Abstract
Apicomplexan parasites such as Toxoplasma gondii contain a primitive plastid, the apicoplast, whose genome consists of a 35-kb circular DNA related to the plastid DNA of plants. Plants synthesize fatty acids in their plastids. The first committed step in fatty acid synthesis is catalyzed by acetyl-CoA carboxylase (ACC). This enzyme is encoded in the nucleus, synthesized in the cytosol, and transported into the plastid. In the present work, two genes encoding ACC from T. gondii were cloned and the gene structure was determined. Both ORFs encode multidomain proteins, each with an N-terminal extension, compared with the cytosolic ACCs from plants. The N-terminal extension of one isozyme, ACC1, was shown to target green fluorescent protein to the apicoplast of T. gondii. In addition, the apicoplast contains a biotinylated protein, consistent with the assertion that ACC1 is localized there. The second ACC in T. gondii appears to be cytosolic. T. gondii mitochondria also contain a biotinylated protein, probably pyruvate carboxylase. These results confirm the essential nature of the apicoplast and explain the inhibition of parasite growth in cultured cells by herbicides targeting ACC.
Keywords: fatty acid biosynthesis, apicoplast, plastid targeting
The presence of a plastid-like, nonphotosynthetic organelle, the apicoplast, surrounded by four membranes and containing its own genome, is a characteristic cellular feature of apicomplexan parasites (1, 2). The apicoplast performs metabolic functions that are essential for parasite survival (3). Understanding these processes has great potential to identify specific targets for new therapies. Molecular characteristics of these processes are likely to be distinct from those found in mammals. Many powerful tools to study gene expression and protein import into the Toxoplasma gondii and Plasmodium falciparum apicoplast have been developed (1, 4–6). These studies are most advanced for T. gondii and P. falciparum, but they could also contribute significantly to the development of novel therapeutic approaches for diseases caused by Cryptosporidium, Eimeria, Babesia, Theileria, Neospora, and Sarcocystis.
The 35-kb apicoplast genome has a limited coding capacity (1, 2). Most proteins needed for apicoplast function therefore must be imported. A distinct bipartite signal, identified at the N terminus of all apicoplast-targeted proteins to date, is sufficient to transport reporter proteins to the apicoplast (1, 5, 6). This transport is similar to the plastid-targeting mechanism found for nuclear-encoded proteins imported into three- or four-membrane plastids in other species (7–9). In plants, protein import into plastids is mediated by a 25- to 125-aa N-terminal transit peptide with no clear sequence consensus among the many proteins imported. The mechanism of protein import to plant chloroplasts has been reviewed recently (10). A transit peptide is required for both selective targeting of proteins to chloroplasts and for translocation across its double-membrane envelope. The transit peptide is removed by a specific protease during transport across these two membranes.
Some key enzymes of the fatty acid biosynthetic pathway were shown to be imported to the apicoplast (5, 6), providing a strong argument for the organelle as a site of de novo fatty acid biosynthesis. We have shown previously that T. gondii encodes two isozymes of acetyl-CoA carboxylase (ACC), a biotin-dependent carboxylase catalyzing the first step of fatty acid biosynthesis (11). Both isozymes are of the multidomain type found in the cytosol of all eukaryotes as well as in plastids of grasses and some other plants. We also have shown that one class of grass-specific inhibitors, which do not affect mammals, aryloxyphenoxypropionate herbicides, inhibits T. gondii ACC activity. This inhibition measured in vitro correlates well with inhibition of the parasite growth in human fibroblasts (11).
Here, we show that of the two nuclear-encoded ACCs of T. gondii, one is imported into the apicoplast whereas the other most likely stays in the cytosol. We propose that ACC1 is an essential enzyme participating in apicoplast fatty acid biosynthesis. Its inhibition by aryloxyphenoxypropionates is likely to be responsible for the parasite growth inhibition by these herbicides (11).
Materials and Methods
Isolation and Analysis of T. gondii ACC Genomic and cDNA Clones.
