Abstract
A consortium of the newly isolated bacterial strains Arthrobacter sp. strain G1 and Ralstonia sp. strain H1 utilized 4-fluorocinnamic acid for growth under aerobic conditions. Strain G1 converted 4-fluorocinnamic acid into 4-fluorobenzoic acid and used the two-carbon side chain for growth, with some formation of 4-fluoroacetophenone as a dead-end side product. In the presence of strain H1, complete mineralization of 4-fluorocinnamic acid and release of fluoride were obtained. Degradation of 4-fluorocinnamic acid by strain G1 occurred through a β-oxidation mechanism and started with the formation of 4-fluorocinnamoyl-coenzyme A (CoA), as indicated by the presence of 4-fluorocinnamoyl-CoA ligase. Enzymes for further transformation were detected in cell extract, i.e., 4-fluorocinnamoyl-CoA hydratase, 4-fluorophenyl-β-hydroxy propionyl-CoA dehydrogenase, and 4-fluorophenyl-β-keto propionyl-CoA thiolase. Degradation of 4-fluorobenzoic acid by strain H1 proceeded via 4-fluorocatechol, which was converted by an ortho-cleavage pathway.
Fluorinated organic compounds are of growing industrial importance, with applications such as agrochemicals, pharmaceuticals, and performance materials (23, 24, 28, 41). The safe use of such compounds, as well as appropriate disposal and treatment of wastes, will benefit from knowledge about their biodegradation. However, little information is available about the microbial metabolism of fluorinated organic compounds compared to other halogenated chemicals. Most studies on the bacterial degradation of fluorinated organics describe fluorobenzoic acids, which under aerobic conditions can be converted into the corresponding fluorocatechols (3, 17, 33). Papers about the degradation of fluorophenols have also appeared (11, 25, 51).
4-Fluorocinnamic acid (4-FCA) is used in industry for the synthesis of flavors and pharmaceuticals (8), and polymers of 4-FCA are applied in electronics (14). It was proposed that under aerobic conditions in nonacclimated industrial activated sludge, 4-FCA could be converted into 4-fluorobenzoic acid (4-FBA) via the formation of 4-fluoroacetophenone (4-FAP) (8, 32). In another study, using activated sludge from a wastewater treatment plant, 4-FCA was suggested to be transformed into an epoxide that is converted to 4-FAP. This compound would then be converted into 4-FBA, but no products were detected from the breakdown of 4-FBA (5).
The conversion of nonhalogenated cinnamic acids to benzoic acids, such as the transformation of ferulic acid to vanillic acid, has been described previously (2, 4, 20, 31, 39). Cinnamic acid, coumaric acid, and ferulic acid are transformed by Streptomyces setonii (47) and Rhodopseudomonas palustris to benzoic acid or the corresponding derivatives (19). Alcanivorax borkumensis MBIC 4326 (9) and Papillibacter cinnamivorans (7) transformed cinnamic acid into benzoic acid. The metabolism of these compounds thus proceeds with side chain degradation prior to ring cleavage. Side chain degradation is carried out either by β-oxidation or by direct deacetylation mechanisms, which leads to elimination of two carbon units from the unsaturated side chain in bacteria, yeasts, and fungi (40).
Since no clear information on the degradation route of 4-FCA is available, we have isolated two pure bacterial strains to study the complete microbial metabolism of the compound, and in this paper, we propose a degradation pathway.
MATERIALS AND METHODS
Growth conditions.
Cells of strains G1 and H1 were grown aerobically at 30°C under rotary shaking or in a fermentor. Growth medium (MMY) contained (per liter) 5.37 g of Na2HPO4·12H2O, 1.36 g of KH2PO4, 0.5 g of (NH4)2SO4, and 0.2 g of MgSO4·7H2O supplemented with a trace element solution (5 ml l−1) containing (per liter) 780 mg of Ca(NO3)2·4H2O, 200 mg of FeSO4·7H2O, 20 mg of Na2SeO4·10H2O, 10 mg of ZnSO4·7H2O, 10 mg of H3BO3, 10 mg of CoCl2·6H2O, 10 mg of CuSO4·5H2O, 4 mg of MnSO4·H2O, 3 mg of Na2MoO4·2H2O, 2 mg of NiCl2·6H2O, and 2 mg of Na2WO4·2H2O. MMY medium also contained 10 mg/ml yeast extract (Difco Laboratories).
Enrichment and isolation of 4-FCA- and 4-FBA-degrading organisms.
Soil samples collected from a site in the Netherlands contaminated mainly with chlorobenzene and halogenated aliphatic compounds were used as the initial inocula for the 4-FCA and 4-FBA enrichment cultures. Flasks contained 40 ml MMY and 5 mM 4-FCA or 5 mM 4-FBA as the sole source of carbon and energy. The cultures were incubated at room temperature on a rotary shaker (150 rpm), and 40% of the suspension was transferred to a new flask containing fresh medium every 15 days. Growth (optical density [OD] at 450 nm) and liberation of fluoride were monitored. Samples of the enrichment cultures were periodically spread onto nutrient broth (NB) plates and MMY agar plates containing 5 mM 4-FCA or 5 mM 4-FBA, and pure cultures were obtained by repetitive streaking on these plates and testing for growth in liquid medium containing 5 mM 4-FCA or 4-FBA. Growth and fluoride release were again monitored to verify 4-FCA and 4-FBA degradation. Pure cultures that were capable of 4-FCA and 4-FBA degradation were selected and designated strains G1 and H1, respectively. The organisms were deposited at DSMZ (DSM 23642 for strain H1 and DSM 23643 for strain G1).
Identification of 4-FCA- and 4-FBA-degrading cultures.
