar]
- 9.Cao, X. 2009. New DNA-sensing pathway feeds RIG-I with RNA. Nat. Immunol. 10:1049-1051. [DOI] [PubMed] [Google Scholar]
- 10.Desai, S. Y., et al. 1995. Activation of interferon-inducible 2′-5′ oligoadenylate synthetase by adenoviral VAI RNA. J. Biol. Chem. 270:3454-3461. [DOI] [PubMed] [Google Scholar]
- 11.Fang, J., S. Bredow, P. Taishi, J. A. Majde, and J. M. Krueger. 1999. Synthetic influenza viral double-stranded RNA induces an acute-phase response in rabbits. J. Med. Virol. 57:198-203. [PubMed] [Google Scholar]
- 12.Fejer, G., et al. 2008. Key role of splenic myeloid DCs in the IFN-alphabeta response to adenoviruses in vivo. PLoS Pathog. 4:e1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwizdek, C., et al. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278:5505-5508. [DOI] [PubMed] [Google Scholar]
- 14.Hornung, V., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 15.Hornung, V., and E. Latz. 2010. Intracellular DNA recognition. Nat. Rev. Immunol. 10:123-130. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, K. J., et al. 2008. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451:725-729. [DOI] [PubMed] [Google Scholar]
- 17.Jechlinger, W. 2006. Optimization and delivery of plasmid DNA for vaccination. Expert Rev. Vaccines 5:803-825. [DOI] [PubMed] [Google Scholar]
- 18.Kobiyama, K., et al. 2009. A signaling polypeptide derived from an innate immune adaptor molecule can be harnessed as a new class of vaccine adjuvant. J. Immunol. 182:1593-1601. [DOI] [PubMed] [Google Scholar]
- 19.Köhler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761-773. [DOI] [PubMed] [Google Scholar]
- 20.Koyama, S., et al. 2009. Innate immune control of nucleic acid-based vaccine immunogenicity. Expert Rev. Vaccines 8:1099-1107. [DOI] [PubMed] [Google Scholar]
- 21.Kumagai, Y., O. Takeuchi, and S. Akira. 2008. Pathogen recognition by innate receptors. J. Infect. Chemother. 14:86-92. [DOI] [PubMed] [Google Scholar]
- 22.Kumagai, Y., O. Takeuchi, and S. Akira. 2008. TLR9 as a key receptor for the recognition of DNA. Adv. Drug Deliv. Rev. 60:795-804. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, H., T. Kawai, and S. Akira. 2009. Pathogen recognition in the innate immune response. Biochem. J. 420:1-16. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, H., S. Koyama, K. J. Ishii, T. Kawai, and S. Akira. 2008. Cutting edge: cooperation of IPS-1 and TRIF-dependent pathways in poly IC-enhanced antibody production and cytotoxic T cell responses. J. Immunol. 180:683-687. [DOI] [PubMed] [Google Scholar]
- 25.Kuwabara, T., et al. 2001. Significantly higher activity of a cytoplasmic hammerhead ribozyme than a corresponding nuclear counterpart: engineered tRNAs with an extended 3′ end can be exported efficiently and specifically to the cytoplasm in mammalian cells. Nucleic Acids Res. 29:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahira, A., P. Das, and D. Chakravortty. 2008. Engagement of TLR signaling as adjuvant: towards smarter vaccine and beyond. Vaccine 26:6777-6783. [DOI] [PubMed] [Google Scholar]
- 27.Lamb, R. A., and P. W. Choppin. 1983. The gene structure and replication of influenza virus. Annu. Rev. Biochem. 52:467-506. [DOI] [PubMed] [Google Scholar]
- 28.Lei, M., Y. Liu, and C. E. Samuel. 1998. Adenovirus VAI RNA antagonizes the RNA-editing activity of the ADAR adenosine deaminase. Virology 245:188-196. [DOI] [PubMed] [Google Scholar]
- 29.Li, D., et al. 2007. Adjuvant effects of plasmid-generated hairpin RNA molecules on DNA vaccination. Vaccine 25:6992-7000. [DOI] [PubMed] [Google Scholar]
- 30.Li, J., R. Kjeken, I. Mathiesen, and D. H. Barouch. 2008. Recruitment of antigen-presenting cells to the site of inoculation and augmentation of human immunodeficiency virus type 1 DNA vaccine immunogenicity by in vivo electroporation. J. Virol. 82:5643-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, S., and B. R. Cullen. 2004. Adenovirus VA1 Noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J. Virol. 78:12868-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luke, J., A. E. Carnes, C. P. Hodgson, and J. A. Williams. 2009. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine 27:6454-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, Y., and M. B. Mathews. 1993. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. J. Virol. 67:6605-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma, Y., and M. B. Mathews. 1996. Secondary and tertiary structure in the central domain of adenovirus type 2 VA RNAI. RNA 2:937-951. [PMC free article] [PubMed] [Google Scholar]
- 35.Marques, J. T., et al. 2006. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 24:559-565. [DOI] [PubMed] [Google Scholar]
- 36.Mathews, M. B., and A. M. Francoeur. 1984. La antigen recognizes and binds to the 3′ oligouridylate tail of a small RNA. Mol. Cell. Biol. 4:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melchjorsen, J., et al. 2005. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 79:12944-12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mestas, J., and C. C. W. Hughes. 2004. Of mice and not men: differences between mouse and human immunology. J. Immunol. 172:2731-2738. [DOI] [PubMed] [Google Scholar]
- 39.Mibayashi, M., et al. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muruve, D. A., et al. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103-107. [DOI] [PubMed] [Google Scholar]
- 41.Nociari, M., O. Ocheretina, J. W. Schoggins, and E. Falck-Pedersen. 2007. Sensing infection by adenovirus: Toll-like receptor-independent viral DNA recognition signals activation of the interferon regulatory factor 3 master regulator. J. Virol. 81:4145-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada, C., et al. 2009. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326:1275-1279. [DOI] [PubMed] [Google Scholar]