Several clones containing fragments of the ACC1 and ACC2 genes isolated from a T. gondii [strain RH(EP)] genomic library (11) were analyzed by restriction and by PCR, and their fragments were subcloned and sequenced. The same library was rescreened with additional probes derived from the 5′ and 3′ ends of some of the clones found in the first screen to obtain overlapping fragments of genomic DNA. Continuity of the cloned fragments of genomic DNA was verified by PCR by using T. gondii genomic DNA as a template. The sequence of a 19.6-kb fragment of the ACC1 gene (GenBank accession no. AF157612) as well as 13- and 4.8-kb fragments of the ACC2 gene (GenBank accession nos. AF157613 and AF330143, respectively) were assembled from these partial sequences. A T. gondii (strain RH) cDNA library obtained from the AIDS Reagents Program (www.aidsreagent.org) then was screened with ACC1 and ACC2 gene-specific probes. Thirteen and five positive clones were found for ACC1 and ACC2, respectively, among ≈3.5 × 105 plaques. Five and four clones obtained for each gene were purified and sequenced. The longest ACC1 and ACC2 cDNA fragments contained 4.6 kb (ACC1 Fragment V) and 3.3 kb (ACC2 Fragment IV), respectively (GenBank accession nos. AF330144 and AF330145).
Reverse Transcription–PCR (RT-PCR) and 5′ Rapid Amplification of cDNA Ends (RACE) Analysis of T. gondii ACC mRNA.
T. gondii RNA and single-strand cDNA prepared with random hexamers were kindly provided by R. McLeod (University of Chicago). A GIBCO/BRL 5′ RACE system was used according to the manufacturer's protocol. The High Fidelity PCR System (Roche) was used for DNA amplification. The first-strand cDNAs for some RT-PCR experiments were prepared by using gene-specific primers. Nested gene-specific primers were used for the subsequent DNA amplification (Table 1). The first-strand cDNA for 5′ RACE was prepared by using a gene-specific primer: GTTTCCCTGTGATCTATCGGC. Anchor primer (AAP; GIBCO/BRL) and a gene-specific primer, AAGCGGACAAAAACCCACTA, were used in the first round of PCR of dC-tailed cDNA. Universal primer (AUAP; GIBCO/BRL) and a nested gene-specific primer, CTGCCAGGGGAAGACTGTC, were used for reamplification. RT-PCR, and 5′ RACE products were cloned into the vector pGEM-T Easy (Promega) and sequenced.
Table 1.
ACC1 and ACC2 gene-specific primers used for RT-PCR
| cDNA | Primers for cDNA synthesis | Forward PCR primers | Reverse PCR primers |
|---|---|---|---|
| ACC1 | |||
| Fragment I | Random hexamers | GTGCGTAGTGGGTTTTTGTCC | GCTGCCATGCCGTTGTTGGCGATG |
| Fragment II | CAGGTGCATTTTGACTTCGATTT | TTCAGGCGTCAACCACGGAATCGA | AGAGCGAGAGTCCCCAACACATCG |
| Fragment III | CAGGTGCATTTTGACTTCGATTT | GGTAGAAGCGATGAAGATGATAATG | GAACGTGGACTGCGGCCAGGCGGA |
| Fragment IV | Random hexamers | ATCGATCTGCTCTCCCACTT | CAGGTGCATTTTGACTTCGATTT |
| ACC2 | |||
| Fragment I | AATGAGATGCAGATTGGCGTAGTT | TCCTGCGTGTGTGTTCGG | AGTCTGCTTCAGCAATAAATTCAG |
| and AGATTGGCGTAGTTGTTCGAGTTC | |||
| Fragment II | CCGATTCTTCCGCTTCAGAG | ATGCTTCCTATCTCGGAGGC | GTGGCTTCGCTGCAAAGAC |
| Fragment III | TGTTCCCATCTGAAAGGTGAC | TTTTCCCGGGTCCCTCACCCAATGCC | CGTCGTTGCCTATCAGAATGA |
| and CGTCGTTGCCTATCAGAATGA | |||
Maintenance and Transformation of T. gondii.
The RH strain of T. gondii was maintained by serial passage in primary human foreskin fibroblasts grown in 5% CO2 as described previously, and parasite transfection was performed by using a BTX 600 electroporation system (12).
Localization of Biotinylated Peptides in Fixed T. gondii.
Human foreskin fibroblasts cells were grown to confluence on coverslips and infected with 5 × 105 parasites. After 16–24 h of infection, the cells were fixed in 3% paraformaldehyde and permeabilized with 0.25% Triton X-100 in phosphate-buffer saline. Biotin within fixed parasites was detected by using FITC-streptavidin (Sigma) at a 1:500 dilution. Rabbit anti-acyl carrier protein (ACP) polyclonal antibodies used at a 1:500 dilution were detected by using rhodamine-labeled anti-rabbit Ig used at a 1:200 dilution (5). For mitochondrial colocalization, Mitotracker Red CMX Ros (Molecular Probes) was added to infected cell cultures before fixing.