16S rRNA genes were amplified from genomic DNA with the universal bacterial primers 27F and 1492R (26). The final PCR mixture (20 μl) contained 50 ng genomic DNA, 1× Pfx50 buffer (Invitrogen), 0.25 μM each deoxynucleoside triphosphate (dNTP), 0.1% (wt/vol) bovine serum albumin, and 0.25 μM each primer. After initial incubation for 11 min at 94°C, amplification was started by the addition of 0.5 U of Pfx50 DNA polymerase (Invitrogen), followed by 25 cycles of 94°C for 1 min, 55°C for 1 min, and 68°C for 1 min, and a final extension step at 68°C for 10 min. The PCR products were purified (QIAquick PCR purification kit; Qiagen Inc.), cloned into the pZero-2 vector (Invitrogen), and sequenced.
Whole-cell transformation and accumulation of intermediates in batch culture.
Cells of strains G1 and H1 were grown on 5 mM (each) 4-FCA and 4-FBA, respectively, and harvested at mid-log phase at an optical density at 450 nm of approximately 0.5. Following centrifugation at 4,000 × g for 10 min, the cells were washed twice with 100 mM potassium phosphate buffer (pH 6.8) and resuspended in the same buffer. The cell suspensions of strains G1 and H1 were separately added to 1-liter flasks containing 250 ml of MMY medium supplemented with 5 or 10 mM 4-FCA or 4-FBA. The cultures were incubated in a rotary shaker at 30°C and 150 rpm. Samples were taken at suitable intervals, centrifuged at 16,000 × g, and analyzed immediately by HPLC, liquid chromatography-mass spectrometry (LC-MS), and ion chromatography.
Analytical methods.
Concentrations of 4-FCA, 4-FBA, and their metabolites in culture supernatants were determined by reverse-phase HPLC (Jasco PU-2086 pump and Jasco AS-2051 autosampler), using a Lichrosorb C18 column (250 by 4.6 mm; 5-μm particle size). The mobile phase contained 70% 0.02 mM ammonium acetate, pH 4.5, and 30% methanol. An injection volume of 10 μl was used. The flow rate was 0.8 ml min−1 with detection at 254 nm with a UV absorbance detector (Jasco UV-2075).
Transformation products of 4-FCA were also analyzed using a Thermo Fisher Scientific LCQ Fleet MS ion trap mass spectrometer with Xcalibur software (version 1.3). Separation was performed on an Alltima HP C18 (2.1- by 50-mm; 3-μm) column. The flow rate was set to 200 μl min−1. Elution solvent A consisted of 10:90 acetonitrile/water, and solvent B was pure acetonitrile, both containing 15 mM ammonium hydroxide. Elution solvent C, which consisted of 30:70:1 water/acetonitrile/formic acid, was used to elute nonspecifically bound compounds from the HPLC column to achieve long-term separation stability. The chromatographic conditions were as follows: 100% A to 55% A/45% B in 8 min; 8 to 10 min, isocratic 55% A/45% A; 55% A/45% B to 100% C in 2 min; 100% C for 5 min; and 100% C to 100% A in 2 min. The equilibration time with A was set to 8 min. Separation was performed at room temperature. The injection volume was 10 μl.
Positive electrospray ionization-tandem mass spectrometry (ESI-MS/MS) and negative atmospheric pressure chemical ionization (APCI)-MS/MS were performed using a source voltage of 5 kV. The heated capillary temperature was set at 300°C. Nitrogen was used for nebulization (60 liters h−1). A capillary voltage of 30 V was used. A vaporizing temperature of 350°C was used in APCI mode.
Fluoride measurements were performed on a DX 120 ion chromatograph (Dionex, Sunnyvale, CA) connected to an autosampler and equipped with an Alltech A-2 anion column (100 by 4.6 mm; 7 μm) and an Alltech guard column (50 by 4 mm). An injection volume of 50 μl was used. The column temperature was set to 30°C. The eluent used was a mixture of NaHCO3 and Na2CO3 in deionized water at a flow rate of 1.2 ml min−1.
Preparation of cell extracts.
Cells of strain G1 or H1 were cultivated in MMY medium supplemented with 5 mM 4-FCA or 4-FBA, respectively. Cells were harvested by centrifugation (6,000 × g and 4°C for 30 min), washed twice with 30 ml of 100 mM potassium phosphate buffer (pH 6.8), and resuspended in 15 ml of the same buffer. The cells were subsequently broken using a Vibra-Cell sonicator (twice for 5 min with a 5-min interval at 50% duty cycle) (Sonics and Materials Inc., Danbury, CT). The lysate was then centrifuged (40,000 × g; 4°C; 60 min) to remove cell debris and unbroken cells. The supernatant was used for enzyme analysis.
Partial purification of proteins involved in the degradation of 4-FCA.
Cell extract of strain G1 induced with 4-FCA was loaded onto a Q Sepharose column (50 ml; Pharmacia, Uppsala, Sweden) preequilibrated with 100 mM potassium phosphate buffer (pH 7.5). After the column was washed with 2 column volumes of buffer, elution was carried with a linear gradient from 0 to 100% of 0.5 M (NH4)2SO4 in the same buffer. Active fractions were collected, separately concentrated by using 10-kDa Millipore filters (Amicon), and tested for coenzyme A (CoA)-ligase, hydratase, dehydrogenase, and thiolase activities.
Enzyme assays.
All enzyme assays were carried out spectrophotometrically at 25°C. One unit of activity was defined as the amount of enzyme required to convert 1 μmol of substrate per min at 25°C. Specific activities were expressed as units per mg of protein. Since β-hydroxy and β-keto derivatives of 4-FCA were not commercially available, the specific activities of their corresponding enzymes were expressed in arbitrary units (units of absorbance change per min) per ml of reaction mixture.
