Abstract
Imprinted genes are expressed monoallelically because one of the two copies is silenced epigentically in a parent-of-origin pattern. This pattern of expression is controlled by differential marking of parental alleles by DNA methylation and chromatin modifications, including both suppressive and permissive histone acetylation and methylation. Suppressive histone modifications mark silenced alleles of imprinted genes, while permissive histone modifications mark the active alleles, suggesting the possibility that imprinted genes would show upregulation in gene expression. However, it is currently unknown whether imprinted genes show such upregulation. To address this question in mice, we estimated the intensity of expression of 59 genes relative to the rest of the genome by analyzing microarray data. Expression levels of 24 genes were validated using quantitative real-time PCR (qPCR). Expression of imprinted genes was found to be upreguled in various adult and embryonic mouse tissues. Consistent with their functions in growth and development, imprinted genes were found to be highly expressed in extraembryonic tissues and progressively upregulated during early embryonic development. In conclusion, upregulation of imprinted genes found in this study is similar to the dosage compensation (twofold upregulation) recently reported for X-linked genes. It has been proposed that the twofold upregulation of X-linked genes has been coupled with low transcriptional variation (noise) which could lead to deleterious effects on the organism. Results of this study suggest a general need for imprinted genes in the mouse to be upregulated to certain levels in order to avoid deleterious effects of variation in gene expression.
Keywords: imprinting, expression, dosage compensation, upregulation, mouse
Introduction
Genomic imprinting is a parent-of-origin-specific monoallelic expression of a subset of genes in placental mammals; some genes are maternally expressed while others are paternally expressed. A growing list of approximately 80 imprinted genes—belonging to gene families with different biological functions—have been identified in humans and mice.1 Manipulation of copy number of murine imprinted genes has provided compelling evidence that these genes control growth and development in a dose-dependent manner and that their aberrant expression is associated with defects in development, growth and behavior.2-4 Similarly, disrupting the balanced expression of human imprinted genes results in a broad spectrum of diseases such as Angelman, Prader-Willi and Beckwith-Wiedeman syndromes, and cancers (reviewed in refs. 5–7). For example, overexpression of the insulin-like growth factor 2 (IGF2) gene and reduced expression of H19 have been reported to be associated with Wilms’ tumor, adrenocortical carcinoma, and rhabdomyosarcoma.8 Paternally expressed gene 10 (PEG10) has been reported to be highly expressed in B-cell chronic leukemia patients.9 Also, delta-like 1 homolog (Drosophila) (DLK1) has been reported to be upregulated in human hepatocellular carcinoma cell lines implying a possible role of this gene in the oncogenesis of this tumor.10
X-linked genes are also monoallelically expressed and epigenetically controlled in placental mammals. Furthermore, several structural and functional characteristics are common between imprinted and X-linked genes, which have led to the suggestion that they are evolutionarily linked.11 X-linked gene expression is dose-dependent, and disruption of normal expression is associated with different human diseases including cancers.12,13 For example, skewed X chromosome inactivation has been shown to have significant associations with both breast and ovarian cancers in humans.14
The two alleles of imprinted genes and those of X-linked genes are differentially marked by DNA methylation and histone (H) modifications.11,15,16 Interestingly, imprinted genes show not only repressive histone modifications, but active histone modifications as well.16,17 For instance, it has been demonstrated that trimethylation at histone H3 lysine 4 (H3K4me3; active histone mark) and trimethylation at histone H3 lysine 9 (H3K9me3; repressive histone mark) overlap at imprinting control regions and secondary differentially methylated regions of imprinted genes.16 Furthermore, imprinted genes show allele-specific histone modification patterns; inactive alleles are bound by suppressive modifications, including H3K9me3, while expressed alleles are associated with active histone modifications, including H3K4me2, H3K4me3, acetylated histone H3 (H3ac) and H4ac.17 Similar to imprinted genes, monoallelically-expressed X-linked genes also show active histone modifications. For example, using chromatin immunoprecipitation (ChIP) to quantify H3K4me2 status (active histone mark) of X-linked and autosomal genes, Rougeulle et al.18 reported that X-linked genes and imprinted genes are distinguished from most autosomal genes by their promoters’ enrichment of H3K4me2.