Analysis of the Subcellular Targeting of ACC in T. gondii Using a Green Fluorescent Protein (GFP) Reporter.
Two cDNA cassettes with engineered BglII and AvrII sites containing the 5′ end fragments of the ACC1 and ACC2 ORFs were prepared by PCR using forward and reverse primers AAAAAGATCTAAAATGCCGACACGCACACAC and AAAACCTAGGGTTGTTGGCGATGAGGACG, and AAAAAGATCTAAAATGACACGACAGCAAGCAGTC and AAAACCTAGGGTTGTTGGCAATGAGGATGC, respectively. The RT-PCR-cloned fragments of cDNA described above were used as templates. The BglII-AvrII fragments were inserted between the BglII and AvrII sites of vector ptubACP-GFP/sag-CAT to replace the ACP cassette (5) to create artificial genes encoding putative ACC-targeting signal/GFP fusions. The structure of the final construct was verified by sequencing. GFP localization was assessed by direct fluorescence microscopy of live parasites 24 h after transient transfection with the construct described above. Mouse monoclonal anti-GFP antibodies (CLONTECH) were used at a 1:1,000 dilution. Rabbit anti-ACP polyclonal antibodies were used as described above. Secondary FITC-conjugated anti-mouse antibodies and rhodamine-conjugated anti-rabbit antibodies were used at a dilution of 1:64 and 1:100, respectively.
Fluorescent Microscopy.
Indirect fluorescence microscopy was done as described previously (12). Fluorescence was visualized on a Zeis IM-35 Axiovert illuminated with a mercury lamp (4, 13, 14), using FITC (450–480 excitation/515–565 emission) or Texas red (530–585 excitation/615 long-pass emission) filter sets. Images were collected by using an interline chip-cooled charge-coupled device camera (Orca 9545; Hamamatsu, Middlesex, NJ). Contrasts and overlays were performed by using openlab software (Improvision, Lexington, MA) and photoshop (Adobe Systems, Mountain View, CA).
Results
T. gondii ACCs and Their Genes.
Fragments (430 bp) of cDNA encoding two different T. gondii ACC isozymes cloned by RT-PCR were used as probes to screen a T. gondii genomic library. Several overlapping lambda clones were isolated and sequenced for each gene. The sequence of the ACC1 gene and two large fragments of the ACC2 gene were assembled from these partial sequences. Overlapping cDNA fragments encoding both ACCs were cloned by RT-PCR and 5′ RACE and by cDNA library screening (ACC1 Fragments I–V and ACC2 Fragments I–IV, Table 1).
Comparison of the cDNA and genomic sequences revealed 14 introns in the ACC1 gene and at least 28 introns in the ACC2 gene (GenBank accession nos. AF157612, AF157613, and AF330143). Two products of alternative splicing were detected for the ACC2 gene transcript by sequencing different clones of the ACC2 Fragment II (Table 1): exons 16 and 17 are missing in one type of mature mRNA. Two transcription termination/polyadenylation sites were found for both genes by sequencing cDNA clones isolated from a T. gondii cDNA library (GenBank accession nos. AF330144 and AF330145.). Introns in the two wheat multidomain ACC genes are located at equivalent positions, except for two introns, which are absent from the cytosolic ACC genes (15, 16). None of the introns found in the ACC1 and ACC2 genes of T. gondii has the same location as the plant introns. The intron positions are different for ACC1 and ACC2 genes of T. gondii.
The approximate position of the transcription start of the ACC1 gene was determined in two steps. First, a series of RT-PCRs was performed by using a downstream primer targeted to the exon encoding the N terminus of the biotin carboxylase domain (identified by amino acid sequence similarity) and a series of gene-specific upstream primers located at different distances from the downstream primer. Second, the position of the transcription start site was deduced from the sequence of the longest specific cDNA fragment cloned by 5′ RACE. The ACC1 leader sequence is about 700 nt long and contains one ATG codon in a reading frame different from that of the ACC1 ORF. The translational start codon is preceded by a stop codon present in the same reading frame about 130 nt upstream. The sequence of the ACC2 leader, identified by RT-PCR, is about 500 nt long. The translation start codon is preceded by a stop codon in the same reading frame about 300 nt upstream. Two ATG codons are present in the ACC2 leader. The first intron is located at the beginning of the coding sequence of both genes.
