Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping
In the absence of the telomere capping protein Cdc13, budding yeast telomeres erode, resulting in checkpoint arrest. This study shows that the helicase Pif1, known as a telomerase inhibitor, also has a direct role in the resection of uncapped telomeres, acting in parallel to the nuclease Exo1.
Keywords: Cdc13, DNA damage response, Exo1, Pif1, uncapped telomeres
Abstract
Essential telomere ‘capping' proteins act as a safeguard against ageing and cancer by inhibiting the DNA damage response (DDR) and regulating telomerase recruitment, thus distinguishing telomeres from double-strand breaks (DSBs). Uncapped telomeres and unrepaired DSBs can both stimulate a potent DDR, leading to cell cycle arrest and cell death. Using the cdc13-1 mutation to conditionally ‘uncap' telomeres in budding yeast, we show that the telomere capping protein Cdc13 protects telomeres from the activity of the helicase Pif1 and the exonuclease Exo1. Our data support a two-stage model for the DDR at uncapped telomeres; Pif1 and Exo1 resect telomeric DNA <5 kb from the chromosome end, stimulating weak checkpoint activation; resection is extended >5 kb by Exo1 and full checkpoint activation occurs. Cdc13 is also crucial for telomerase recruitment. However, cells lacking Cdc13, Pif1 and Exo1, do not senesce and maintain their telomeres in a manner dependent upon telomerase, Ku and homologous recombination. Thus, attenuation of the DDR at uncapped telomeres can circumvent the need for otherwise-essential telomere capping proteins.
Introduction
Telomeres consist of double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA), bound by dsDNA- and ssDNA-binding proteins (Blackburn et al, 2006; Lydall, 2009). This nucleoprotein ‘cap' has at least two functions: to shield the telomeric DNA from stimulating the DNA damage response (DDR) and to regulate elongation of the telomere by telomerase. In human senescent cells, dysfunctional telomeres induce a sustained DDR (d'Adda di Fagagna et al, 2003). In both budding yeast and mice, nuclease activities that attack dysfunctional telomeres contribute to telomere-driven senescence (Maringele and Lydall, 2004; Schaetzlein et al, 2007). Therefore, understanding the regulation of nuclease activities at dysfunctional telomeres in yeast is likely to be informative about similar processes occurring at mammalian telomeres and the human ageing process.
dsDNA-binding proteins and accessory factors are required at both human telomeres (TRF1, TRF2, TIN2, TPP1, RAP1) and budding yeast telomeres (Rap1, Rif1, Rif2) to prevent DDRs (Wotton and Shore, 1997; de Lange, 2005; Celli and de Lange, 2005; Marcand et al, 2008; Bonetti et al, 2010; Vodenicharov et al, 2010). In budding yeast, telomeric ssDNA is bound by Cdc13 with accessory proteins Stn1 and Ten1, whereas in human cells, it is bound by POT1 (de Lange, 2005; Gao et al, 2007). Cdc13–Stn1–Ten1 forms an evolutionarily conserved complex (the CST complex) that has telomeric roles in most organisms studied so far (Miyake et al, 2009; Surovtseva et al, 2009). POT1 binds telomeric ssDNA and is connected to the dsDNA-binding proteins of the telomere cap by TPP1 and TIN2 (de Lange, 2009). Inactivation of POT1 or Cdc13 induces ‘telomere uncapping' and has similar consequences—initiation of a DDR and resection of the telomeric DNA by nuclease activities (Garvik et al, 1995; Baumann and Cech, 2001; Pitt and Cooper, 2010).
The response to telomere uncapping is readily studied in budding yeast by inactivation of Cdc13 using the thermosensitive allele cdc13-1 (Garvik et al, 1995). Following Cdc13 inactivation, a potent DDR is initiated; telomeric DNA is resected by nucleases, which degrade the AC (5′) strand to generate extensive TG (3′) ssDNA that stimulates activation of the DNA damage checkpoint, in a manner analogous to that at DNA double-strand breaks (DSBs) (Figure 1A) (Garvik et al, 1995; Lydall and Weinert, 1995; Vodenicharov and Wellinger, 2006). There is relatively little understanding of the nuclease activities responsible for generating ssDNA at uncapped telomeres (Zubko et al, 2004). In contrast, there has been much recent progress identifying nuclease activities that function at DSBs (Gravel et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008).
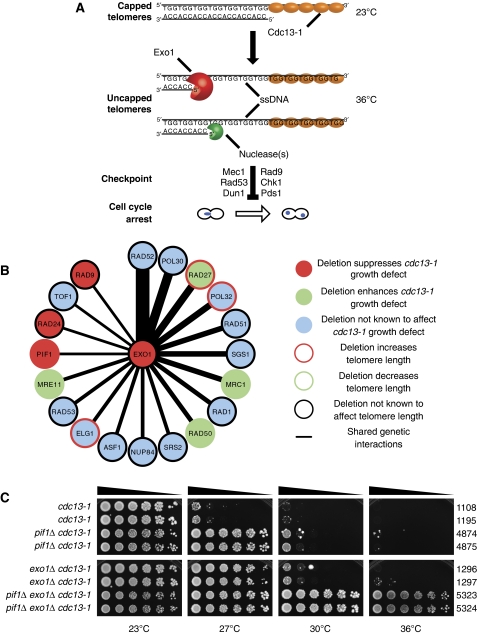
Figure 1.
Pif1 and Exo1 inhibit growth of cdc13-1 mutants. (A) Inactivation of Cdc13 by use of the temperature-sensitive allele cdc13-1 leads to telomere uncapping. Exo1 and additional nuclease(s) generate ssDNA at uncapped telomeres, which is the stimulus for Mec1-dependent checkpoint activation and cell cycle arrest. (B) Ranked diagram of genes that share genetic interactions with EXO1 and are important in the context of telomeres. (Thicker lines mean more shared genetic interactions). (C) Strains of the genotypes shown were serially diluted across agar plates and grown at the temperatures indicated for 3 days. In this and other figures, strain numbers (DLYs) are shown adjacent.
Exo1 is the only nuclease known to generate ssDNA at uncapped telomeres in budding yeast (Maringele and Lydall, 2002). Exo1 is a 5′ to 3′ dsDNA exonuclease involved in DSB resection and in mismatch repair (Tsubouchi and Ogawa, 2000; Gravel et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008). In the absence of Exo1, ssDNA is still generated following Cdc13 inactivation, demonstrating that other nuclease activities must also function at uncapped telomeres. The determinant(s) of this Exo1-independent ssDNA generation have not so far been identified, but at least two hypothetical nuclease activities have been proposed (ExoX and ExoY) (Zubko et al, 2004).
We sought to identify additional nuclease activities functioning at uncapped telomeres following inactivation of Cdc13. Bioinformatic analysis of genetic interactions found the helicase Pif1 to be a candidate for contributing to nuclease activity. Consistent with this hypothesis, we found that Pif1 and Exo1 are required for different nuclease activities that generate ssDNA and activate the DNA damage checkpoint following Cdc13 inactivation. Furthermore, deletion of both PIF1 and EXO1 permits yeast cells to tolerate complete loss of the essential telomere capping protein Cdc13.
Results
PIF1 and EXO1 define parallel pathways that inhibit growth of cdc13-1 mutants
To identify potential nuclease(s) active in cdc13-1 mutants, we reasoned that genes responsible for such activities would interact with similar genes to those that EXO1 interacts with. We used the BioGRID database to create a ranked list of genes that had similar genetic interactions to EXO1 (Figure 1B) (Stark et al, 2006). Of these, 9/19 affected cdc13-1 growth or telomere length. Deletion of EXO1 suppresses cdc13-1 growth defects, so we focussed on those genes that also suppressed cdc13-1 growth defects. By these criteria, two previously characterized checkpoint genes (RAD9 and RAD24) and the helicase-encoding PIF1 behaved similarly to EXO1 (Figure 1B). Rad9 and Rad24 do indeed regulate nuclease activities at uncapped telomeres and are required for checkpoint activation (Garvik et al, 1995; Lydall and Weinert, 1995; Zubko et al, 2004). Pif1 has been shown to inhibit growth of cdc13-1 mutants (Downey et al, 2006), whereas overexpression of Pif1 has been shown to enhance growth defects seen in cdc13-1 mutants, but the contribution of Pif1 to the nuclease activity and checkpoint activation in cdc13-1 mutants had not been assessed (Vega et al, 2007; Chang et al, 2009).