The assay mixture for CoA ligase activity contained, in a total volume of 1 ml, 200 mM potassium phosphate buffer (pH 7.0), 0.2 mM 4-FCA, 0.5 mM ATP, 0.2 mM coenzyme A, 0.5 mM MgCl2, and a suitable amount of partially purified CoA ligase. The extinction coefficient for 4-fluorocinnamoyl-CoA (FC-CoA) was determined by measuring the final absorbance after complete conversion of 25 and 50 μM substrate. The ligase activity was determined by monitoring the conversion of 4-FCA to its CoA thioester at 312 nm (extinction coefficient [ɛ] = 18,800 M−1 cm−1) using a Lambda Bio 10 UV/visible-light spectrophotometer (Perkin Elmer). Activity was also determined by measuring the disappearance of 4-FCA by HPLC.
Conversion of FC-CoA was tested by adding partially purified hydratase, dehydrogenase, and NAD (0.2 mM) to the reaction mixture. This was expected to initiate the conversion of FC-CoA to 4-fluorophenyl-β-hydroxy propionyl-CoA and subsequently to 4-fluorophenyl-β-keto propionyl-CoA. The conversion was followed in a spectrophotometer at 314 nm and 356 nm.
CoA-dependent conversion of 4-fluorophenyl-β-keto propionyl-CoA to 4-fluorobenzoyl-CoA was tested by adding coenzyme A (0.2 mM) and partially purified thiolase to the reaction mixture. The formation of 4-fluorobenzoic acid was measured by HPLC.
The assay mixture for catechol 1,2-dioxygenase activity contained, in a total volume of 1 ml, 100 mM potassium phosphate buffer (pH 7.0), 0.1 mM 4-fluorocatechol (4-FC), and a suitable amount of cell extract of Ralstonia sp. H1.
Protein concentrations were determined using Coomassie brilliant blue with bovine serum albumin as the standard.
Nucleotide sequence accession numbers.
The 16S rRNA sequences of strains G1 and H1 were deposited at GenBank (accession numbers EU342346 and EU342347, respectively).
RESULTS
Bacterial strains growing on 4-FCA and 4-FBA.
The bacterial strains G1 and H1, which can grow on 4-FCA and 4-FBA, respectively, were isolated by selective enrichment. Strain G1 was identified as an Arthrobacter sp. and strain H1 as a Ralstonia sp. The 16S rRNA gene sequence of strain G1 shows 99% identity to the rRNA gene sequence of Arthrobacter nitroguajacolicus (GU385852.1) and Arthrobacter boritolerans strain BTM-3c (AB288060.1). Strain H1 16S rRNA had 99% sequence identity to those of Ralstonia campinensis strain WS2 (AF312020.1) and strain LMG 20576 (AY040355.1). Ralstonia metallidurans and Ralstonia basilensis are environmental organisms known for their metal resistance and ability to degrade a wide range of recalcitrant xenobiotics (16, 46).
Strain G1 could grow on catechol, phenol, 4-hydroxycinnamic acid, 4-hydroxy-3-methoxycinnamic acid, 4-fluorocinnamic acid, and 4-chlorocinnamic acid. With the last two compounds, there was no release of halide. Strain H1 could grow on 2 mM 4-fluorobenzoic acid or 4-fluorobenzaldehyde (4-FBD) with >90% release of fluoride. Strain H1 could also grow with catechol (C), 4-chlorocatechol (4-CC), and 4-fluorocatechol. No growth of strains G1 and H1 was observed with 4-fluorophenyl acetic acid, 4-fluorophenoxy acetic acid, 4-fluorophenol, 4-fluoroacetophenone, α-fluorocinnamic acid, 2-fluorophenyl acetic acid, 3-fluorophenyl acetic acid, α-fluorophenyl acetic acid, fluorobenzene, 4-chlorophenol, 4-bromophenol, 4-iodophenol, 4-nitrocatechol, and 4-fluorophenyl acetic acid.
Degradation of 4-FCA by strain G1 in shake flasks.
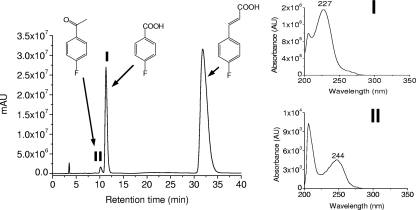
During growth of strain G1 with 10 mM 4-FCA as the sole source of carbon and energy, the formation of two stable products was observed, and they were identified as 4-fluorobenzoic acid and 4-fluoroacetophenone by comparing their HPLC retention times with those of authentic standards, as well as from the absorbance maxima (227 nm for 4-FBA and 244 nm for 4-FAP) (Fig. 1) and mass spectra ([M − H]− at m/z 137 of 4-FAP and [M − H]− at m/z 139 of 4-FBA).
FIG. 1.
HPLC elution profile of the aqueous supernatant after 75 h of incubation of Arthrobacter sp. strain G1 with 10 mM 4-FCA. The UV-visible spectra of identified metabolites (I and II) are displayed as insets.
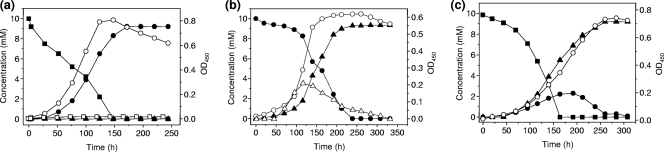
Strain G1 degraded 10 mM 4-FCA in 150 h, during which the optical density at 450 nm increased to 0.8 (Fig. 2 a). As soon as the concentration of 4-FCA started to decrease, 4-FBA was formed and accumulated in the medium. 4-FAP was formed as a side product and accumulated in the medium up to a concentration of 61 μM. Between 52 and 124 h, growth was exponential, and a maximal growth rate (μmax) of 0.025 h−1 was calculated. Release of fluoride was not observed during the growth of strain G1 on 4-FCA.