The amino acid sequence of ACC1 is 30–40% identical to other eukaryotic multidomain ACCs. Parts of the sequence outside of the conserved functional domains (biotin carboxylase, biotin carrier, and carboxyltransferase) were difficult to align reliably. As noted before (11), the most closely related sequence is that of the diatom Cyclotella cryptica ACC, which is approximately 50% identical. A similar comparison with T. gondii ACC2 and P. falciparum ACC is not yet possible because full-length gene sequence and the complete intron–exon structure of these genes have not yet been determined. The sequence of the P. falciparum fragment of the ACC1 gene cloned before (11) was identical to the sequence of a gene found on chromosome 14 (TIGR contig 137, Malaria Genome Project). Sequence comparisons of the biotin carboxylase domain showed that the ACC of P. falciparum is more closely related to ACC1 of T. gondii and ACC of C. cryptica than to any other multidomain ACC, including ACC2 of T. gondii (11). The ACC gene of P. falciparum is most likely an orthologue of the T. gondii ACC1 gene. A homolog of the T. gondii ACC2 gene in P. falciparum has not been identified.
Subcellular Localization of T. gondii ACCs.
Two major biotinylated proteins were detected in T. gondii by Western blot analysis with labeled streptavidin: a 240-kDa subunit of ACC and a 130-kDa subunit of pyruvate carboxylase. Partially purified protein extracts prepared from tachyzoites showed a significant substrate (acetyl-CoA)-dependent ACC activity (11). To test the hypothesis that one ACC isozyme is localized in the apicoplast and that PC, a mitochondrial enzyme in other species (17), is localized in T. gondii mitochondria as well, we took advantage of the strong binding of streptavidin to biotin-containing proteins. FITC-conjugated streptavidin detected biotin-containing enzymes associated with the apicoplast as well as the mitochondrion (Fig. 1). However, a significantly lower level of streptavidin was associated with the mitochondrion.
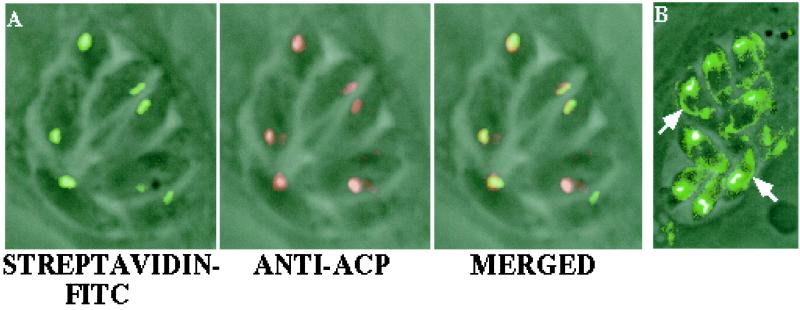
Figure 1.
Biotin-containing proteins are present in the T. gondii apicoplast and mitochondrion. Incubation of fixed parasites with FITC-conjugated streptavidin detects a biotin-dependent protein in the apicoplast (A), as indicated by colocalization with ACP. ACP was shown previously to be localized in the apicoplast (5). Overexposure of streptavidin-FITC-labeled parasites indicates the presence of another biotin-containing protein, presumably pyruvate carboxylase, in the mitochondria (arrows), as confirmed by colocalization with the mitochondrial-specific marker Mitotracker Red (not shown) (B).
The structure of the 5′ end of T. gondii ACC ORFs was of special interest because it was expected to yield a clue to protein subcellular localization and the structure of the targeting signal. The highly conserved biotin carboxylase domain is located near the N terminus of all eukaryotic-type multidomain ACCs (15, 18). In plant cytosolic ACCs, this domain is preceded by only 20 aa. Other cytosolic ACCs, fungal (e.g., yeast) and animal (e.g., rat), have longer extensions, approximately 50 and 100 aa, respectively, at their N termini (Fig. 2A). Plant plastid ACCs such as wheat have a 120-aa sequence at their N terminus, which contains the plastid transit peptide (15, 19). Finally, ACC from C. cryptica, which is proposed to be targeted across four membranes to the chloroplast (20), contains a putative signal peptide within its 80 N-terminal amino acid residues. However, the presence of such an amino acid extension at the N terminus by itself is not a sufficient indicator of protein subcellular localization.
Figure 2.
Apicoplast-targeting signal in T. gondii ACC. Amino acid sequence comparison of the N terminus of ACC1 and ACC2 of T. gondii with those of cytosolic and plastid multidomain ACCs from other species (A). The first methionine residues of each ORF are underlined. Asterisks indicate the first conserved amino acid residues of the BC domain. The putative transmembrane helix-like motif (amino acids 85–107) and a signal peptide cleavage site (between A106 and G107) found in T. gondii ACC1 are double-underlined. (B) Structure of constructs for the expression of the putative ACC signal/transit peptide–GFP fusions in T. gondii. ptub, promoter of the T. gondii TUB1 gene; sag-CAT, selectable marker (chloramphenicol resistance) for T. gondii transformation; M, first methionine of ACC ORF.