Pif1 is a helicase with both mitochondrial and nuclear functions (Van Dyck et al, 1992; Schulz and Zakian, 1994). In the nucleus, Pif1 has been implicated in negative regulation of telomerase, generation of long flaps during Okazaki fragment processing, unwinding of G-quadruplexes and disassembly of stalled replication forks (Zhou et al, 2000; Boule et al, 2005; Budd et al, 2006; Chang et al, 2009; George et al, 2009; Makovets and Blackburn, 2009; Pike et al, 2009; Ribeyre et al, 2009; Zhang and Durocher, 2010). To test the hypothesis that Pif1 contributes to a nuclease activity at uncapped telomeres in cdc13-1 mutants, we compared the effects of Pif1 and Exo1 on cell growth after Cdc13-1 inactivation. At the permissive temperature (23°C), Cdc13-1 is functional and efficiently caps the telomeres, permitting growth of cdc13-1 mutants. At the non-permissive temperature (36°C), Cdc13-1 is completely defective and cdc13-1 mutants are unable to grow (Figure 1A and C). At semi-permissive temperatures (25–29°C), moderate Cdc13-1 inactivation occurs and growth of cdc13-1 mutants is inhibited (Figure 1C). As previously reported, cdc13-1 pif1Δ and cdc13-1 exo1Δ mutants are able to grow at 27°C, whereas cdc13-1 mutants are not (Figure 1C) (Zubko et al, 2004; Downey et al, 2006). These effects on growth are consistent with the hypothesis that Pif1, like Exo1, contributes to nuclease activity at uncapped telomeres.
Pif1 and Exo1 inhibit growth of cdc13-1 mutants, possibly by contributing to nuclease activity at uncapped telomeres. To test whether the two proteins worked in the same pathway/complex or in different pathways, we examined the effect Pif1 on growth of cdc13-1 exo1Δ mutants. cdc13-1 exo1Δ mutants were unable to grow at 30°C, whereas cdc13-1 exo1Δ pif1Δ mutants were able to grow at 30 and 36°C (Figure 1C). Remarkably, at 36°C, the growth of cdc13-1 exo1Δ pif1Δ mutants was barely distinguishable from that of CDC13+ exo1Δ pif1Δ mutants (Supplementary Figure S1A). We confirmed that this effect was due to the pif1Δ and exo1Δ mutations and not due to second site suppressors arising in our strains by crossing a cdc13-1 mutant able to grow at 36°C, with a cdc13-1 strain and confirming that all cdc13-1 exo1Δ pif1Δ progeny could all grow at 36°C (Supplementary Figure S1B). We conclude that Pif1 and Exo1 inhibit growth of cdc13-1 mutants through different pathways, and inactivation of these pathways may eliminate the requirement for telomere capping by Cdc13.
At DSBs, parallel nuclease activities dependent upon Exo1, the helicase Sgs1 and nuclease Dna2 generate extensive ssDNA (Gravel et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008). We hypothesized that, as with Exo1, elimination of Sgs1 or Dna2 in cells lacking Pif1 might improve the growth of cdc13-1 mutants and perhaps even permit growth at 36°C. However, we found that cdc13-1 pif1Δ dna2Δ mutants grew less well than cdc13-1 pif1Δ mutants (Supplementary Figure S2A). We were unable to examine the effect of dna2Δ on the growth of cdc13-1 PIF1+ mutants, as DNA2 is an essential gene unless PIF1 is deleted (Budd et al, 2006). We also found that cdc13-1 pif1Δ sgs1Δ mutants grew slightly less well than cdc13-1 pif1Δ mutants and that cdc13-1 sgs1Δ mutants grew slightly less well than cdc13-1 mutants (Supplementary Figure S2B), consistent with other work from our laboratory (Ngo and Lydall, 2010). We conclude that Exo1 inhibits the growth of cdc13-1 mutants with uncapped telomeres, whereas Sgs1 and Dna2 contribute to the vitality of such cells. Therefore, we chose to focus on the roles of Pif1 and Exo1 at uncapped telomeres.
Elimination of Pif1 and Exo1 permits telomere maintenance following inactivation of Cdc13
Yeast cells can overcome the requirement for Cdc13 by altering telomere structure, as observed in rare variants, which can be selected for after inactivation of telomerase or after attenuation of nuclease/checkpoint activities at uncapped telomeres (Larrivee and Wellinger, 2006; Zubko and Lydall, 2006). To test whether elimination of Pif1 and Exo1 caused alterations in telomere structure that could explain the growth of cdc13-1 cells at 36°C, we performed Southern blots to examine telomere structure, probing for Y′ sequences (Figure 2B), which are components of the majority of yeast telomeres (Supplementary Figure S3A and B). The Y′ probe contained G-rich sequences and weakly cross-hybridized to telomeres that did not contain Y′ sequences, so we also probed for TG repeat sequences to detect telomeres that lacked Y′ elements (Supplementary Figure S4).
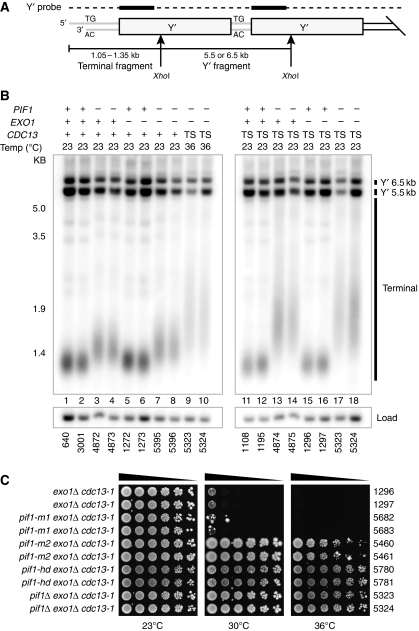
Figure 2.
Exo1 and nuclear, helicase activity of Pif1 prevent telomere maintenance following inactivation of Cdc13. (A) Cartoon of yeast telomeres, indicating the fragments detected by telomere Southern blots using Y′ probe. Arrows represent XhoI cut sites. (B) Genomic DNA was prepared from two independent CDC13+ (+) or cdc13-1 (TS) strains, grown at 23 or 36°C, digested with XhoI and Southern blotted to detect telomeric Y′ and terminal fragments. Blots were reprobed to detect CDC15 as a loading control. Also see Supplementary Figure S4. (C) cdc13-1 exo1Δ mutants defective in nuclear Pif1 (pif1-m2), mitochondrial Pif1 (pif1-m1) or carrying the helicase-deficient allele of Pif1 (pif1-hd) were serially diluted across agar plates and grown at the temperatures indicated for 3 days.
pif1Δ mutants have long telomeres (Schulz and Zakian, 1994) and consistent with this, CDC13+exo1Δ pif1Δ mutants have longer telomeres than CDC13+EXO1+PIF1+ strains (compare lanes 1–2, 3–4 and 7–8; Figure 2B). The telomeres of cdc13-1 exo1Δ pif1Δ mutants grown at 36°C were longer than those of CDC13+exo1Δ pif1Δ mutants grown at 23°C (compare lanes 9–10 with lanes 7–8, Figure 2B) but indistinguishable from those of cdc13-1 exo1Δ pif1Δ mutants grown at 23°C (compare lanes 9–10 with lanes 17–18, Figure 2B). This demonstrates that no gross alterations in telomere structure occur when cdc13-1 exo1Δ pif1Δ mutants are grown at 36°C. Furthermore, cdc13-1 exo1Δ pif1Δ mutants are able to grow at 36°C, whereas cdc13-1 pif1Δ mutants are not, but are indistinguishable in telomere structure (compare lanes 13–14 with lanes 17–18, Figure 2B). We conclude that alterations in telomere structure most likely do not account for the growth of cdc13-1 exo1Δ pif1Δ mutants at 36°C.
Pif1 exists as both nuclear and mitochondrial isoforms (Schulz and Zakian, 1994). Therefore, we wanted to know whether the nuclear or mitochondrial function of Pif1 inhibited growth of cdc13-1 exo1Δ mutants at 36°C. The pif1-m2 allele, lacking nuclear Pif1, permitted growth of cdc13-1 exo1Δ mutants at 36°C, whereas the pif1-m1 allele, lacking mitochondrial Pif1, did not (Figure 2C). We note that the growth of cdc13-1 exo1Δ pif1-m2 mutants at 36°C is less than cdc13-1 exo1Δ pif1Δ mutants (Figure 2C). This is consistent with other reports that low levels of nuclear Pif1 activity persist in pif1-m2 mutants (Schulz and Zakian, 1994; Ribeyre et al, 2009). We also confirmed that helicase activity of Pif1 inhibited growth of cdc13-1 exo1Δ mutants because the pif1-hd allele, deficient in helicase activity, also permitted growth at 36°C (Figure 2C) (Zhou et al, 2000; Ribeyre et al, 2009). We conclude that nuclear, helicase-dependent activity of Pif1 inhibits growth of telomere capping-defective cdc13-1 mutants.