FIG. 2.
(a) Growth of strain G1 in a shake flask in MMY medium supplemented with 10 mM 4-FCA. (b) Growth of strain H1 in MMY medium in a shake flask supplemented with 10 mM 4-FBA. (c) Growth of a bacterial consortium of strains G1 and H1 in MMY in a shake flask supplemented with 10 mM 4-FCA. ▪, 4-FCA; •, 4-FBA; ○, optical density at 450 nm; ▴, fluoride; □, 4-FAP; ▵, 4-fluorocatechol.
Degradation of 4-FBA by strain H1 in shake flasks.
The growth of strain H1 with 10 mM 4-FBA was apparent from an increase in optical density and a decrease in the 4-FBA concentration. Up to 94% of the added fluorine was released as fluoride, and the optical density increased to 0.57 (Fig. 2b). From 70 to 160 h, growth was exponential and a μmax of 0.022 h−1 was calculated. During degradation of 4-FBA, the color of the medium changed from light pink to dark brown. This suggested that 4-fluorocatechol was produced. The formation of three different metabolites was observed with HPLC, and one of them was indeed identified as 4-FC by comparison with an authentic standard.
Complete mineralization of 4-FCA by a mixed culture.
Strain G1 could degrade 4-FCA only by cleavage of the side chain, releasing 4-FBA as a major product. Degradation of 4-FBA was observed only when strain H1 was present in the culture, together with strain G1. The degradation of 4-FBA seemed to be a slow process, and degradation of 10 mM 4-FCA required 160 h. Release of fluoride occurred as soon as conversion of 4-FBA by strain H1 started and was complete in 255 h, although the rate of fluoride release did not increase over time (Fig. 2c). From the exponential phase (70 to 230 h), an apparent μmax of 0.013 h−1 was calculated. The optical density of the cocultures remained lower than expected, possibly due to inhibitory effects of intermediates or because strain G1 was already in the decay phase before H1 consumed all 4-FBA that was formed.
Initial steps in the degradation of 4-FCA.
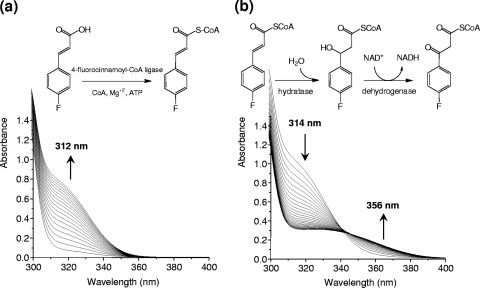
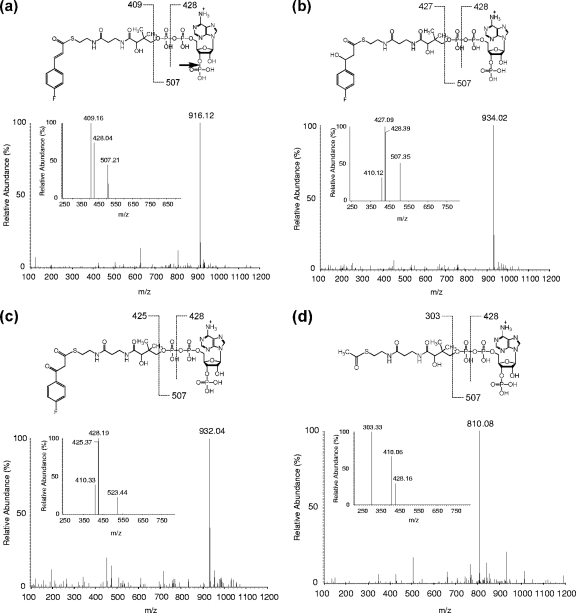
The conversion of 4-FCA by cell extracts prepared from cultures of strain G1 grown on 4-FCA was determined. In the presence of coenzyme A, ATP, and MgSO4, 4-FCA was converted to FC-CoA, as indicated by the increase in absorbance at 312 nm (Fig. 3 a). The rate of CoA adduct formation corresponds to a specific activity of 0.28 U·mg of protein−1. The cell extract also converted cinnamic acid, ferulic acid, and coumaric acid into the corresponding CoA adducts. No activity was detected without the addition of CoA or ATP to the reaction mixture. The formation of FC-CoA was confirmed by LC-MS. Upon ESI-MS, the ion that had the highest intensity in positive ionization mode MS spectra was identified as FC-CoA ([M + H]+ at m/z 916.12) (Fig. 4 a). An ESI-MS/MS analysis of this most abundant peak yielded as the most predominant peak a fragment corresponding to 4-fluorocinnamoylpantetheine (m/z 409) (inset in Fig. 4a) (6, 35). In-source fragmentation sometimes gave another ion at m/z 507 (insets in Fig. 4a and b), corresponding to phosphoadenosine diphosphate (27). Further fragmentation of the m/z 507 fragment gave a peak of m/z 428 (35), representing loss of HPO3 from the phosphoadenosine moiety of the CoA ester. The peak at m/z 428 represents either a product formed by cleavage within the phosphate backbone of phosphoadenosine diphosphate (6) or loss of the phosphate that is attached to the ribose (indicated with an arrow in the inset of Fig. 4a).
FIG. 3.