It has been demonstrated that suppressive histone modifications are involved in allelic silencing of imprinted genes.19,20 However, it is currently unknown whether imprinted genes are normally upregulated. Nevertheless, consistent with this notion, recent studies have demonstrated that the expression of X-linked genes relative to that of autosomal genes (biallelic expression), in both male and female, is globally upregulated at least twofold in various cells and tissues of placental mammals, including humans and mice.21-23
Taken together, it is appealing to hypothesize that—similar to X-linked genes—imprinted genes show global upregulation relative to the rest of the genome. To test this hypothesis, we analyzed the intensity of expression of imprinted genes relative to the average level of gene expression of the rest of the genome in embryonic and adult mouse tissues. Microarray data of expression of imprinted and nonimprinted genes were retrieved and analyzed, then qPCR was performed to confirm microarray results. Imprinted genes were considered upregulated if their expression relative to control genes was higher than 0.5 (equivalent to -1 on the log base 2 scale (log2)). In this study, we report that imprinted genes show global upregulation of their expression in various adult and embryonic mouse tissues.
Results and Discussion
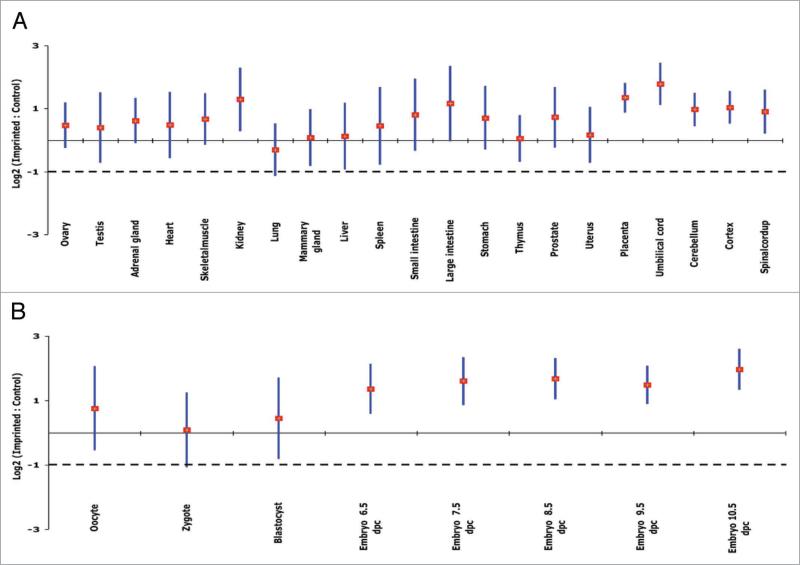
The expression profiles of 36,182 transcripts for 29 adult and embryonic mouse tissues were obtained from a publicly available microarray data set.24 This microarray data set represents the global gene expression of mixed samples (four males and three females) from different adult (10-to-12-week old) C57BL/6 mouse tissues. A total of 58 different Affymetrix slides were analyzed in this study. The log2 transformed data followed a normal distribution. Out of the 80 known imprinted genes,1 the intensity of expression of 59 (~73%) genes was investigated in this experiment. The mean expression ratios of imprinted/nonimprinted genes indicated upregulation of imprinted genes in most adult mouse tissues with remarkable upregulation in kidney, placenta and umbilical cord (Fig. 1A). The overall average of imprinted/nonimprinted gene expression ratios was log2 = 0.63, with various tissues exceeding the null hypothesis of log2 = -1 (≈linear 0.5), indicating that expression of imprinted genes is upregulated at least twofold (Fig. 1A).
Figure 1.
Upregulation of imprinted genes in mouse tissues, based on microarray results.24 For the indicated tissue, each vertical bar represents the confidence interval (95%) of imprinted/control ratios (imprinted genes, n = 59; control genes, all nonimprinted genes); the diamond marks the mean. Gene expression levels are on the log2 scale. The solid and broken lines show the expected values for two-fold upregulation and for no upregulation, respectively. (A) Adult tissues. Imprinted genes are upregulated significantly relative to all nonimprinted genes, with means at least two times higher than the rest of the genome across various tissue types except for lung. (B) Embryonic tissues at specific developmental stages. Imprinted genes are upregulated significantly throughout early developmental stages except for the fertilized oocyte, with means at least two times higher than the nonimprinted genome.