Pif1- and Exo1-dependent nucleases initiate the DDR following Cdc13 inactivation
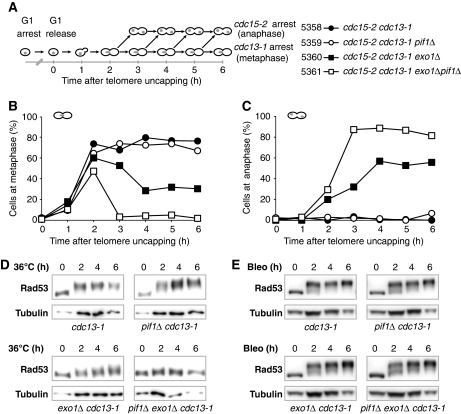
Upon Cdc13 inactivation, nuclease activities generate ssDNA, which stimulates checkpoint kinase cascades and induces metaphase arrest (Figure 1A) (Garvik et al, 1995). To test the role of Pif1 in cell cycle arrest, cdc13-1 mutants were synchronized in G1 using α factor at 23°C, then released to 36°C to assess metaphase arrest (Figure 3A). All strains also harboured the cdc15-2 mutation so that any cells that overcame cdc13-1-induced metaphase arrest would arrest in late anaphase due to cdc15-2 and be unable to enter another cell cycle (Figure 3A) (Lydall and Weinert, 1995; Zubko et al, 2004). As expected, cdc13-1 mutants accumulated at metaphase and did not pass through to anaphase (Figure 3B and C). cdc13-1 exo1Δ mutants accumulated at metaphase with similar kinetics to cdc13-1 mutants but, as previously reported, a subpopulation of cdc13-1 exo1Δ cells escaped metaphase arrest and accumulated in late anaphase due to the cdc15-2 mutation (Figure 3B and C) (Zubko et al, 2004). cdc13-1 pif1Δ mutants behaved like cdc13-1 mutants and did not pass through to anaphase (Figure 3B and C). Interestingly, cdc13-1 exo1Δ pif1Δ mutants did not accumulate in metaphase at all and passed readily through to anaphase (Figure 3B and C). Taken together, these results show that Pif1 has no effect on metaphase arrest of cdc13-1 mutants at 36°C when Exo1 is present, but it is responsible for the arrest of a subpopulation of cells when Exo1 is absent.
Figure 3.
Pif1 and Exo1 stimulate cell cycle arrest following inactivation of Cdc13. (A) Synchronous culture experiments to examine the effect of Pif1 and Exo1 on cell cycle arrest following telomere uncapping. cdc15-2 cdc13-1 cells were synchronized in G1 using α-factor then released at 36°C. Cells arrest at metaphase from telomere uncapping (cdc13-1) or at anaphase from Cdc15 inactivation (cdc15-2). Samples taken at the time points indicated were stained with DAPI and >100 cells of each genotype scored for cell cycle position (Zubko et al, 2006). (B) Percentage of cells at metaphase. (C) Percentage of cells at late anaphase. (D) Western blots of Rad53 following shift to 36°C. Upper and lower panels were run in parallel on separate gels, but transferred, detected and imaged simultaneously. (E) Western blots of Rad53 following treatment with bleomycin. Corresponding scoring of cells at metaphase and anaphase given as Supplementary Figure S5.
Following inactivation of Cdc13, Mec1-dependent checkpoint activation occurs, leading to activation and hyperphosphorylaiton of the kinase Rad53 (Figure 1A) (Sweeney et al, 2005; Morin et al, 2008). We used the synchronous cultures to examine Rad53 phosphorylation by western blot (Figure 3A). cdc13-1 and cdc13-1 pif1Δ mutants exhibited strong Rad53 phosphorylation, indicated by a marked upward mobility shift of Rad53 (upper panels, Figure 3D). A reduction in Rad53 phosphorylation was seen in cdc13-1 exo1Δ mutants, correlating with the recovery from metaphase arrest displayed by cdc13-1 exo1Δ mutants following telomere uncapping (Figure 3C and D). Interestingly, no discernable change in the mobility of Rad53 could be seen in cdc13-1 exo1Δ pif1Δ mutants, consistent with their complete failure to arrest cell division at 36°C (Figure 3C and D). We conclude that in the absence of Pif1 and Exo1, the checkpoint kinase Rad53 is not activated after telomere uncapping in cdc13-1 mutants.
To see whether cdc13-1 exo1Δ pif1Δ strains were defective in the DDR after other types of DNA damage as well as after telomere uncapping, we treated cells with bleomycin to induce DSBs. At both DSBs and uncapped telomeres, ssDNA is an important stimulus for the Mec1-dependent checkpoint. We treated the same set of strains examined in Figure 3B–D, with bleomycin at 23°C after release from G1 arrest. cdc13-1, cdc13-1 pif1Δ, cdc13-1 exo1Δ and cdc13-1 exo1Δ pif1Δ mutants all behaved similarly, phosphorylating Rad53 and arresting at methaphase (Figure 3E; Supplementary Figure S4). We conclude that a functional DDR pathway operates in (cdc13-1) exo1Δ pif1Δ cells but that these cells are specifically defective in responding to telomere uncapping.
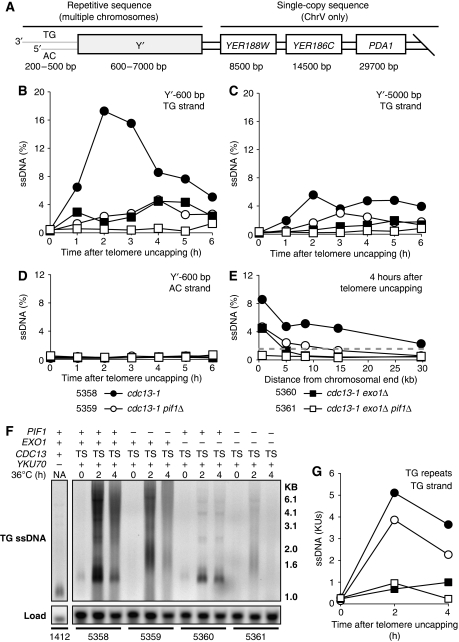
Telomeric ssDNA stimulates metaphase arrest following telomere uncapping (Garvik et al, 1995). We used synchronous cultures (Figure 3A) and quantitative amplification of ssDNA (QAOS) to measure subtelomeric ssDNA in repetitive Y′ elements (present on Chromosome V and most other chromosome ends) following Cdc13 inactivation (Figure 4A) (Booth et al, 2001). cdc13-1 mutants with uncapped telomeres generated ssDNA at both the Y′600 and Y′5000 loci (Figure 4B and C). cdc13-1 exo1Δ mutants generated less ssDNA following telomere uncapping at the Y′ loci, as previously reported (Maringele and Lydall, 2002; Zubko et al, 2004). Interestingly, cdc13-1 pif1Δ mutants, like cdc13-1 exo1Δ mutants showed reduced ssDNA generation in the Y′600 and Y′5000 loci following telomere uncapping. Furthermore, cdc13-1 exo1Δ pif1Δ mutants generated no detectable ssDNA at Y′600 or Y′5000 loci following telomere uncapping (Figure 4B and C). We confirmed that ssDNA generation occurred on the TG (3′) strand due to degradation of the AC (5′) strand, as we were unable to detect ssDNA on the AC strand (Figure 4D). We conclude that Pif1, like Exo1, is important for ssDNA generation after telomere uncapping in cdc13-1 mutants and appears to regulate a nuclease activity, which functions in parallel to Exo1 at chromosome ends.
Figure 4.
Pif1 and Exo1 generate ssDNA at uncapped telomeres. (A) Physical map of the right telomere of Chromosome V. Synchronous cultures were subjected to telomere uncapping as in Figure 3A, and samples were taken to measure ssDNA. (B) TG-strand ssDNA 600 bp from the end of the telomere (Y′600) measured by QAOS. (C) TG-strand ssDNA 5000 bp away from the end of the telomere (Y′5000). (D) AC-strand ssDNA 600 bp away from the end of the telomere. (E) TG-strand ssDNA generated at loci indicated in (A), 4 h after telomere uncapping. Dashed line represents 1.56% (1/64), corresponding to the value expected for 1 single-stranded locus per yeast cell in G2. (F) TG-strand ssDNA in the telomeric TG repeats detected by in-gel assay, comparing cdc13-1 mutants (TS) to a yku70Δ control. Loading determined by Southern hybridization with a CDC15 probe. Lanes were run and detected on the same gel and cropped for presentation purposes. (G) Quantification of the ssDNA signal in each lane in (F). ssDNA is measured in KUs (Ku units). In all, 1 Ku Unit is the ssDNA signal in an asynchronously dividing yku70Δ control at 23°C.