A distinct bipartite signal/transit peptide is present at the N terminus of T. gondii and P. falciparum proteins that are known to be targeted to the apicoplast (1, 5, 6). Analysis of the predicted coding region of T. gondii ACC1 and ACC2 genes revealed long N-terminal extensions in both proteins. The ORFs extend ≈300 and ≈200 aa upstream of the respective biotin carboxylase domains (Fig. 2A). There is no significant sequence similarity among signal/transit peptides (proven or putative) found in different apicoplast-targeted proteins. However, the extreme N terminus of unprocessed T. gondii ACC1 is predicted to contain a transmembrane helix-like motif (amino acids 85–107) and a putative signal peptide cleavage site (between amino acids 106 and 107), both suggesting that the N-terminal extension contains a signal peptide. tmhmm and signalp software, available at www.genome.cbs.dtu.dk, were used for this analysis. Such motifs are found in other nuclear-encoded apicoplast proteins such as ACP and ribosomal protein S9 (5), although closer to the N terminus (amino acid residues 11–33). The ACC2 ORF does not have any such motifs, suggesting a nonapicoplast localization of this isozyme.
To determine the subcellular localization of the T. gondii ACC isozymes, synthetic genes were constructed in which DNA fragments containing the 5′ ends of ACC1 and ACC2 ORFs were fused in-frame to a GFP ORF. This fusion was inserted into a T. gondii expression vector under control of the TUB1 promoter (Fig. 2B). Several amino acid residues of the conserved biotin carboxylase domain (Fig. 2A) were included to ensure that all residues present at the N terminus that could be essential for proper targeting were present in these fusions. The N-terminal extension of ACC1 was found to direct GFP to the apicoplast in transiently transfected parasites (Fig. 3A). The identity of this compartment was confirmed to be the apicoplast by immunofluorescence microscopy by using anti-GFP and anti-ACP antibodies in which ACP and GFP colocalized to the same compartment (Fig. 3B). ACP is known to be imported to the apicoplast (5). On the other hand, fluorescence of the GFP reporter fused to the ACC2 N-terminal extension was localized only in the cytosol (not shown); none was associated with the apicoplast. None of the N-terminal extensions tested directed GFP to the mitochondrion. These experiments indicate that ACC1 and ACC2 encode the plastid and cytosolic isozymes, respectively.
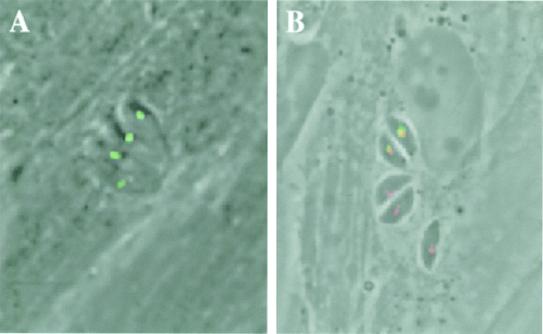
Figure 3.
T. gondii ACC1 is localized in the apicoplast. Fusing GFP to the putative N-terminal-targeting signal of ACC1 was sufficient to direct this reporter to the apicoplast in living parasites (A). Subcellular localization of GFP was assessed by direct fluorescence microscopy of live parasites 24 h after transient transfection. The apicoplast ACC1 localization was confirmed in fixed cells by localization of ACP with anti-ACP antibodies and GFP with anti-GFP antibodies, followed by detection with rhodamine- and FITC-conjugated secondary antibodies, respectively (B). Three parasitophorous vacuoles are visible in B: the upper vacuole (containing two parasites) was transiently transfected with ACC1-GFP; the yellow signal indicates precise colocalization with ACP.
Discussion
T. gondii Acetyl-CoA Carboxylases.
ACC catalyzes the first committed step of de novo fatty acid synthesis, the carboxylation of acetyl-CoA, to produce malonyl-CoA by using bicarbonate as a source of the carboxyl group, biotin as a cofactor, and ATP as a source of energy. It also provides malonyl-CoA for the synthesis of very long chain fatty acids and for secondary metabolism. In plants, the contribution of plastid ACC to the control of flux through the fatty acid pathway is significant, and this is reflected in the response of whole plants to ACC-targeting herbicides (21–23). The multidomain ACC localized in the plastids of grasses is strongly inhibited by aryloxyphenoxypropionate and cyclohexanedione herbicides (24). Sensitive plants are killed as a result of inhibition of their de novo fatty acid biosynthesis. We have shown recently that ACC activity detected in Toxoplasma extracts is also sensitive to aryloxyphenoxypropionates. Significantly, Clodinafop, an aryloxyphenoxypropionate tested in our study, inhibits intracellular Toxoplasma growth in human fibroblasts (11).