While some genes are imprinted across different tissue types,25 others are imprinted in a tissue-specific as well as a development-specific manner.26 To investigate the variation of expression among different tissue types at the same developmental stage, we performed a student's t-test on the microarray data set. No significant differences were found among tissues, in spite of apparent tissue-to-tissue expression variation (Fig. 1A). To study the expression of imprinted genes throughout fertilization and early embryonic development, we analyzed the expression level of these genes before fertilization and through day 10.5 of embryonic development using microarray data. After fertilization, imprinted genes show progressive upregulation until they reach a plateau at day 8.5 of development (Fig. 1B). It is known that embryonic development and growth in placental mammals is controlled, in part, by imprinted genes.2,3 In addition, accelerated growth in placental mammals takes place at very early developmental stages,27 and then slows down postnatally.28 Using microarray and real-time PCR approaches29,30 demonstrated that expression of many imprinted genes that regulate fetal growth show postnatal downregulation in different mouse organs. In contrast, our finding suggests that progressive upregulation of imprinted genes could be explained by the involvement of these genes in embryonic growth at very early stages of fetal development.
Microarray data were also used to choose internal control genes for validation of upregulation results in the qPCR experiments. The use of suitable internal control genes is a key factor in reliable estimates of relative gene expression in qPCR experiments.31 Our approach was to select internal control genes that represent the average global expression of all nonimprinted genes across various tissue types. After ranking all genes in the microarray data set based on their expression values (see Methods), a total of seven genes were selected because their expression levels were close to the average of all nonimprinted genes. After analysis of the seven control genes using qPCR, we chose AU042671 and Cry2 genes as internal controls because their expression across various tissues was the most consistent compared to all other genes.
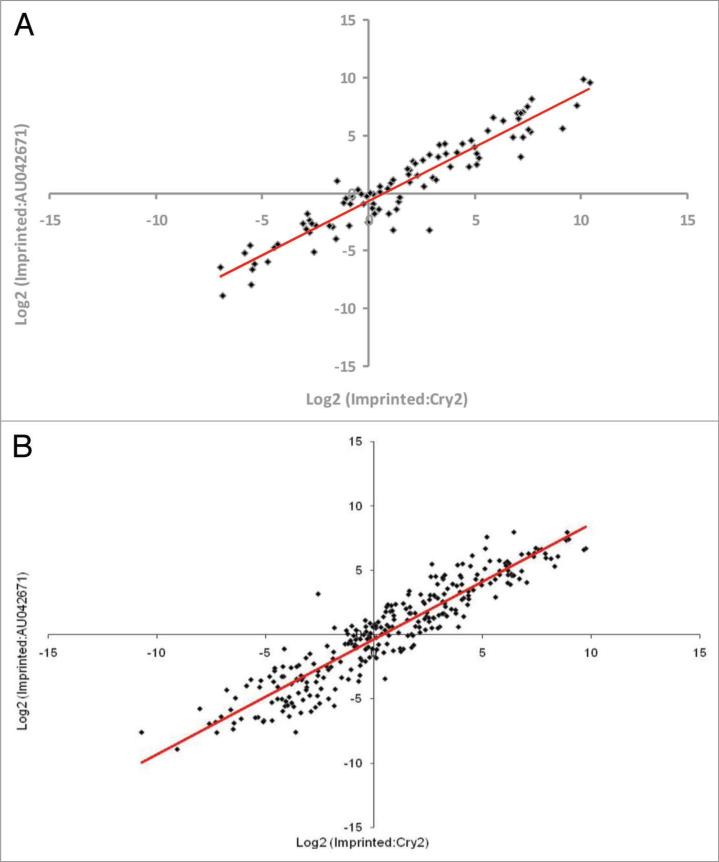
To confirm the microarray results and to further investigate the upregulation of imprinted genes during embryonic development, the expression of 24 imprinted genes in various tissue types obtained from two developmental stages was analyzed using qPCR. Whole embryo, yolk sac, umbilical cord and placenta were collected from 10.5-dpc conceptuses. Brain, liver, heart, umbilical cord, trophectoderm and gonads were collected from 14.5-dpc conceptuses. RNA was extracted from each tissue and the expression of the 24 imprinted and the two control genes was analyzed by qPCR. The imprinted/AU042671 expression ratios were plotted against the imprinted/Cry2 expression ratios for all examined tissues. For tissues of both 10.5 dpc (Fig. 2A) and 14.5 dpc (Fig. 2B), there was a clear linear relationship between the two internal control genes with correlations (r) of 0.946 for 10.5-dpc conceptuses and 0.935 for 14.5-dpc conceptuses. Consequently, the calculated values relative to AU042671 and to Cry2, were averaged for each imprinted gene in the