To examine how Pif1 and Exo1 affect ssDNA accumulation further from chromosome ends, we measured ssDNA at single-copy loci on Chromosome V after Cdc13 inactivation. At 4 h, cdc13-1 mutants generated ssDNA at all loci examined, with the amount of ssDNA decreasing at loci further from the chromosome end, as previously reported (Zubko et al, 2004). cdc13-1 exo1Δ mutants generated less ssDNA in the Y′ repeats and no ssDNA in single-copy loci on Chromosome V, also as previously reported (Figure 4E) (Zubko et al, 2004). cdc13-1 pif1Δ mutants generated similar amounts of ssDNA to cdc13-1 exo1Δ mutants in the Y′ repeats. However, at more distal, single-copy loci, cdc13-1 pif1Δ mutants generated less ssDNA than cdc13-1 mutants but more than exo1Δ cdc13-1 mutants. The higher levels of ssDNA generated further from the chromosome end in cdc13-1 pif1Δ mutants, in comparison with cdc13-1 exo1Δ mutants, most likely accounts for their sustained metaphase arrest following telomere uncapping (Figure 3B and C). Furthermore, the ssDNA generated by cdc13-1 and cdc13-1 pif1Δ mutants <10 kb from the chromosome end is >1.6% (1/64) (Figure 4E). Assuming 1 single-stranded telomere per cell is sufficient to stimulate arrest, this suggests that ssDNA extending <10 kb on one of the 64 G2 telomeres in Saccharomyces cerevisiae is sufficient to stimulate metaphase arrest (Figure 4E) (Sandell and Zakian, 1993; Vaze et al, 2002; Zubko et al, 2004).
No checkpoint activation was detected in cdc13-1 exo1Δ pif1Δ mutants following telomere uncapping, and no ssDNA was detected in the Y′ elements (Figures 3D and 4B). However, yku70Δ mutants at 23°C have detectable ssDNA in the telomeric TG repeats but do not undergo checkpoint activation (Gravel et al, 1998; Polotnianka et al, 1998; Maringele and Lydall, 2002). Thus, we hypothesized that cdc13-1 exo1Δ pif1Δ mutants might still generate detectable ssDNA in the TG repeats. We used synchronous cultures (Figure 3A) and measured ssDNA by in-gel assay to measure ssDNA in the TG repeats in cdc13-1 mutants (Figure 4F and G). cdc13-1 mutants generated large amounts of TG ssDNA at 2 and 4 h following telomere uncapping (Figure 4F), corresponding to an approximately five-fold increase in signal compared with yku70Δ mutants (Figure 4G). cdc13-1 exo1Δ pif1Δ mutants also generated detectable ssDNA 2 h following telomere uncapping, but at a level approximately equal to that of a yku70Δ mutant (Figure 4F and G). However, the ssDNA generated in cdc13-1 exo1Δ pif1Δ mutants was transient and was no longer detectable 4 h after telomere uncapping (Figure 4F and G). Surprisingly, cdc13-1 pif1Δ mutants displayed only a modest decrease in ssDNA generation in the TG repeats following telomere uncapping, whereas cdc13-1 exo1Δ mutants generated very little ssDNA (Figure 4F and G). We conclude that cdc13-1 exo1Δ pif1Δ mutants generate limited, transient ssDNA that is insufficient to stimulate checkpoint activation and that Exo1 is much more important than Pif1 for ssDNA generation in the TG repeats following telomere uncapping.
Pif1 has important functions in cells lacking telomerase
It has been suggested that increased levels of telomerase at the telomeres of cdc13-1 pif1Δ cells shields uncapped telomeres from nuclease activities (Vega et al, 2007). However, this is somewhat inconsistent with our observation that Pif1 has relatively little effect on ssDNA generation in the telomeric TG repeats, where telomerase presumably binds (Figure 4G). Therefore, we wanted to know whether the ability of the pif1Δ mutation to improve the growth of cdc13-1 mutants was dependent upon the telomerase template component (TLC1) or catalytic subunit (Est2). Interestingly, we found that cdc13-1 tlc1Δ pif1Δ and cdc13-1 tlc1Δ exo1Δ mutants grew better at 25°C than cdc13-1 tlc1Δ mutants (compare rows 7–8 and 11–12 with 3–4, Figure 5A; Supplementary Figure S6). We also found that cdc13-1 est2Δ pif1Δ and cdc13-1 est2Δ exo1Δ mutants were able to grow at 25°C, whereas cdc13-1 est2Δ mutants were not (Supplementary Figure S7). We conclude that Pif1 has a telomerase (TLC1, Est2) independent effect at uncapped telomeres. However, we note that est2Δ cdc13-1 and tlc1Δ cdc13-1 mutants grow worse than cdc13-1 mutants, demonstrating that telomerase contributes to telomere capping following inactivation of Cdc13.
Figure 5.
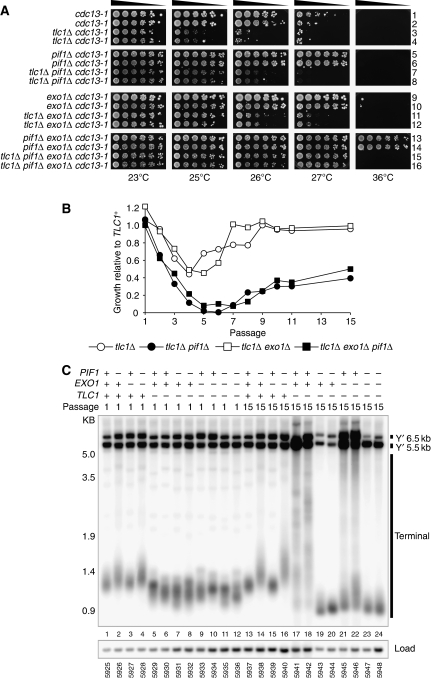
Pif1 has telomerase-independent effects at telomeres. (A) A cdc13-1/CDC13+tlc1Δ/TLC1+exo1Δ/EXO1+pif1Δ/PIF1+ diploid (DLY1628 x DLY5324) was sporulated, dissected and germinated at 23°C to generate strains of the indicated genotype at 23°C. These were taken from the germination plate, grown to saturation, then serially diluted across agar plates and grown at the temperatures indicated for 3 days. (B) Strains of the genotypes indicated were passaged repeatedly by restreaking at 30°C for 3 days along with TLC1+ controls. At the passages indicated, strains were assayed for growth, which was then quantified (Supplementary Figure S9). Growth at each passage is given as a fraction of the growth of the relevant TLC1+ strain and the mean of two independent strains is shown. (C) At passages 1 and 15, strains assayed for growth in (B) had genomic DNA isolated and were Southern blotted with Y′ probe, as in Figure 2B. See Supplementary Figure S10 for detection with TG probe.
Pif1 is responsible for the residual checkpoint activation in cdc13-1 exo1Δ mutants (Figure 3D) and inhibits growth of cdc13-1 mutants lacking telomerase (Figure 5A). We hypothesized that Pif1 would contribute to ssDNA generation at uncapped telomeres and subsequent checkpoint activation, even in cdc13-1 mutants lacking telomerase. To test this, we measured Rad53 phosphorylation (Supplementary Figure S8A) and telomeric TG repeat ssDNA (Supplementary Figure S8B and C) in cdc13-1 and cdc13-1 tlc1Δ mutants, before and after telomere uncapping. In cdc13-1 tlc1Δ exo1Δ pif1Δ mutants, there was a decrease in Rad53 phosphorylation compared with cdc13-1 tlc1Δ exo1Δ mutants (Supplementary Figure S8A). We also found that cdc13-1 tlc1Δ pif1Δ and cdc13-1 tlc1Δ exo1Δ pif1Δ mutants generated less ssDNA than cdc13-1 tlc1Δ and cdc13-1 tlc1Δ exo1Δ mutants, respectively, following telomere uncapping (Supplementary Figure S8B and C). We conclude that Pif1 has a contribution to ssDNA generation and checkpoint activation following telomere uncapping that is independent of telomerase. However, we note that 40% of cdc13-1 tlc1Δ cells were at metaphase at 23°C compared with 30% of cdc13-1 tlc1Δ pif1Δ cells (Supplementary Figure S8D). Thus, we cannot exclude the possibility that reduced ssDNA generation in cdc13-1 tlc1Δ pif1Δ mutants is due to altered kinetics of accumulation at metaphase.