The molecular mechanism of ACC inhibition by aryloxyphenoxypropionates and cyclohexanediones is not yet known. We recently have localized the herbicide sensitivity determinant to a 400-aa fragment of the carboxyltransferase domain (24) and showed that the second ACC half-reaction is affected (unpublished results). In contrast, all multisubunit enzymes in bacteria and chloroplasts of dicot plants as well as multidomain cytosolic enzymes in plants, fungi, and animals are resistant to these herbicides, providing the basis for using these herbicides in agriculture and their potential future use in medicine.
ACC of P. falciparum (ref. 11; Malaria Genome Project), for which the existence of only one intron has been confirmed so far experimentally (unpublished results), lacks introns in those parts of the gene that could be analyzed by comparisons with coding sequences of T. gondii ACC1 and ACCs from other species. Additional introns are likely to be present in the P. falciparum gene, based on the analysis of the coding capacity of the gene. Differences in intron number and localization attest to the ancient origin of the ACC genes in Apicomplexa.
Bacterial-type multisubunit ACC genes, similar to those found in the plastid DNA of many dicot plants and algae, are not present in the apicoplast genome (GenBank accession no. U87145) (1, 2). The only ACCs detected in T. gondii (11) were of the multidomain type encoded in the nuclear genome of eukaryotes. The structure of the T. gondii ACCs (multidomain enzyme) and their genes (presence of multiple introns) as well as the phylogenetic analysis based on amino acid sequences all agree with their eukaryotic origin. The results of phylogenetic analysis based on the sequence of full-length ACC1 and larger fragments of ACC2 are consistent with the results based on the biotin carboxylase domain sequence reported previously (11). It is postulated that a plastid of an eukaryotic alga introduced to an ancestral eukaryote by secondary endosymbiosis was the ancestor of the apicoplast (1, 2). Thus, several possibilities that may explain the origin of the ACC genes in Apicomplexa exist, all of which require the acquisition of a signal/transit peptide by at least one ACC isozyme for targeting it to the modern apicoplast. First, the apicoplast ACC gene could have arisen by divergence of one copy of a duplicated cytosolic gene of the ancestral eukaryote. Second, it is possible that a nuclear-encoded multidomain ACC already functioned in the plastid of the ancestral eukaryotic alga and was transferred to the host nucleus after endosymbiosis but before the endosymbiont nucleus was eliminated. Third, it is also possible that the ACC function of the ancestral apicoplast was taken over by a new isozyme whose gene originated from the cytosolic algal gene transferred to the host nuclear genome after the secondary endosymbiosis. Such transfer of genes to the nuclear genome and replacement of plastid prokaryotic genes by duplicated cytosolic genes that acquired plastid-targeting signals are well documented in the plant kingdom (25). ACC gene duplication and acquisition of new subcellular specificity occurred independently in different plant lineages such as grasses, some Brassicaceae, and some Geraniaceae (19, 21). Currently, it is difficult to determine which of the above hypotheses is correct because of the lack of sufficient information for in-depth phylogenetic analysis. For example, it is impossible at the moment to determine which and whether apicomplexan ACC isozymes originated from the cytosolic ACC of the alga or the cytosolic ACC of the ancestral eukaryote that engulfed it.
Fatty Acid Biosynthesis and Source of Acetate/Acetyl-CoA in Apicomplexa.
Where are fatty acids made in Toxoplasma and other Apicomplexa? Apicoplast localization of ACC (this work) and fatty acid synthase (5, 6) supports the hypothesis that the organelle is a site of de novo fatty acid biosynthesis. Such compartmentalization raises the question of the source of acetyl-CoA used by the pathway. Acetyl-CoA cannot cross membranes and, therefore, it has to be made in each compartment or has to be transported as free acetate or another metabolite that could easily be converted into acetyl-CoA. Plants are a good reference point to analyze this problem because of their dual-compartment localization of fatty acid biosynthesis (26). There is probably no single source of acetyl-CoA used as a substrate for fatty acid biosynthesis in plants (26, 27). A variety of sources contribute to the pool, differing in each compartment, tissue, and developmental stage. On the other hand, significant differences between plant and parasite can be expected. For example, there is no intercellular transport of fatty acids in plants; each cell is autonomous in this respect. But apicomplexan parasites are quite likely to import some fatty acids from the host cell. Imported fatty acids might be used directly or degraded all the way to acetyl-CoA via mitochondrial β-oxidation. This acetyl-CoA, after transport to other compartments, could be used for de novo fatty acid biosynthesis.