successive experiments. Expression values of the 24 imprinted genes relative to the two control genes (AU042671 and Cry2) in 10.5-dpc and 14.5-dpc mouse conceptuses, using qPCR, are shown in Tables 1 and 2, and Supplementary Table 1. Expression ratios of ~70% of the examined imprinted genes were above the -1.0 value (loge2 scale; 0.5 linear scale) that would be expected if the imprinted gene does not show upregulation relative to the control genes. With the exception of Igf2, which was highly expressed in 10.5-dpc conceptuses, expression ratios were scattered over a range of approximately 270 folds across the investigated tissues of the two developmental stages. Expression levels of X-linked genes relative to that of autosomal genes (X/A ratio; log2 scale) have been reported to vary over a large fold range (200 folds) in mouse embryonic stem cells as well, of which X-linked genes show dosage compensation.32
Figure 2.
Scatter graph of log2 (imprinted:AU042671) ratios versus log2 (imprinted:Cry2) ratios from mouse conceptuses, based on quantitative real-time PCR. Each data point represents the expression of one single imprinted gene (n = 24) relative to two internal control genes in a specific tissue type. Gene expression levels are on the log2 scale. (A) Relative expression of imprinted genes in four different tissue types from 10.5-dpc conceptuses (n = 8) are shown. The distribution of the data show a clear linear relationship between the two internal control genes with high correlation (r = 0.946). (B) Relative expression of imprinted genes in six different tissue types from each sex (n = 2) of 14.5-dpc conceptuses (n = 8). The distribution of the data show a clear linear relationship between the two internal control genes with high correlation (r = 0.934).
Table 1.
Expression of 24 imprinted genes relative to the two control genes AU042671 and Cry2 in 10.5-dpc mouse conceptuses, based on quantitative real-time PCR results
| Gene | Expressed allele | Embryo | Placenta | Umbilical cord | Yolk sac |
|---|---|---|---|---|---|
| Asb4 | Maternal | 1.978 | 1.445 | 4.357 | -0.047 |
| Cd81 | Maternal | -4.433 | -3.104 | -2.858 | -2.767 |
| Cdkn1c | Maternal | 4.488 | 7.357 | 3.783 | 5.177 |
| Dcn | Maternal | -2.538 | 0.977 | -0.738 | -0.462 |
| Gtl2 | Maternal | 1.964 | 1.932 | 3.537 | 3.533 |
| H19 | Maternal | -2.327 | -0.332 | 0.072 | -0.725 |
| Igf2r | Maternal | 3.731 | 3.817 | 5.525 | 4.132 |
| Mash2 | Maternal | -5.523 | 4.715 | -2.285 | -3.825 |
| Osbpl5 | Maternal | -0.163 | -0.184 | -0.542 | -1.013 |
| Ppp1r9a | Maternal | -5.013 | -6.037 | -4.562 | -7.872 |
| Tssc3 | Maternal | -2.653 | 6.317 | -0.208 | 5.828 |
| Zim1 | Maternal | 0.323 | 1.142 | 2.719 | -1.882 |
| Air | Paternal | -0.059 | -0.593 | 3.205 | 0.352 |
| Dio3 | Paternal | -6.685 | 1.780 | -5.320 | -1.300 |
| Igf2 | Paternal | 6.464 | 9.983 | 9.988 | 8.668 |
| Impact | Paternal | -2.655 | 0.042 | -0.905 | 1.628 |
| Nap1l5 | Paternal | -0.992 | -5.753 | -4.362 | -6.707 |
| Ndn | Paternal | 3.113 | -3.020 | 2.422 | 0.570 |
| Nnat | Paternal | 0.578 | -2.327 | -0.185 | -0.403 |
| Peg1 | Paternal | 7.495 | 7.006 | 6.947 | 6.532 |
| Peg10 | Paternal | -0.567 | 7.035 | 3.949 | 4.245 |
| Peg3 | Paternal | 4.747 | 6.746 | 7.918 | 6.077 |
| Slc38a4 | Paternal | 3.078 | 7.128 | 6.206 | 2.200 |
| Zac1 | Paternal | 0.648 | 2.382 | NA | 2.182 |
NA, not available.