Although Pif1 clearly demonstrates a telomerase-independent effect in cdc13-1 mutants, we note that cdc13-1 tlc1Δ exo1Δ mutants could grow at 27°C, whereas cdc13-1 tlc1Δ pif1Δ mutants could not (compare rows 11–12 with 7–8, Figure 5A). In contrast, cdc13-1 TLC1+exo1Δ mutants and cdc13-1 TLC1+pif1Δ mutants could both grow similarly at 27°C (compare rows 9–10 with 5–6, Figure 5A). This shows that Pif1 is less potent than Exo1 at inhibiting growth of cdc13-1 tlc1Δ mutants than cdc13-1 TLC1+ mutants.
To help clarify the role of Pif1 in cdc13-1 tlc1Δ and cdc13-1 TLC1+ mutants, we examined the effect of Pif1 in CDC13+ tlc1Δ mutants. Yeast strains that cannot recruit telomerase (e.g. tlc1Δ) lose telomeric DNA with each cell division until the cultures senesce and lose proliferative capacity, much like mammalian fibroblasts. Senescent, telomerase-deficient yeast cultures usually recover, using telomerase-independent, recombination-dependent mechanisms of telomere maintenance, leading to clear changes in telomere structure (Lundblad and Blackburn, 1993; Teng and Zakian, 1999). Therefore, we germinated spores containing combinations of null mutations in telomerase (TLC1), PIF1 and EXO1, and assessed growth at various passages (Figure 5B). As expected, tlc1Δ mutants grew well at passage 1, poorly from passages 2–5 (senescence) and grew well again from passage 7 (recovery) (Figure 5B). tlc1Δ exo1Δ mutants showed a similar pattern of growth to tlc1Δ mutants but grew slightly better prior to senescence, as previously reported (passages 3 and 4, Figure 5B) (Maringele and Lydall, 2004). In contrast, tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ strains showed a rapid decline in growth from passages 2–6 (senescence) and exhibited a protracted senescence period but slowly recovered by passage 15 (Figure 5B). We conclude that Pif1 inhibits entry into senescence and promotes recovery. Taken with the data discussed earlier, Pif1 contributes to good growth of tlc1Δ mutants from passage 2 onwards, yet inhibits growth of cdc13-1 mutants (Figures 1C and 5A). In contrast, Exo1 has only a small effect on the growth of tlc1Δ mutants, yet inhibits growth of cdc13-1 mutants (Figures 1C and 5A). Thus, it is likely that the relatively poor growth of cdc13-1 tlc1Δ pif1Δ compared with cdc13-1 tlc1Δ exo1Δ mutants is due to the poor growth of tlc1Δ pif1Δ mutants compared with tlc1Δ exo1Δ mutants.
After long cultivation (passage 15, 45 days growth) tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ mutants could be distinguished from those of tlc1Δ and tlc1Δ exo1Δ mutants by their poor growth (Figure 5B). Telomerase-deficient survivors typically show clear alterations in telomere structure—type I survivors undergo dramatic amplification of Y′ elements and retain short TG overhangs, whereas type II survivors show modest amplification of Y′ elements but amplify their TG overhangs (Teng and Zakian, 1999). Therefore, we examined the telomeres of tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ mutants. At passage 1, before entry into senescence, tlc1Δ, tlc1Δ exo1Δ, tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ mutants all had short telomeres (compare lanes 1–4 with lanes 5–12, Figure 5C). At passage 15, tlc1Δ mutants and tlc1Δ exo1Δ mutants had amplified Y′ elements and terminal fragments to generate type II (lanes 17–18, Figure 5D) or type I (lanes 21–22, Figure 5D) survivors. In contrast, tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ mutants at passage 15 had shorter telomeres than at passage 1 and had undergone a reduction in Y′ elements, but did not appear to have generated type I or type II survivor telomere structures (compare lanes 7–8 with 19–20 and lanes 11–12 with 23–24, Figure 5D). We noted that tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ mutants resembled type I survivors in that our TG probe did not detect any individual telomeres further up the gel (marked with arrows, compare lanes 7–8 with 19–20 and lanes 11–12 with 23–24, Supplementary Figure S10), indicating that all telomeres in these strains had acquired a terminal Y′ fragment. However, the terminal fragments of tlc1Δ pif1Δ and tlc1Δ pif1Δ exo1Δ were even shorter than those of type I survivors and they had undergone a reduction, not an amplification in Y′ elements, clearly distinguishing them from typical type I survivors (compares lanes 19–20, 23–24 with lanes 21–22, Figure 5D). We conclude that Pif1 is required for the generation of type I and type II survivors and that in the absence of Pif1, cells lacking telomerase can improve growth following senescence without adopting typical type I or type II survivor structures.
Telomerase is crucial for survival in the absence of Cdc13
Cdc13 has two crucial functions at telomeres—one, to shield telomeres from nuclease activities and the second to recruit telomerase to the telomere (Nugent et al, 1996). Although elimination of Pif1 and Exo1 permits cdc13-1 mutants to grow at 36°C (presumably when cdc13-1 is completely inactivated), this does not permit cdc13-1 tlc1Δ mutants to grow at 36°C (compare rows 13–14 with 15–16, Figure 5A), demonstrating that telomerase has a function in cdc13-1 mutants at 36°C. Therefore, we hypothesized that at 36°C, Cdc13-1 might retain the ability to recruit telomerase, and Cdc13-dependent telomerase activity might be essential for growth. Alternatively, telomerase might be recruited to telomeres in a Cdc13-independent manner.
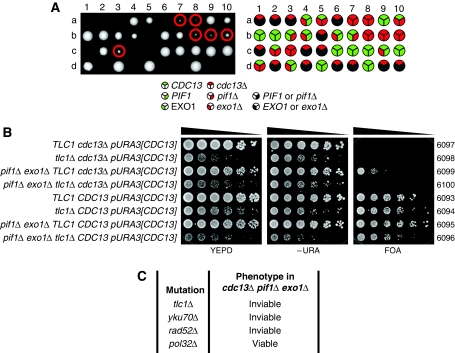
To test whether Cdc13-1 retained the ability to recruit telomerase in cdc13-1 exo1Δ pif1Δ cells at 36°C, we decided to delete CDC13. We generated diploid strains heterozygous for pif1Δ, exo1Δ and cdc13Δ mutations. Diploids were sporulated, tetrads dissected and vegetative cells containing combinations of the three deletion mutations were allowed to form colonies. Figure 6A shows a representative image of one tetrad dissection plate. Importantly, we found that cdc13Δ exo1Δ pif1Δ strains were viable and had a germination efficiency of ∼90% compared with CDC13 exo1Δ pif1Δ strains (Figure 6A; Supplementary Table SI). However, it was clear that cdc13Δ exo1Δ pif1Δ mutants grew less well than CDC13+exo1Δ pif1Δ mutants on the tetrad dissection plates (Figure 6A). No viable colonies were formed from cdc13Δ cells that were either PIF1+ or EXO1+. We conclude that neither Cdc13-dependent telomerase activity nor Cdc13-dependent capping activities are required for growth following elimination of Pif1 and Exo1.
Figure 6.
cdc13Δ exo1Δ pif1Δ cells are viable but depend upon telomerase and homologous recombination for survival. (A) cdc13Δ/CDC13 exo1Δ/EXO1 pif1Δ/PIF1 diploids (DDY341) were dissected. Viable cdc13Δ exo1Δ pif1Δ meiotic progeny are circled. Spores were incubated for 5 days at 23°C before genotypes were determined by replica plating to selective medium. (B) Strains of the genotypes indicated, all carrying a pURA3[CDC13] plasmid (pDL1012) were germinated, grown to saturation, then serially diluted across YEPD, −URA and FOA agar plates. Plates were incubated at 30°C for 4 days, YEPD plate shown after 2 days, −URA plate shown after 3 days and FOA plate shown after 4 days. (C) Summary of the roles of various telomere-related genes and their role in the survival of cdc13Δ pif1Δ exo1Δ mutants as assessed in Supplementary Figures S11 and S12.