Are fatty acids made in other compartments in Apicomplexa? In plants, very long chain fatty acids are synthesized in the cytosol (28) and lipoic acid is synthesized in mitochondria (29). In both cases, malonyl-CoA made in the cytosol by the cytosolic ACC isozyme is used as a substrate. The presence of cytosolic ACC in Toxoplasma suggests that this compartment also could participate in fatty acid biosynthesis in the parasite. Identification of cytosolic fatty acid synthase or fatty acid elongase would be helpful in validating this hypothesis.
Metabolic Functions of the Apicoplast.
What does the apicoplast do? The apicoplast genome itself encodes no proteins likely to play a direct role in parasite metabolic pathways. However, recent work has identified numerous nuclear-encoded genes that appear likely to be targeted to the apicoplast. These observations suggest that the apicoplast, like the plastid in plants, performs important functions but relies on nuclear genes to provide the necessary enzymes. A specialized transport system has evolved to deliver these proteins to the organelle across four membranes (1, 5, 6). At least some of its functions are essential for the parasite and are responsible for its conservation during evolution. Macrolide (and other) antibiotics targeting apicoplast ribosomes (30–32) and fluoroquinolone antibiotics eliminating the apicoplast genome (33) kill the parasite. Thiolactomycin, an inhibitor of fatty acid elongation specific for the multisubunit enzyme found in the apicoplast, has been shown to prevent the growth of P. falciparum (5). Inhibitors of the isoprenoid pathway are potential antimalarial drugs (34). Aryloxyphenoxypropionates inhibit intracellular Toxoplasma growth in human fibroblasts (11) by targeting the apicoplast ACC isozyme (unpublished results). These latter results strongly suggest that key enzymes of the fatty acid biosynthetic pathway are valid targets for inhibitors, leading to the development of new therapeutic treatment. These observations warrant a thorough analysis of all of the processes performed by the apicoplast as well as the organelle biogenesis itself as potential targets.
Genome and expressed sequence tag sequencing efforts undertaken for various apicomplexan parasites (Toxoplasma, Plasmodium, Cryptosporidium, Eimeria) will significantly accelerate research in this field (35–37). Genome-based reconstruction of metabolic pathways will aid identification and biochemical analysis of new potential targets. ACC is an extremely good target in grasses, and our recent results on the T. gondii enzyme (11) warrant its full exploration as a target of antiparasitic treatment. Other key enzymes that are known to be essential in other species should be investigated even if no good inhibitors have been found yet. From the biotin-dependent carboxylase family, pyruvate carboxylase is an essential mitochondrial enzyme with pleiotropic functions (17) and cytosolic ACC is essential in yeast because of its involvement in nuclear membrane biogenesis (38).
Acknowledgments
This work was supported by a gift from Monsanto.