Table 2.
Expression of 24 imprinted genes relative to two control genes AU042671 and Cry2 in 14.5-dpc male mouse conceptuses, based on quantitative real-time PCR results
| Gene | Expressed allele | Brain | Gonads | Heart | Liver | Trophectoderm | Umbilical cord |
|---|---|---|---|---|---|---|---|
| Asb4 | Maternal | -0.137 | 1.824 | 0.879 | -0.170 | -1.512 | 2.252 |
| Cd81 | Maternal | -4.581 | -2.167 | 0.002 | -3.703 | -3.322 | -1.280 |
| Cdkn1c | Maternal | 1.147 | 1.201 | 3.755 | 1.377 | 4.238 | 2.421 |
| Dcn | Maternal | 1.286 | 2.250 | 0.425 | 1.683 | 7.097 | 5.228 |
| Gtl2 | Maternal | -0.858 | 0.315 | -0.215 | 0.915 | -0.232 | 2.892 |
| H19 | Maternal | -7.047 | -4.827 | -3.013 | -4.390 | -3.593 | -0.280 |
| Igf2r | Maternal | 1.165 | 2.375 | 5.891 | 2.514 | 3.812 | 5.540 |
| Mash2 | Maternal | -6.220 | -0.910 | -6.662 | -5.540 | 1.918 | -2.879 |
| Osbpl5 | Maternal | -0.502 | 1.887 | 0.743 | -3.695 | 0.428 | 0.727 |
| Ppp1r9a | Maternal | -3.334 | -5.880 | -5.938 | -6.892 | -3.498 | -4.719 |
| Tssc3 | Maternal | -7.257 | NA | -4.739 | 0.358 | 2.879 | -2.519 |
| Zim1 | Maternal | -0.715 | 2.842 | 2.773 | 1.532 | 4.410 | 5.418 |
| Air | Paternal | -4.248 | 0.121 | 0.512 | -2.217 | -0.582 | 1.788 |
| Dio3 | Paternal | -5.653 | NA | NA | -9.148 | -1.877 | -6.918 |
| Igf2 | Paternal | 1.375 | 5.585 | 5.727 | 4.082 | 4.720 | 8.438 |
| Impact | Paternal | -2.920 | NA | 0.372 | -3.585 | -0.854 | 1.278 |
| Nap1l5 | Paternal | -1.516 | -3.419 | -1.460 | -2.618 | -3.290 | -3.020 |
| Ndn | Paternal | 4.638 | 3.572 | 2.739 | -0.642 | 1.682 | 4.499 |
| Nnat | Paternal | 4.292 | -0.852 | -4.643 | -3.193 | -3.170 | 0.678 |
| Peg1 | Paternal | 5.917 | 6.715 | 8.233 | 5.342 | 7.100 | 7.214 |
| Peg10 | Paternal | -0.612 | -2.323 | -5.873 | -0.912 | 5.525 | 1.782 |
| Peg3 | Paternal | 3.785 | 6.821 | 5.533 | 3.568 | 6.872 | 8.198 |
| Slc38a4 | Paternal | -0.483 | 4.026 | 5.814 | 3.815 | 8.103 | 5.378 |
| Zac1 | Paternal | -0.613 | -0.983 | 2.082 | -0.575 | 1.923 | 2.845 |
NA, not available.