The viability of cdc13Δ exo1Δ pif1Δ strains was surprising in the light of the requirement for telomerase in cdc13-1 exo1Δ pif1Δ strains. We hypothesized that Cdc13-independent telomerase activity was essential for the viability of cdc13Δ exo1Δ pif1Δ strains. To test this, we generated diploid strains heterozygous for pif1Δ, exo1Δ, cdc13Δ and tlc1Δ mutations, containing a plasmid that carried a wild-type copy of CDC13. Diploids were sporulated and tetrad dissection was performed to generate strains containing combinations of the four deletion mutations, in addition to the wild-type copy of CDC13. These strains were diluted across agar plates either fully supplemented (YEPD), lacking uracil (−URA) or containing FOA (FOA). FOA is toxic to cells with an active uracil biosynthetic pathway so only cells able to survive in the absence of the URA3- and CDC13-containing plasmid would be able to grow. As expected, cdc13Δ TLC1+ and cdc13Δ tlc1Δ mutants were able to grow on YEPD and –URA, but not on FOA medium, demonstrating that CDC13 was essential for survival of these cell types (Figure 6B). cdc13Δ TLC1+exo1Δ pif1Δ strains were able to grow on FOA, YEPD and –URA, demonstrating that CDC13 was not essential in this background (Figure 6B). However, cdc13Δ TLC1+exo1Δ pif1Δ cells grew much more poorly on FOA than on –URA and YEPD consistent the poor growth of cdc13Δ exo1Δ pif1Δ cells on the tetrad dissection plate(s) (Figure 6A). Importantly, cdc13Δ tlc1Δ exo1Δ pif1Δ cells were able to grow on YEPD and –URA but not on FOA, demonstrating that they could not survive in the absence of Cdc13. We conclude that telomerase is essential for the survival of cdc13Δ exo1Δ pif1Δ mutants, suggesting that Cdc13-independent recruitment of telomerase is essential for their survival.
We used the same plasmid-based method to assess the requirement for various proteins involved in telomere maintenance to the growth of cdc13Δ exo1Δ pif1Δ mutants. We confirmed that TLC1 was required for the viability of cdc13Δ pif1Δ exo1Δ as cdc13Δ pif1Δ exo1Δ tlc1Δ mutants could not lose a plasmid carrying CDC13 (p[URA3]CDC13) (Figure 6C). We also found that Yku70 (a component of the Ku complex, which binds TLC1 to aid in recruitment of telomerase to the telomere) and Rad52 (required for homologous recombination and the generation of type I and type II survivor telomere structures) were required for the viability of cdc13Δ pif1Δ exo1Δ mutants (Figure 6C). However, we found that Pol32 (subunit of Polymerase δ, required for the generation of type I and type II survivor telomere structures) was dispensable for the viability of cdc13Δ pif1Δ exo1Δ mutants (Figure 6C), although elimination of Pol32 did reduce the frequency at which cdc13Δ pif1Δ exo1Δ mutants were able to lose the pURA3[CDC13] (Supplementary Figure S12). We conclude that cdc13Δ pif1Δ exo1Δ mutants are distinct from type I and type II survivors, as they do not require Pol32, and their telomeres are maintained through a combination of homologous recombination, Ku and telomerase activity.
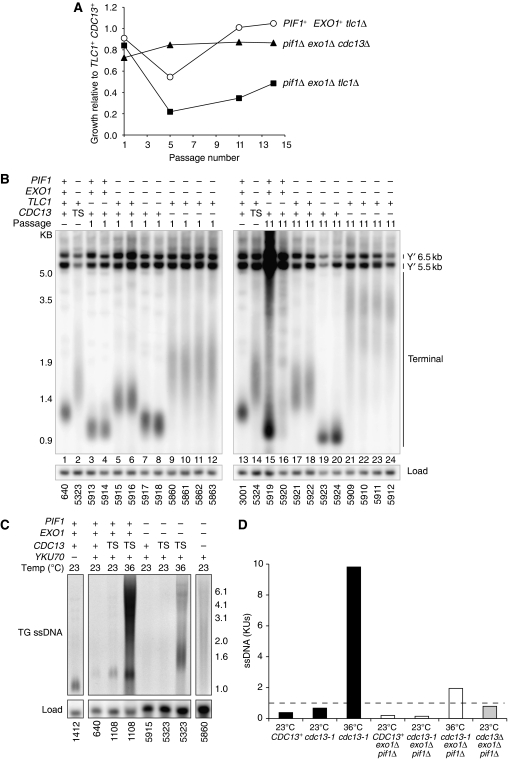
If cdc13Δ exo1Δ pif1Δ mutants are able to recruit telomerase then they may not senesce or undergo the rearrangements in telomere structure characteristic of telomerase-deficient strains. To test this hypothesis, we compared the growth and telomere structure of cdc13Δ exo1Δ pif1Δ strains with tlc1Δ and tlc1Δ exo1Δ pif1Δ strains. As expected, telomerase-deficient tlc1Δ mutants senesced and recovered (Figure 7A) and by passage 11 they had generated type I (lane 15, Figure 7B) and type II (lane 16, Figure 7B) survivors. Interestingly, cdc13Δ exo1Δ pif1Δ strains showed a slight growth defect at passage 1 (Figure 7A), consistent with the small colonies formed by cdc13Δ exo1Δ pif1Δ mutants on the tetrad dissection plate (Figure 6A) and the growth defect of cdc13Δ TLC1+exo1Δ pif1Δ pURA3[CDC13] mutants grown on FOA (Figure 6B). However, by passage 5 and in all subsequent passages cdc13Δ exo1Δ pif1Δ mutants had improved in growth (Figure 7A). The telomeres of cdc13Δ exo1Δ pif1Δ mutants were long with more variation in length, and by passage 11, the median telomere length and variance in length increased (lanes 9–12, 21–24, Figure 7B). This is consistent with previous work showing that hypomorphic alleles of Cdc13 can cause increased telomere length and variance in telomere length (Chandra et al, 2001). No clear alterations in the telomere structure of cdc13Δ exo1Δ pif1Δ mutants were observed, and at passage 1, their telomeres most closely resembled those of cdc13-1 exo1Δ pif1Δ mutants with capped telomeres, which notably do not senesce (compare lanes 2, 14 with lanes 9–12, Figure 7B). The growth and telomere structure of cdc13Δ exo1Δ pif1Δ mutants was clearly distinct from that of telomerase-deficient tlc1Δ exo1Δ pif1Δ mutants, which rapidly senesced and slowly recovered (Figure 7A), while maintaining a relatively normal telomere structure (compare lanes 7–8 with lanes 19–20, Figure 7B). We conclude that cdc13Δ exo1Δ pif1Δ mutants do not undergo senescence and maintain their telomeres for at least 11 passages (44 days). This is consistent with our notion that cdc13Δ exo1Δ pif1Δ mutants are able to maintain telomeres in a telomerase-dependent manner, even in the absence of Cdc13.
Figure 7.
Cells lacking Cdc13 maintain telomeres and do not senesce. (A) Strains of the genotypes indicated were passaged repeatedly at 23°C for 4 days along with TLC1+CDC13+ controls. At the passages indicated, strains were assayed for growth (Supplementary Figure S13). Growth at each passage is given as a fraction of the growth of the relevant TLC1+ CDC13+ strain. The mean of two tlc1Δ strains or four cdc13Δ strains is shown. (B) At passages 1 and 11, strains that were assayed for growth in (A) had genomic DNA isolated and were Southern blotted, as in Figure 2B. PIF1+EXO1+CDC13+ and exo1Δ pif1Δ cdc13-1 (TS) strains were grown independently. See Supplementary Figure S14 for original blot probed with Y′ and TG probe. (C) CDC13+ (+), cdc13Δ (−) and cdc13-1 (TS) strains of the genotypes indicated were either grown exponentially at 23 or 36°C for 4 h before their DNA was isolated and telomeric TG ssDNA was detected by in-gel assay as in Figure 4F. (D) Quantification of ssDNA from the in-gel assay shown in (C), as in Figure 4F.
Finally, although we observed that cdc13-1 exo1Δ pif1Δ mutants generated ssDNA only in the TG repeats, transiently over the course of a single cell cycle, we wished to know whether repeated cell division in the absence of telomere capping would lead to accumulation of ssDNA. By in-gel assay, we found that cdc13-1 exo1Δ pif1Δ mutants grown at 36°C for 4 h (∼2 population doublings with uncapped telomeres) and cdc13Δ exo1Δ pif1Δ mutants from Passage 1 (∼50 population doublings with uncapped telomeres) generated comparable levels of ssDNA in the TG repeats to a yku70Δ mutant (Figure 7C and D). This was comparable to the transient level of ssDNA seen in cdc13-1 exo1Δ pif1Δ mutants 2 h after telomere uncapping, within a single cell cycle (Figure 4G). We conclude that continued growth following telomere uncapping in exo1Δ pif1Δ mutants does not lead to ssDNA accumulation. This suggests that no residual nuclease activities continue to resect uncapped telomeres in the absence of Pif1 and Exo1.