- 43.Pichlmair, A., et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 44.Pichlmair, A., et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plumet, S., et al. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS 2:e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poeck, H., et al. 2008. 5′-Triphosphate-siRNA: turning gene silencing and RIG-I activation against melanoma. Nat. Med. 14:1256-1263. [DOI] [PubMed] [Google Scholar]
- 47.Poeck, H., et al. 2010. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 11:63-69. [DOI] [PubMed] [Google Scholar]
- 48.Pullen, G. R., M. G. Fitzgerald, and C. S. Hosking. 1986. Antibody avidity determination by ELISA using thiocyanate elution. J. Immunol. Methods 86:83-87. [DOI] [PubMed] [Google Scholar]
- 49.Rahman, A., P. Malhotra, R. Dhar, T. Kewalramani, and B. Thimmapaya. 1995. Effect of single-base substitutions in the central domain of virus-associated RNA I on its function. J. Virol. 69:4299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samanta, M., D. Iwakira, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki, S., R. R. Amara, W. S. Yeow, P. M. Pitha, and H. L. Robinson. 2002. Regulation of DNA-raised immune responses by cotransfected interferon-regulatory factors. J. Virol. 76:6652-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlee, M., et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shirota, H., L. Petrenko, T. Hattori, and D. M. Klinman. 2009. Contribution of IRF-3 mediated IFN-β production to DNA vaccine dependent cellular immune responses. Vaccine 27:2144-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, R., S. Gupta, and R. Reddy. 1990. Capping of mammalian U6 small nuclear RNA in vitro is directed by a conserved stem-loop and AUAUAC sequence: conversion of a noncapped RNA into a capped RNA. Mol. Cell Biol. 10:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strat, A., et al. 2006. Specific and nontoxic silencing in mammalian cells with expressed long dsRNAs. Nucleic Acids Res. 34:3803-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumpter, R., et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suthar, M. S., et al. 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 6:e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unterholzner, L., et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaine, M., S. Wang, A. Hackett, J. Arthos, and S. Lu. 2010. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine 28:2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vennström, B., U. Pettersson, and L. Philipson. 1978. Two initiation sites for adenovirus 5.5S RNA. Nucleic Acids Res. 5:195-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vitali, P., and A. D. J. Scadden. 2010. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 17:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Q., and G. G. Carmichael. 2004. Effect of length and location on the cellular response to double-stranded RNA. Microb. Mol. Biol. Rev. 68:432-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, W., M. Cella, S. Gilfillan, and M. Colonna. 2010. Cutting edge: polinosinic:polycytidylic acid boosts the generation of memory CD8 cells through melanoma differentiation-associated protein 5 expressed in stromal cells. J. Immunol. 184:2751-2755. [DOI] [PubMed] [Google Scholar]
- 64.Whitmore, M. M., et al. 2004. Synergistic activation of innate immunity by double-stranded RNA and CpG DNA promotes enhanced antitumor activity. Cancer Res. 64:5850-5860. [DOI] [PubMed] [Google Scholar]
- 65.Williams, J. A., A. E. Carnes, and C. P. Hodgson. 2009. Plasmid DNA vaccine vector design: impact on efficacy, safety and upstream production. Biotechnol. Adv. 27:353-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams, J. A., C. P. Hodgson, and A. E. Carnes. 2009. Rapid process development for high yielding plasmid DNA fed-batch fermentation. BioPharm Int. 22(11):46-51. [Google Scholar]
- 67.Williams, J. A., J. Luke, L. Johnson, and C. P. Hodgson. 2006. pDNAVACCultra vector family: high throughput intracellular targeting DNA vaccine plasmids. Vaccine 24:4671-4676. [DOI] [PubMed] [Google Scholar]
- 68.Williams, J. A., et al. 2009. Generic plasmid DNA production platform incorporating low metabolic burden seed-stock and fed-batch fermentation processes. Biotechnol. Bioeng. 103:1129-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoneyama, M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 70.Yu, C. Y., R. L. Chiang, T. H. Chang, C. L. Liao, and Y. L. Lin. 2010. The interferon stimulator mitochondrial antiviral signaling protein facilitates cell death by disrupting the mitochondrial membrane potential and by activating caspases. J. Virol. 84:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng, Y., and B. R. Cullen. 2004. Structural requirements for pre-microRNA binding and nuclear export by exportin 5. Nucleic Acids Res. 32:4776-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng, X., and P. C. Bevilacqua. 2004. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, J., X. Huang, and Y. Yang. 2007. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and-independent pathways. J. Virol. 81:3170-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu, J., X. Huang, and Y. Yang. 2009. The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J. Clin. Invest. 119:2388-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou, J., M. Chang, P. Nie, and C. J. Secombes. 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zuker, M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]