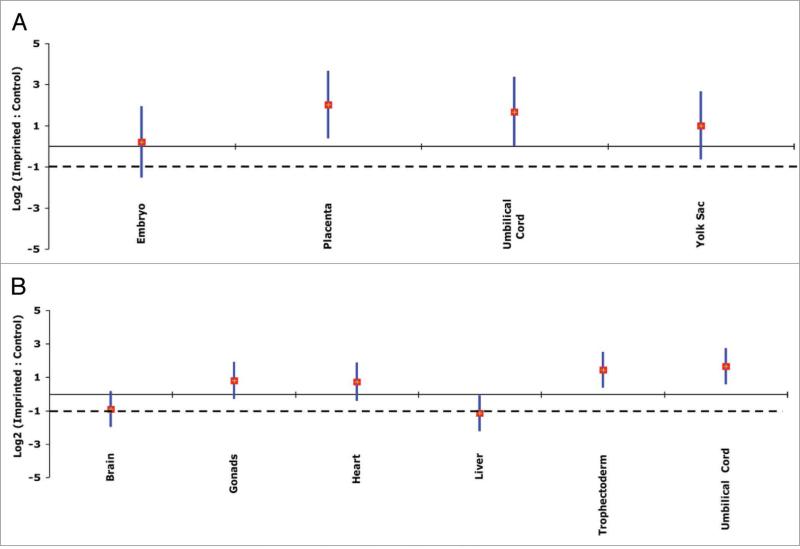
The qPCR analysis revealed that imprinted genes showed overall upregulation in whole embryo, placenta, umbilical cord and yolk sac tissues of 10.5-dpc conceptuses (Fig. 3A; Table 1) and in gonads, heart, trophectoderm and umbilical cord of 14.5-dpc conceptuses (Fig. 3B; Table 2; Suppl. Table 1). In contrast, overall upregulation was not as prominent in brain or in liver tissues at 14.5 dpc, although expression levels of the majority of the imprinted genes in these tissues were above the -1.0 value (loge2 scale; 0.5 linear scale) (Fig. 3B; Table 2; Suppl. Table 1). Proliferation rate and maturity time are different for different organs.33 Also, genomic imprinting is involved in coordinating embryonic growth in order to maintain body proportions.3 Collectively, the need for expression of imprinted genes should be different in different organs, which is consistent with our results. For example, the high expression of imprinted genes observed in extraembryonic tissues could be due to the need to maintain a healthy and functional placenta during fetal growth and development. Thus, the qPCR results confirmed those of the microarray. There were no statistically significant differences of imprinted/control expression ratios between males and females.
Figure 3.
Upregulation of imprinted genes in mouse conceptuses, based on quantitative real-time PCR results. Each vertical bar represents a confidence interval (95%) of imprinted/control ratios (imprinted n = 24; control = AU042671 and Cry2: these control genes were chosen to represent the average gene expression of all nonimprinted genes in mouse); the diamonds mark the means for the indicated tissue. Gene expression levels are on the log2 scale. The solid and broken lines show the expected values for two-fold upregulation and for no upregulation, respectively. (A) 10.5-dpc mouse conceptuses. Results of four different tissue types are shown (cDNA of each tissue was pooled from eight individuals). Imprinted genes are upregulated significantly relative to control genes in various tissue types except for whole embryo. (B) 14.5-dpc conceptuses. Results from six different tissue types from eight conceptuses are shown. Imprinted genes are upregulated significantly relative to control genes, with their means at least two times higher than the rest of the genome in various tissue types except for whole brain and liver tissues.