Discussion
We have shown that Pif1 and Exo1 are responsible for extensive ssDNA generation at uncapped telomeres in cdc13-1 mutants and also that Pif1 has telomerase-independent functions at telomeres. This leads us to propose a model where Pif1 can initiate the DDR at uncapped telomeres by controlling nuclease activity. Furthermore, and remarkably, cells lacking Pif1 and Exo1 are viable and grow well in the absence of the usually essential telomere capping protein Cdc13.
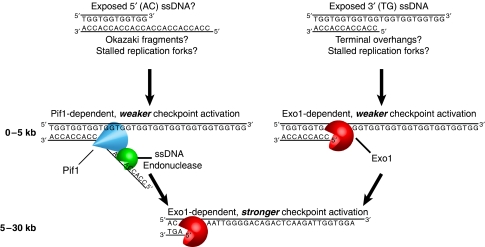
In our model (Figure 8), Pif1 unwinds telomeric duplex DNA, generating ssDNA that is cleaved by an unidentified ssDNA endonuclease in a manner analogous to the function of Pif1 in Okazaki fragment processing and at stalled replication forks (Budd et al, 2006; Chang et al, 2009; George et al, 2009; Pike et al, 2009). We propose that close to the chromosome end (<5 kb) both Pif1- and Exo1-dependent activities generate ssDNA, causing weak initial checkpoint activation. We propose that Exo1 subsequently generates ssDNA >5 kb from the chromosome end, leading to stronger checkpoint activation that is sufficient to arrest all cells with uncapped telomeres. Our model is consistent with Pif1 being ExoY, a hypothetical nuclease proposed to function in parallel to Exo1 at uncapped telomeres (Zubko et al, 2004). Pif1, like ExoY, is more important for ssDNA generation at the end of the chromosome than further away (Figure 4E).
Figure 8.
A model for the roles of Pif1 and Exo1 in the DDR at uncapped telomeres. Following telomere uncapping, 5′ and 3′ ssDNA is exposed within 5 kb of the chromosome end, serving as a substrate for resection. 5′ ssDNA is unwound by Pif1 and presumably cleaved by an unidentified ssDNA endonuclease, whereas the junction between 3′ ssDNA and dsDNA is recognized and resected by the exonuclease Exo1. This low-level processing leads to weak checkpoint activation. Resection of 3′ ssDNA overhangs beyond 5 kb requires Exo1. This further processing causes robust checkpoint activation.
In our model, Exo1 recognizes the junction between 3′ (TG) ssDNA and duplex DNA, at native telomeric overhangs as previously suggested (Maringele and Lydall, 2002) or at stalled replication forks (Segurado and Diffley, 2008). We propose that Pif1 binds and unwinds 5′ (AC) overhangs because Pif1 is a 5′–3′ helicase (Lahaye et al, 1991; Zhou et al, 2000; Pike et al, 2009). If Pif1 does engage telomeric dsDNA and convert it to ssDNA, 5′ (AC) ssDNA presumably exists (Supplementary Figure S15A). Interestingly, 5′ telomeric ssDNA has been observed both in mammalian cells and in Caenorhabditis elegans (Cimino-Reale et al, 2003; Raices et al, 2008) and but not so far in S. cerevisiae. However, 5′ ssDNA overhangs could in principle occur at stalled replication fork structures (Supplementary Figure S15B) or Okazaki fragments.
DSBs that can be repaired by homologous recombination and DSB-induced shortened telomeres are processed by nucleases dependent upon Sgs1/Dna2, Exo1 and MRX/Sae2 (Gravel et al, 2008; Zhu et al, 2008; Mimitou and Symington, 2009). Other work recently published from our laboratory demonstrates that Sgs1 also contributes to resection of uncapped telomeres, but elimination of Sgs1 and Exo1 is insufficient to prevent the resection of uncapped telomeres in cdc13-1 mutants (Ngo and Lydall, 2010). The work presented here demonstrates that elimination of Pif1 and Exo1 prevents resection of uncapped telomeres in cdc13-1 mutants. However, at DSBs that can be repaired by homologous recombination or at DSB-induced shortened telomeres, Pif1 has little effect on resection (Zhu et al, 2008; Bonetti et al, 2009). Interestingly, Pif1 has been shown to have a critical role repair of DSBs where break-induced replication (BIR) is the main repair pathway (Chung et al, 2010). A major challenge will be to determine which substrates are exposed at DSBs, shortened telomeres and uncapped telomeres and how nuclease activities are coordinated to process them.
Pif1 contributes to the vitality of cells lacking telomerase, both before and after recovery from senescence (Figure 5B). Interestingly, pif1Δ cells improve their growth following senescence without adopting typical survivor-like telomeric DNA structures (Figure 5C). Usually following senescence, survivors are generated by homologous-recombination- and BIR-dependent alterations in telomere structure (Teng and Zakian, 1999; Lydeard et al, 2007). If BIR is eliminated, cells lacking telomerase senesce and undergo a complete loss in viability (Lydeard et al, 2007). The relatively unaltered telomere structure and poor growth following senescence in cells lacking Pif1 and telomerase is consistent with the impaired BIR seen in cells lacking Pif1 (Chung et al, 2010). Therefore, reduced BIR in pif1Δ cells may be sufficient to maintain comparatively normal telomere structure in telomerase-deficient cells but insufficient to permit the typical amplification of Y′ elements or terminal TG repeats seen in survivors. The absence of telomeric repeat amplification could prevent these cells from achieving the high levels of post-senescence growth seen in other telomerase-deficient mutants.
We have demonstrated that attenuation of the DDR at uncapped telomeres, by elimination of Pif1 and Exo1 permits telomere maintenance in a Cdc13-indpendent but telomerase and Ku-dependent manner. This is surprising because Cdc13 is considered crucial for efficient recruitment of telomerase and thus to prevent senescence (Nugent et al, 1996). We propose that in the absence of Cdc13, Yku80 binds TLC1, the telomerase RNA, to help recruit telomerase to the telomere (Peterson et al, 2001). The requirement for Rad52 for the survival of cdc13Δ exo1Δ pif1Δ mutants is surprising. It will be interesting to investigate whether telomeric repeats from extremely long telomeres in cdc13Δ exo1Δ pif1Δ mutants (Figure 7B) can be distributed to shorter telomeres by homologous recombination, thus preventing short telomeres from becoming critically short. Finally, it will also be paramount to determine whether the requirement for telomerase in cdc13Δ exo1Δ pif1Δ mutants is a consequence of the increased utilization of telomerase that has been reported at the telomeres of cells lacking Pif1 (Boule et al, 2005).
We show that following inactivation of Cdc13 or telomerase, telomeric DNA can be stabilized by elimination of Pif1, eliminating resection of uncapped telomeres in cells lacking Cdc13 or permitting telomere maintenance without the generation of typical type I or type II survivor structures in cells lacking telomerase. As Pif1 and Exo1 are conserved from yeast, to mice, to humans, Pif1 might also contribute to the premature mortality caused by telomere dysfunction in telomerase knockout mice (Lahaye et al, 1991; Huang and Symington, 1993; Wei et al, 2003; Mateyak and Zakian, 2006; Snow et al, 2007). pif1−/− and exo1−/− mice have previously been examined (Wei et al, 2003; Snow et al, 2007), and it might be interesting to combine these mutations in a telomerase knockout background to investigate the consequences of telomere dysfunction in pif1−/− exo1−/− mice, as EXO1 contributes to the telomere dysfunction and premature mortality seen in telomerase knockout mice (Schaetzlein et al, 2007).
Materials and methods
Bioinformatics/analysis
A genetic interaction network was created in Cytoscape using S. cerevisiae genetic interactions from BioGRID (v2.0.53) (Stark et al, 2006; Cline et al, 2007). Genes that had genetic interactions with EXO1 were identified (first neighbours of EXO1). A ranked list of all known genes was created, according to how many of the first neighbours of EXO1 they had a genetic interaction with. The top 10% of this ranked list were then shown. Nodes for each gene were coloured according to whether the genes affected cdc13-1 growth defects or telomere length (Askree et al, 2004; Zubko et al, 2004; Downey et al, 2006; Gatbonton et al, 2006; Tsolou and Lydall, 2007; Addinall et al, 2008; Ungar et al, 2009).