Abbreviations
- ACC
acetyl-CoA carboxylase
- ACP
acyl carrier protein
- RT-PCR
reverse transcription–PCR
- GFP
green fluorescent protein
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Roos D S, Crawford M J, Donald R G K, Kissinger J C, Klimczak L J, Striepen B. Curr Opin Microbiol. 1999;2:426–432. doi: 10.1016/S1369-5274(99)80075-7. [DOI] [PubMed] [Google Scholar]
- 2.Wilson R J M, Williamson D H. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFadden G I, Roos D S. Trends Microbiol. 1999;7:328–333. doi: 10.1016/s0966-842x(99)01547-4. [DOI] [PubMed] [Google Scholar]
- 4.Hager K M, Stripen B, Tilney L G, Roos D S. J Cell Sci. 1999;112:2631–2638. doi: 10.1242/jcs.112.16.2631. [DOI] [PubMed] [Google Scholar]
- 5.Waller R F, Keeling P J, Donald R G K, Striepen B, Handman E, Lang-Unnasch N, Cowman A F, Besra G S, Roos D S, McFadden G I. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waller R F, Reed M B, Cowman A F, McFadden G I. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang M, Apt K E, Kroth P G. J Biol Chem. 1998;273:30973–30978. doi: 10.1074/jbc.273.47.30973. [DOI] [PubMed] [Google Scholar]
- 8.Schwartzbach S D, Osafune T, Löffelhardt W. Plant Mol Biol. 1998;38:247–263. [PubMed] [Google Scholar]
- 9.Sulli C, Fang Z-W, Muchhal U, Schwartzbach S D. J Biol Chem. 1999;274:457–463. doi: 10.1074/jbc.274.1.457. [DOI] [PubMed] [Google Scholar]
- 10.Keegstra K, Cline K. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuther E, Johnson J J, Haselkorn R, McLeod R, Gornicki P. Proc Natl Acad Sci USA. 1999;96:13387–13392. doi: 10.1073/pnas.96.23.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roos D S, Donald R G, Morrissette N S, Moulton A L. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 13.Striepen B, He C Y X, Matrajt M, Soldati D, Roos D S. Mol Biochem Parasitol. 1998;92:325–338. doi: 10.1016/s0166-6851(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 14.Striepen B, Crawford M J, Shaw M K, Tilney L G, Seeber F, Roos D S. J Cell Biol. 2000;151:1423–1433. doi: 10.1083/jcb.151.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. Proc Natl Acad Sci USA. 1997;94:14179–14185. doi: 10.1073/pnas.94.25.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podkowinski J, Sroga G E, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1996;93:1870–1874. doi: 10.1073/pnas.93.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jitrapakdee S, Wallace J C. Biochem J. 1999;340:1–16. doi: 10.1042/bj3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gornicki P, Podkowinski J, Scappino L A, DiMaio J, Ward E, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulte W, Topfer R, Stracke R, Schell J, Martini N. Proc Natl Acad Sci USA. 1997;94:3465–3470. doi: 10.1073/pnas.94.7.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roessler P G, Ohlrogge J B. J Biol Chem. 1993;268:19254–19259. [PubMed] [Google Scholar]
- 21.Christopher J T, Holtum J A M. Aust J Plant Phys. 2000;27:845–850. [Google Scholar]
- 22.Golz A, Focke M, Lichtenthaler H K. J Plant Physiol. 1994;143:426–433. [Google Scholar]
- 23.Konishi T, Sasaki Y. Proc Natl Acad Sci USA. 1994;91:3598–3601. doi: 10.1073/pnas.91.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolskaya T, Zagnitko O, Tevzadze G, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1999;96:14647–14651. doi: 10.1073/pnas.96.25.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegava M, Kowallik K V. Nature (London) 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 26.Ohlrogge J B, Jaworski J G, Post-Beittenmiller D. Lipid Metabolism in Plants. Boca Raton, FL: CRC; 1993. pp. 3–32. [Google Scholar]
- 27.Ke J, Behal R H, Back S L, Nikolau B J, Wurtele E S, Oliver D J. Plant Physiol. 2000;123:497–508. doi: 10.1104/pp.123.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Post-Beittenmiller D. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:405–430. doi: 10.1146/annurev.arplant.47.1.405. [DOI] [PubMed] [Google Scholar]
- 29.Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignon J. J Biol Chem. 2000;275:5016–5025. doi: 10.1074/jbc.275.7.5016. [DOI] [PubMed] [Google Scholar]
- 30.Beckers C J M, Roos D S, Donald R G K, Luft B J, Schwab J C, Cao Y, Joiner K A. J Clin Invest. 1995;95:367–376. doi: 10.1172/JCI117665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fichera M E, Bhopale M K, Roos D S. Antimicrob Agents Chemother. 1995;39:1530–1537. doi: 10.1128/aac.39.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfefferkorn E R, Nothnagel R F, Borotz S E. Antimicrob Agents Chemother. 1992;36:1091–1096. doi: 10.1128/aac.36.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fichera M E, Roos D S. Nature (London) 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 34.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Ebert M, Zeidler J, Lichtenthaler H K, et al. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 35.Ajioka J W, Boothroyd J C, Brunk B P, Hehl A, Hiller L, Manger I D, Marra M, Overton G C, Roos D S, Wan K-L, et al. Genome Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- 36.Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher C M, Craig A, Davies R M, Devlin K, Feltwell T, et al. Nature (London) 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- 37.Gardner M J, Tettelin H, Carucci D J, Cummings L M, Aravind L, Koonin E V, Shallom S, Mason T, Yu K, Fujii C, et al. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 38.Schneiter R, Hitomi M, Ivessa A S, Fasch E-V, Kohlwein S D, Tartakoff A M. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]