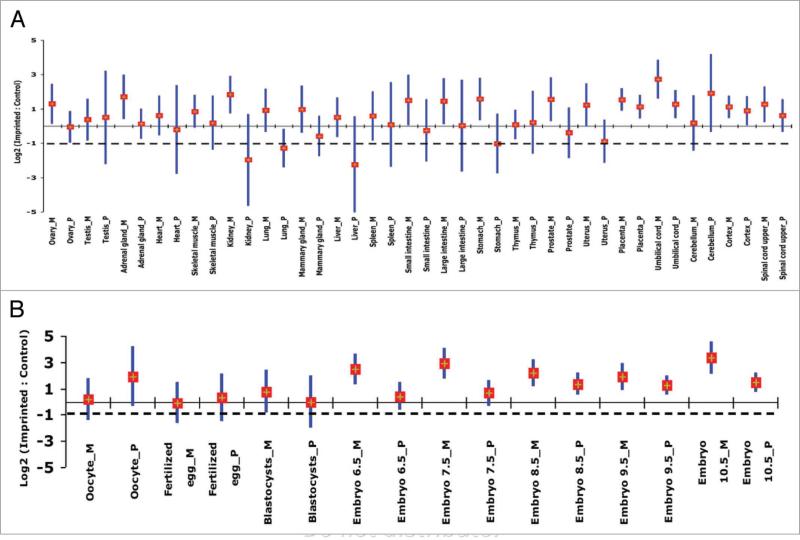
Maternally- and paternally-expressed genes could function differently in embryonic and extraembryonic tissues.34-36 For example, as demonstrated by chimeric studies, the paternal genome is more involved in the development of the hypothalamus, while the maternal genome is more involved in the development of the neocortex.36,37 Moreover, single gene studies have shown that genomic imprinting in the brain is spatially and developmentally regulated. For example, the Ube3a gene was found to be maternally expressed in Purkinje cells, hippocampal neurons, and the mitral cells of the olfactory bulb in mouse, while in other brain tissues it showed biallelic expression.38 To investigate whether or not upregulation is influenced by the parent-of-origin of imprinted genes, the expression of maternally- and paternally-expressed genes relative to control genes was analyzed separately, using microarray and qPCR data. Expression levels showed that both maternally- and paternally-expressed genes were upregulated in the majority of adult and embryonic tissues at different developmental stages (Fig. 4A and B; Tables 1 and 2; Suppl. Table 1). However, the average expression levels of maternally and paternally expressed genes tend to vary in some tissues. For instance, the mean expression of paternally-expressed genes was found to be high in the cerebellum of adults, microarray data, (Fig. 4A) and in the whole brain at 14.5 dpc, qPCR data (Table 2; Suppl. Table 1), when compared to maternally expressed-genes. Furthermore, the average of the paternally-expressed genes in whole brain of embryonic 14.5 dpc embryos showed at least twofold upregulation, while the average of the maternally-expressed genes showed at least twofold down-regulation. This differential regulation of paternally- and maternally-expressed genes could be ascribed to differences in silencing-related DNA sequence features. Indeed, previous studies have shown that paternally- and maternally-expressed genes are characterized by different densities of sequence characteristics. For example, CpG content has been found to be different between paternally- and maternally-derived differentially methylated regions.39,40 In addition, it has been reported that short interspersed transposable elements are less abundant in paternally-expressed compared to maternally-expressed genes.41 Thus, the differential regulation of paternally- and maternally-expressed genes could be attributed to some of these sequence features. However, the question of what molecular mechanisms are responsible for such differential regulation requires further investigation.
Figure 4.
Upregulation of maternally-(M) and paternally-expressed (P) genes in adult and embryonic mouse tissues based on microarray results. Each vertical bar represents the confidence interval (95%) of imprinted/control ratios; a diamond marks the mean for the indicated tissue. Gene expression levels are on the log2 scale. The solid and broken lines show the expected values for two-fold upregulation and for no upregulation, respectively. (A) Adult (10–12 weeks old) mouse tissues and two extraembryonic tissues. (B) Embryonic tissues at specific developmental stages. Although not significantly different, a trend in parent-of-origin effect in expression of imprinted genes is clear. A relative expression of maternally-expressed genes was higher in various adult tissues and in embryos of various ages except in the cerebellum in adults.
Genomic imprinting and X chromosome inactivation are two epigenetic phenomena that share many common features in placental mammals. Differential DNA methylation, cis-acting control centers, and involvement of both suppressive and permissive histone modifications, and their chromatin-associated factors are all characteristics of both X-chromosome inactivation and genomic imprinting (reviewed in ref. 11). Furthermore, the X chromosome in XX females is paternally inactivated/imprinted and maternally expressed in extra-embryonic tissues in eutherians (i.e., mice and cows)42,43 and across tissues in marsupials.44,45 Recent work has shown that the active alleles of X-linked genes are upregulated on average twofold relative to the rest of the genome in various tissues in placental mammals, such as the mice and humans,21,22,46 and up to 1.6-fold in the undifferentiated female and male mouse embryonic stem cells.32 We find that the global upregulation of imprinted genes relative to the rest of the genome across different tissue types parallels that of X-linked genes. Thus, our findings add another element to the common features between X-linked and imprinted genes; both are upregulated compared to the rest of the genome.