Yeast strains
All strains used in this study are RAD5+ and in the W303 genetic background (Strain Table, Supplementary data). Standard genetic procedures of transformation and tetrad analysis were used. New gene deletions were constructed by transforming a diploid with PCR-based deletion modules (Goldstein and McCusker, 1999). Point mutations were generated integrated into the genome as described (Schulz and Zakian, 1994; Ribeyre et al, 2009).
Yeast growth assays
Growth assays were performed as previously described (Zubko et al, 2004). Pooled colonies were inoculated into 2 ml of YEPD and grown to saturation at 23°C. Five-fold serial dilutions were replicated onto agar plates and grown at a range of temperatures. Plates were photographed using an SPimager (S&P Robotics). Levels were adjusted with Photoshop CS4.
Passage experiments and quantification of growth
To passage cultures, multiple individual colonies were pooled and restruck. Where growth was quantified, unmodified images were analysed with Colonyzer (Lawless et al, 2010) and the sum of the trimmed greyscale pixel values for all pixels corresponding to spots and colonies of each strain were used as a measure of growth. Growth was then expressed relative to a TLC1+ or TLC1+ CDC13+ strain included on the same plate.
Telomere length detection
Southern hybridization to examine telomere length and structure was performed similarly to previously described (Maringele and Lydall, 2004). Genomic DNA was extracted, digested with XhoI then run overnight on a 1% agarose gel at 1 V/cm. Southern transfer and detection was then performed using DIG-High Prime Labelling and Detection Kit (Roche) as per the manufacturer's instructions and visualized on a FUJI LAS4000. Telomeric probes were synthesized using PCR DIG Probe Synthesis Kit (Roche). TG probe was ∼180 bp of TG repeats, whereas Y′ probe was ∼820 bp of Y′ sequence, both amplified from pDL987 (pHT128) (Tsubouchi and Ogawa, 2000) using oligos m933 and m934 or m935 and m936, respectively. CDC15 probe was synthesized using oligos m1045 and m1046 as previously described (Foster et al, 2006).
Synchronous cultures, cell cycle scoring and QAOS
Experiments to measure cell cycle progression and ssDNA following telomere uncapping were carried out in bar1Δ cdc15-2 cdc13-1 cells and performed as described (Zubko et al, 2006). Where indicated, cells were treated with Bleomycin at a final concentration of 50 μg/ml (Morin et al, 2008).
Rad53 phosphorylation
Western blotting to detect Rad53 phosphorylation was performed essentially as described (Morin et al, 2008). Antibodies against Rad53 were from Dan Durocher, Toronto. Anti-tubulin antibodies were from Keith Gull, Oxford. Multiple gels were run to process all samples from a single experiment, but were transferred and detected in parallel and imaged simultaneously.
In-gel assay
In-gel assays were performed essentially as previously described (Zubko and Lydall, 2006) using a Cy5-labelled oligonucleotide (m2188) detected on a GE Healthcare Typhoon Trio imager. Following detection of ssDNA, the gel was subjected to Southern transfer and hybridization to detect CDC15. To quantify ssDNA, the fluorescent signal from the 0.7 to 12 kb range on the gel was measured, relative to the intensity of an exponentially dividing yku70Δ mutant on the same gel. All values were then normalized relative to the CDC15 signal as determined by Southern blot.
Supplementary Material
Acknowledgments
We gratefully acknowledge the members of our laboratory for input, particularly H Ngo and A Greenall and especially for A Leake's invaluable assistance. We thank our colleagues in the ONDEX project, particularly J Weile. We thank V Zakian, A Nicolas, H Tsubouchi, V Geli, D Durocher, A Bianchi and K Gull for providing strains, plasmids or antibodies. B Connolly, T Richardson and J Brown provided invaluable advice on fluorescent labelling of oligonucleotide probes and detection by phosphorimager. We especially thank L Harrington and S Makovets for critically reading the paper. This study was supported by BBSRC ONDEX (BB/F006039/1), MRC and Wellcome Trust (075294).
Footnotes
The authors declare that they have no conflict of interest.
References
- Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, Leake A, Hallinan J, Shaw O, James K, Wilkinson DJ, Wipat A, Durocher D, Lydall D (2008) A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180: 2251–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ (2004) A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW (2006) Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP (2010) Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet 6: e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP (2009) Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Booth C, Griffith E, Brady G, Lydall D (2001) Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res 29: 4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL (2006) Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol 26: 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T (2005) DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Luke B, Kraft C, Li Z, Peter M, Lingner J, Rothstein R (2009) Telomerase is essential to alleviate pif1-induced replication stress at telomeres. Genetics 183: 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G (2010) Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet 6: e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino-Reale G, Pascale E, Alvino E, Starace G, D′Ambrosio E (2003) Long telomeric C-rich 5′-tails in human replicating cells. J Biol Chem 278: 2136–2140 [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T (2009) How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, Emili A, Greenblatt JF, Harrington L, Lydall D, Durocher D (2006) A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168 [DOI] [PubMed] [Google Scholar]
- Foster SS, Zubko MK, Guillard S, Lydall D (2006) MRX protects telomeric DNA at uncapped telomeres of budding yeast cdc13-1 mutants. DNA Repair (Amst) 5: 840–851 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V (2007) RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A (2006) Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM (2009) Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res 37: 6491–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Huang KN, Symington LS (1993) A 5′-3′ exonuclease from Saccharomyces cerevisiae is required for in vitro recombination between linear DNA molecules with overlapping homology. Mol Cell Biol 13: 3125–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye A, Stahl H, Thines-Sempoux D, Foury F (1991) PIF1: a DNA helicase in yeast mitochondria. EMBO J 10: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, Wellinger RJ (2006) Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat Cell Biol 8: 741–747 [DOI] [PubMed] [Google Scholar]
- Lawless C, Wilkinson DJ, Young A, Addinall SG, Lydall DA (2010) Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics 11: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- Lydall D (2009) Taming the tiger by the tail: modulation of DNA damage responses by telomeres. EMBO J 28: 2174–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Weinert T (1995) Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270: 1488–1491 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Makovets S, Blackburn EH (2009) DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol 11: 1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I (2008) Multiple pathways inhibit NHEJ at telomeres. Genes Dev 22: 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2004) EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 166: 1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Zakian VA (2006) Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle 5: 2796–2804 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D (2008) Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 27: 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo HP, Lydall D (2010) Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9. PLoS Genet 6: e1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE (2001) The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Pike JE, Burgers PM, Campbell JL, Bambara RA (2009) Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem 284: 25170–25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt CW, Cooper JP, Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res (advance online publication 3 July 2010; doi:10.1093/nar/gkq580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol 8: 831–834 [DOI] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J (2008) C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A (2009) The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA (1993) Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75: 729–739 [DOI] [PubMed] [Google Scholar]
- Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, Schlegelberger B, Schirmacher P, Kunkel TA, Greenberg RA, Edelmann W, Rudolph KL (2007) Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell 130: 863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA (1994) The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155 [DOI] [PubMed] [Google Scholar]
- Segurado M, Diffley JF (2008) Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L (2007) Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol 27: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34 (Database issue): D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA (1999) Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19: 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolou A, Lydall D (2007) Mrc1 protects uncapped budding yeast telomeres from exonuclease EXO1. DNA Repair (Amst) 6: 1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H (2000) Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar L, Yosef N, Sela Y, Sharan R, Ruppin E, Kupiec M (2009) A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Res 37: 3840–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E, Foury F, Stillman B, Brill SJ (1992) A single-stranded DNA binding protein required for mitochondrial DNA replication in S cerevisiae is homologous to E. coli SSB. EMBO J 11: 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385 [DOI] [PubMed] [Google Scholar]
- Vega LR, Phillips JA, Thornton BR, Benanti JA, Onigbanjo MT, Toczyski DP, Zakian VA (2007) Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet 3: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenicharov MD, Laterreur N, Wellinger RJ (2010) Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J 29: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenicharov MD, Wellinger RJ (2006) DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H Jr, Kneitz B, Yang G, Kunkel TA, Kolodner RD, Cohen PE, Edelmann W (2003) Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev 17: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Zhang W, Durocher D (2010) De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev 24: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA (2000) Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289: 771–774 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Guillard S, Lydall D (2004) Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 168: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Lydall D (2006) Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat Cell Biol 8: 734–740 [DOI] [PubMed] [Google Scholar]
- Zubko MK, Maringele L, Foster SS, Lydall D (2006) Detecting repair intermediates in vivo: effects of DNA damage response genes on single-stranded DNA accumulation at uncapped telomeres in budding yeast. Methods Enzymol 409: 285–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.