(a) Spectral changes observed upon transformation of 4-FCA to 4-fluorocinnamoyl-CoA by 4-fluorocinnamoyl-CoA ligase and subsequently to 4-fluorophenyl-β-hydroxy propionyl-CoA and 4-fluorophenyl-β-keto propionyl-CoA by hydratase and dehydrogenase activities of Arthrobacter sp. strain G1. The reaction mixture (1-ml volume) contained 0.2 mM 4-FCA, 0.5 mM ATP, 0.2 mM CoA, and 0.5 mM MgCl2. (b) Scans recorded after addition of NAD. The arrows indicate the direction of scans, which were recorded at 3-min intervals at 25°C after the addition of 50 to 100 μl of fractions collected from Q Sepharose chromatography.
FIG. 4.
ESI-MS and ESI-MS/MS spectra of 4-fluorocinnamoyl-CoA and acetyl-CoA. (a) Spectrum of 4-fluorocinnamoyl-CoA singly charged quasi-molecular ion [M + H]+ at m/z 916.15. Fragmentation of 4-fluorocinnamoyl-CoA is shown as an inset. (b) Spectrum of 4-fluorophenyl-β-hydroxy propionyl-CoA singly charged quasi-molecular ion [M + H]+ at m/z 934.02. Fragmentation of 4-fluorophenyl-β-hydroxy propionyl-CoA is shown in the inset. (c) Spectrum of 4-fluorophenyl-β-keto propionyl-CoA with singly charged quasi-molecular ion [M + H]+ at m/z 932.02. Fragmentation of protonated 4-fluorophenyl-β-keto propionyl-CoA is shown in the inset. (d) Detection of acetyl-CoA in positive-mode ESI-MS with singly charged quasi-molecular ion [M + H]+ at m/z 810.08. Fragmentation of protonated acetyl-CoA is shown in the inset.
Addition of NAD to a reaction mixture of cell extract preincubated with CoA, ATP, and MgCl2 resulted in a rapid decrease in absorbance (Fig. 3b). This indicates transformation of the formed FC-CoA, which is strongly absorbing due to the double bond, into a product with a single bond between the α- and β-carbons, in agreement with a β-substituted intermediate. In view of the β-oxidation pathway, this is expected to be a hydratase reaction.
To determine whether cells of strain G1 grown on 4-FCA indeed contain an NAD-stimulated FC-CoA hydratase, an extract of such cells was separated into fractions by Q Sepharose column chromatography (see Fig. S1 in the supplemental material). First, the fractions that contained the highest 4-FCA-CoA ligase activities were used to transform 4-FCA into FC-CoA. Other fractions were then tested for hydratase and dehydrogenase activities by measuring the decrease in absorbance when samples and NAD were added to the reaction mixture. Various column fractions containing hydratase and dehydrogenase activities were found. When these fractions and NAD were added to the reaction mixture containing FC-CoA, two products were observed in electrospray ionization LC-MS (total ion current [TIC]) at 10.8-min and 11.5-min retention times (chromatogram not shown). The ESI-TIC peak at 10.8 min was analyzed further, revealing that the ion with the highest intensity in positive-ionization mode MS spectra was 4-fluorophenyl-β-hydroxy propionyl-CoA ([M + H]+ at m/z 934.02) (Fig. 4b). The fragmentation of this peak by MS/MS resulted in the formation of 4-fluorophenyl-β-hydroxy pantetheine (m/z 427) (inset in Fig. 4b). The product corresponding to the ESI-TIC peak at 11.5 min was identified as 4-fluorophenyl-β-keto propionyl-CoA by LC-MS (positive ionization, [M + H]+ at m/z 932.04) (Fig. 4c). An ESI-MS/MS analysis of the most abundant peak (m/z 932.04) resulted in the detection of a 4-fluorophenyl-β-keto pantetheine fragment (m/z 425) (inset in Fig. 4c). These results indeed indicate the presence of a hydratase and a dehydrogenase.
4-Fluorobenzoic acid and acetyl-CoA were released from 4-fluorophenyl-β-keto propionyl-CoA in these reaction mixtures when additional CoA fractions that eluted from the Q-Sepharose column were added to the reaction mixture. 4-FBA was detected with HPLC, and acetyl-CoA was detected by LC-MS at m/z 810.08 (positive-mode ionization) (Fig. 4d). The ESI-MS/MS fragmentation of this highest peak gave a peak of m/z 303, characteristic of a pantetheine (inset in Fig. 4d). This suggests a hydrolase.
Degradation pathway of 4-FBA.
In order to study the pathway of 4-FBA by Ralstonia sp. strain H1, cells pregrown in 5-FBA were collected, washed, and resuspended in MMY medium to an OD450 of 1.5. Transformation products that were formed after addition of 5 mM 4-FBA and 5 mM glucose were analyzed by HPLC, LC-MS, and nuclear magnetic resonance (NMR). The formation of two products was observed, and they were identified by APCI-MS in negative mode. The main transformation product was 4-fluorocatechol, which eluted from the HPLC at 3.8 min ([M + H]− at m/z 127.31), and its identity was confirmed by 1H NMR (see Fig. S2 in the supplemental material). This indicates that 4-FBA degradation starts with dioxygenation and not with defluorination.
The dioxygenase-mediated conversion of catechols was tested spectrophotometrically with cell extract prepared from cells grown on 4-FBA. Transformation of C, 4-FC, 4-CC, 3-fluorocatechol (3-FC), and 4-nitrocatechol (4-NC) was observed with rates decreasing in that order (data not shown). Spectral changes during 4-FC transformation were consistent with ring cleavage yielding 3-fluoromuconate, followed by cycloisomerization to form muconolactone (see Fig. S3 in the supplemental material), suggesting that the pathway of 4-FBA degradation in strain H1 is similar to that in other bacteria that metabolize this compound via 4-fluorocatechol (43, 44, 45).
DISCUSSION
In this paper, we describe two bacterial strains that as a consortium can utilize 4-FCA for growth, and we propose a degradation pathway that is similar to that of ferulic acid and some other cinnamic acid derivatives for the initial steps (22). Previously, 4-FCA conversion was proposed to proceed via 4-FAP to 4-FBA (5, 8, 32), but our experiments indicate that this is not the major route in Arthrobacter sp. strain G1. Formation of 4-FPA is more likely due to a side reaction, i.e., spontaneous decarboxylation of intermediates that occur in the functional route of the side chain cleavage pathway. Similarly, accumulation of acetophenone as a side product may occur during the degradation of cinnamic acid by a Pseudomonas sp. (20).
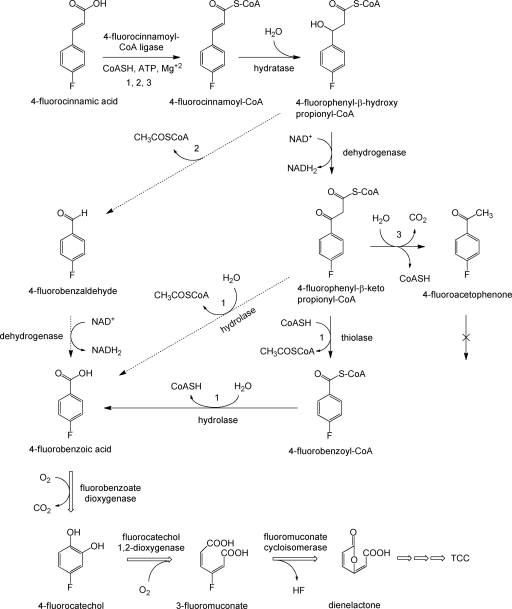
Degradation of 4-FCA in strain G1 starts with formation of a CoA adduct, catalyzed by a 4-fluorocinnamoyl-CoA ligase activity (Fig. 5, route 1). The activation of 4-FCA is similar to the initial degradation step in several organisms that use substituted cinnamic acids (1, 13, 29, 31, 34, 37) but different from the pathway in microorganisms that are capable of direct CoA-independent deacetylation with formation of a benzaldehyde derivative (15, 30, 36, 48). In general, bacterial strains that metabolize cinnamic acid, ferulic acid, or coumaric acid with initial formation of a CoA adduct proceed with direct hydration, followed either by deacetylation in a lyase (aldolase) type of reaction (1, 29, 34, 37) or by oxidation to the β-keto derivative followed by conversion to the carboxylic acid by a thioesterase (12, 13, 22, 49). Unlike what was proposed for the transformation of cinnamic acid, coumaric acid, and ferulic acid (1, 29, 36, 37), we found no evidence for the conversion of the β-hydroxy propionyl-CoA derivative to an aldehyde. Instead, we detected formation of 4-fluorophenyl-β-hydroxy propionyl-CoA and the corresponding β-keto CoA adduct when NAD was added to the ATP- and CoA-containing cinnamoyl-CoA ligase reaction mixture. The route via 4-fluorobenzaldehyde (Fig. 5, route 2) is also unlikely because 4-FBA was not formed from 4-fluorobenzaldehyde in a separate reaction mixture that contained NAD and cell extract. Transformation of the CoA adduct of 4-FCA to 4-fluorophenyl-β-hydroxy propionyl-CoA required addition of NAD, even though it is not a redox reaction. This could be due to the role of a multifunctional β-oxidation enzyme complex that catalyzes both the second and third reactions of the β-oxidation cycle, i.e., 2-enoyl-CoA hydration and 3-hydroxyacyl-CoA dehydrogenation, as has been found in other microorganisms (21, 38, 42, 50).
FIG. 5.
Proposed pathway for the catabolism of 4-FCA (route 1). Pathway 1 is proposed for Arthrobacter sp. strain G1. The dotted arrows indicate routes 2 and 3, corresponding to lyase-mediated deacetylation and decarboxylation leading to benzaldehyde and acetophenone, respectively. The double-lined arrow indicates further degradation by Ralstonia sp. strain H1.
Mineralization of 4-FBA and fluoride release were possible only when strain H1 was present in coculture with strain G1, since Arthrobacter G1 is not able to use 4-FBA as a growth substrate. Microbial cleavage of the aromatic ring of 4-FBA can occur before or after defluorination (3, 10, 17, 33, 43, 45). In the first case, the initial step in 4-FBA degradation is the substitution of fluorine to yield 4-hydroxybenzoic acid, which occurs in Aureobacterium sp. strain RHO25 (33). In the 4-FBA-degrading strain H1, transient accumulation of 4-fluorocatechol in the culture medium indicated that fluoride release occurs only after ring cleavage and starts with formation of 4-fluorocatechol. This is followed by ortho ring cleavage with formation of dienelactone or 4-fluoromuconolactone and then spontaneous elimination of a proton and fluoride (10, 18, 43, 44, 45) (Fig. 5).
Supplementary Material
Acknowledgments
We thank Pieter Wietzes for technical support and Theodora Tiemersma for help in LC-MS/MS analysis and gratefully acknowledge the Higher Education Commission, Government of Pakistan, for financial support.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achterholt, S., H. Priefert, and A. Steinbüchel. 2000. Identification of Amycolatopsis sp. strain HR167 genes involved in the bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 54:799-807. [DOI] [PubMed] [Google Scholar]
- 2.Blakley, E. R., and F. J. Simpson. 1964. Microbial metabolism of cinnamic acid. Can. J. Microbiol. 10:175-185. [DOI] [PubMed] [Google Scholar]
- 3.Boersma, F. G. H., W. C. McRoberts, S. L. Cobb, and C. D. Murphy. 2004. A 19F NMR study of fluorobenzoate biodegradation by Sphingomonas sp. HB-1. FEMS Microbiol. Lett. 237:355-361. [DOI] [PubMed] [Google Scholar]
- 4.Civolani, C., P. Barghini, A. R. Roncetti, M. Ruzzi, and A. Schiesser. 2000. Bioconversion of ferulic acid into vanillic acid by means of a vanillate negative mutant of Pseudomonas fluorescens strain BF13. Appl. Environ. Microbiol. 66:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creaser, C., L. F. dos Santos, D. G. Lamarca, A. New, and J. C. Wolff. 2002. Biodegradation studies of 4-fluorobenzoic acid and 4-fluorocinnamic acid: an evaluation of membrane inlet mass spectrometry as an alternative to high performance liquid chromatography and ion chromatography. Anal. Chim. Act 454:137-145. [Google Scholar]
- 6.Dalluge, J. J., et al. 2002. Separation and identification of organic acid-coenzyme A thioesters using liquid chromatography/electrospray ionization-mass spectrometry. Anal. Bioanal. Chem. 374:835-840. [DOI] [PubMed] [Google Scholar]
- 7.Defnoun, S., M. Labat, M. Ambrosio, J. L. Garcia, and B. K. C. Patel. 2000. Papillibacter cinnamivorans gen. nov., sp nov., a cinnamate-transforming bacterium from a shea cake digester. Int. J. Syst. Evol. Microbiol. 50:1221-1228. [DOI] [PubMed] [Google Scholar]
- 8.dos Santos, L. M. F., et al. 2001. Aerobic biotransformation of 4-fluorocinnamic acid to 4-fluorobenzoic acid. Biodegradation 12:23-29. [DOI] [PubMed] [Google Scholar]
- 9.Dutta, T. K., and S. Harayama. 2001. Biodegradation of n-alkylcycloalkanes and n-alkylbenzenes via new pathways in Alcanivorax sp. strain MBIC 4326. Appl. Environ. Microbiol. 67:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engesser, K. H., and P. Schulte. 1989. Degradation of 2-bromo-, 2-chloro- and 2-fluorobenzoate by Pseudomonas putida CLB 250. FEMS Microbiol. Lett. 51:143-147. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira, M. I., et al. 2009. Analysis of two gene clusters involved in the degradation of 4-fluorophenol by Arthrobacter sp. strain IF1. Appl. Environ. Microbiol. 75:7767-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, W., and P. Oriel. 1999. Degradation of 3-phenylpropionic acid by Haloferax. sp. D1227. Extremophiles 3:45-53. [DOI] [PubMed] [Google Scholar]
- 13.Gasson, M. J., et al. 1998. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J. Biol. Chem. 273:4163-4170. [DOI] [PubMed] [Google Scholar]
- 14.Gerus, I., A. Glushchenko, Y. Kurioz, Y. Reznikov, and O. Tereshchenko. 2004. Sticking of liquid crystal on photosensitive polymer layers. Opto Electron. Rev. 12:281-284. [Google Scholar]
- 15.Ghosh, S., A. Sachan, and A. Mitra. 2005. Degradation of ferulic acid by a white rot fungus Schizophyllum commune. World J. Microbiol. Biotechnol. 21:385-388. [Google Scholar]
- 16.Goris, J., et al. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle. et al. emend. Int. J. Syst. Evol. Microbiol. 51:1773-1782. [DOI] [PubMed] [Google Scholar]
- 17.Harper, D. B., and E. R. Blakley. 1971. The metabolism of p-fluorobenzoic acid by a Pseudomonas sp. Can. J. Microbiol. 17:1015-1023. [DOI] [PubMed] [Google Scholar]
- 18.Harper, D. B., and E. R. Blakley. 1971. The metabolism of p-fluorophenylacetic acid by a Pseudomonas sp. II. Isolation and identification of intermediates. Can. J. Microbiol. 17:645-650. [DOI] [PubMed] [Google Scholar]
- 19.Harwood, C. S., and J. Gibson. 1988. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 54:712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilton, M. D., and W. J. Cain. 1990. Bioconversion of cinnamic acid to acetophenone by a Pseudomonad—microbial production of a natural flavor compound. Appl. Environ. Microbiol. 56:623-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiltunen, J. K., et al. 1992. Peroxisomal multifunctional β-oxidation protein of Saccharomyces cerevisiae—molecular analysis of the FOX2 gene and gene-product. J. Biol. Chem. 267:6646-6653. [PubMed] [Google Scholar]
- 22.Huang, Z., L. Dostal, and J. P. N. Rosazza. 1993. Mechanisms of ferulic acid conversions to vanillic acid and guaiacol by Rhodotorula rubra. J. Biol. Chem. 268:23954-23958. [PubMed] [Google Scholar]
- 23.Hutchinson, J., and G. Sandford. 1997. Elemental fluorine in organic chemistry. Organofluorine Chem. 193:1-43. [Google Scholar]
- 24.Key, B. D., R. D. Howell, and C. S. Criddle. 1997. Fluorinated organics in the biosphere. Environ. Sci. Technol. 31:2445-2454. [Google Scholar]
- 25.Kim, E. J., J. R. Jeon, Y. M. Kim, K. Murugesan, and Y. S. Chang. 2010. Mineralization and transformation of monofluorophenols by Pseudonocardia benzenivorans. Appl. Microbiol. Biotechnol. 87:1569-1577. [DOI] [PubMed] [Google Scholar]
- 26.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 27.Magnes, C., F. M. Sinner, W. Regittnig, and T. R. Pieber. 2005. LC/MS/MS method for quantitative determination of long-chain fatty acyl-CoAs. Anal. Chem. 77:2889-2894. [DOI] [PubMed] [Google Scholar]
- 28.Mann, J. 1987. Modern methods for the introduction of fluorine into organic molecules—an approach to compounds with altered chemical and biological activities. Chem. Soc. Rev. 16:381-436. [Google Scholar]
- 29.Mitra, A., et al. 1999. 4-Hydroxycinnamoyl-CoA hydratase/lyase (HCHL)—an enzyme of phenylpropanoid chain cleavage from Pseudomonas. Arch. Biochem. Biophys. 365:10-16. [DOI] [PubMed] [Google Scholar]
- 30.Muheim, A., and K. Lerch. 1999. Towards a high-yield bioconversion of ferulic acid to vanillin. Appl. Environ. Microbiol. 51:456-461. [Google Scholar]
- 31.Narbad, A., and M. J. Gasson. 1998. Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology 144:1397-1405. [DOI] [PubMed] [Google Scholar]
- 32.New, A. P., L. M. F. dos Santos, G. Lo Biundo, and A. Spicq. 2000. Analytical techniques used for monitoring the biodegradation of fluorinated compounds in waste streams from pharmaceutical production. J. Chromatogr. A 889:177-184. [DOI] [PubMed] [Google Scholar]
- 33.Oltmanns, R. H., R. Müller, M. K. Otto, and F. Lingens. 1989. Evidence for a new pathway in the bacterial degradation of 4-fluorobenzoate. Appl. Environ. Microbiol. 55:2499-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overhage, J., H. Priefert, and A. Steinbüchel. 1999. Biochemical and genetic analysis of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, J. W., W. S. Jung, S. R. Park, B. C. Park, and Y. J. Yoon. 2007. Analysis of intracellular short organic acid-coenzyme A esters from actinomycetes using liquid chromatography-electrospray ionization-mass spectrometry. J. Mass Spectrom. 42:1136-1147. [DOI] [PubMed] [Google Scholar]
- 36.Peng, X., N. Misawa, and S. Harayama. 2003. Isolation and characterization of thermophilic bacilli degrading cinnamic, 4-coumaric, and ferulic acids. Appl. Environ. Microbiol. 69:1417-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plaggenborg, R., A. Steinbüchel, and H. Priefert. 2001. The coenzyme A-dependent, non-β-oxidation pathway and not direct deacetylation is the major route for ferulic acid degradation in Delftia acidovorans. FEMS Microbiol. Lett. 205:9-16. [DOI] [PubMed] [Google Scholar]
- 38.Pramanik, A., S. Pawar, E. Antonian, and H. Schulz. 1979. Five different enzymatic activities are associated with the multienzyme complex of fatty acid oxidation from Escherichia coli. J. Bacteriol. 137:469-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahouti, M., F. Seiglemurandi, R. Steiman, and K. E. Eriksson. 1989. Metabolism of ferulic acid by Paecilomyces variotii and Pestalotia palmarum. Appl. Environ. Microbiol. 55:2391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosazza, J. P. N., Z. Huang, L. Dostal, T. Volm, and B. Rousseau. 1995. Biocatalytic transformations of ferulic acid: an abundant aromatic natural product. J. Ind. Microbiol. 15:457-471. [DOI] [PubMed] [Google Scholar]
- 41.Sandford, G. 2000. Organofluorine chemistry. Philos. Trans. R. Soc. Lond. A Math. Phys. Eng. Sci. 358:455-471. [Google Scholar]
- 42.Sato, S., M. Hayashi, S. Imamura, Y. Ozeki, and A. Kawaguchi. 1992. Primary structure of the genes, faoA and faoB, from Pseudomonas fragi B-0771 which encode the two subunits of the HDT multienzyme complex involved in fatty acid β-oxidation. J. Biochem. 111:8-15. [DOI] [PubMed] [Google Scholar]
- 43.Schlömann, M., E. Schmidt, and H.-J. Knackmuss. 1990. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 172:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlömann, M., P. Fischer, E. Schmidt, and H.-J. Knackmuss. 1990. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J. Bacteriol. 172:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber, A., M. Hellwig, E. Dorn, W. Reineke, and H.-J. Knackmuss. 1980. Critical reactions in fluorobenzoic acid degradation by Pseudomonas sp. B13. Appl. Environ. Microbiol. 39:58-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinle, P., G. Stucki, R. Stettler, and K. W. Hanselmann. 1998. Aerobic mineralization of 2,6-dichlorophenol by Ralstonia sp. strain RK1. Appl. Environ. Microbiol. 64:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutherland, J. B., D. L. Crawford, and A. L. Pometto. 1983. Metabolism of cinnamic, p-coumaric, and ferulic acids by Streptomyces setonii. Can. J. Microbiol. 29:1253-1257. [DOI] [PubMed] [Google Scholar]
- 48.Toms, A., and J. M. Wood. 1970. Degradation of trans-ferulic acid by Pseudomonas acidovorans. Biochemistry 9:337-343. [DOI] [PubMed] [Google Scholar]
- 49.Xiang, L., and B. S. Moore. 2003. Characterization of benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium “Streptomyces maritimus”. J. Bacteriol. 185:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, S.-Y., and H. Schulz. 1983. The large subunit of the fatty acid oxidation complex from Escherichia coli is a multifunctional polypeptide. J. Biol. Chem. 258:9780-9785. [PubMed] [Google Scholar]
- 51.Zhang, C., Q. Zhou, L. Chen, Z. Wu, and B. Xu. 2007. Biodegradation of meta-fluorophenol by an acclimated activated sludge. J. Hazard. Materials 141:295-300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.