Why is upregulation of expression of imprinted genes important to cells in placental mammals? Noise or randomness is defined as variability in gene expression in genetically identical cells or organisms under similar environmental conditions which could result in remarkable phenotypic variability and even in lethality.47-50 It has been proposed that in order to avoid lethality caused by transcriptional noise, essential genes50,51 and haploinsufficient genes52 should show low noise. Indeed, a recent study by Yin and colleagues53 showed that X-linked genes—upregulated twofold because of dosage compensation—and biallelically-expressed genes show less transcriptional noise than non-imprinted monoallelically-expressed genes in humans and mice. If imprinted genes, which are essential by definition, are vulnerable to transcriptional noise, we would expect that either downregulation or upregulation of these genes would be deleterious. Consistent with this notion, lethality has been documented in both knockouts and excess of expression of the imprinted genes Peg11 and Dlk1.3,54 Likewise, disrupting normal expression of Cdkn1c either by knockouts or overexpression in mice was found to be lethal.2,55 Also, it has been shown that down-regulation of CDKN1C in human is associated with pancreatic cancers.56
It has been shown that transcriptional noise and expression level are negatively correlated in eukaryotes.50,51,57 That is, genes expressed at high levels show less transcriptional noise. Yin et al.53 found that autosomal monoallelically-expressed genes—expressed at low levels compared to biallelically-expressed genes—show three fold higher transcriptional noise than do biallelically-expressed genes. Thus, because of their monoallelic nature, we propose that upregulation of imprinted genes has evolved to buffer any deleterious effects that could result from high transcriptional noise. In other words, selection worked to minimize transcriptional noise by upregulating the expression of imprinted genes as suggested for X-linked genes.53 It would be of interest to confirm the hypothesis that imprinted genes display similar noise levels to that shown by both X-linked genes and biallelically-expressed genes in mice in future studies.
In summary, imprinted genes examined in various adult and embryonic mouse tissues showed global upregulation in a broad spectrum of tissue types. Upregulation of imprinted genes observed in microarray data analysis was confirmed in qPCR experiments. Expression of imprinted genes was found to be highly upregulated in extraembryonic tissues. Also, progressive upregulation was observed during embryonic development, independent of sex. These results are consistent with the known role of imprinted genes in development and growth of embryonic and extraembryonic tissues.33,34,58,59 Maternally- and paternally-expressed genes, however, showed different patterns of upregulation in various tissues. Although not statistically significant, the maternally-expressed genes were found to be upregulated in extraembryonic tissues and downregulated in brain tissues.
Materials and Methods
Microarray data
Microarray data were obtained from a public array database (mouse custom-made 25-oligomer array). This array included 36,182 transcripts and was originally designed to cover all known protein-coding transcripts in the gene atlas of mouse protein-coding transcriptomes.24 The data can be downloaded from (http://symatlas.gnf.org) and from the Gene Expression Omnibus (www.ncbi.nih.gov/geo) entry GNF1M. Expression data for 29 adult and embryonic mouse tissues were used in this study. The tissues analyzed from this microarray were adrenal gland, heart, kidney, lung, mammary gland, placenta, skeletal muscle, thymus, liver, large intestine, small intestine, stomach, spleen, cerebellum, cortex, spinal cord, prostate, testis, uterus, umbilical cord, ovary, oocyte, fertilized egg, blastocyst and embryos of 6.5, 7.5, 8.5, 9.5, 10.5 dpc. Adult mouse tissue samples were collected from 4 males and 3 females of C57BL/6 mice at ages of 10 to 12 weeks. The original attempt was to analyze all known imprinted genes that follow specific criteria: (i) the gene must not be a microRNA to avoid overlap with other genes; (ii) the gene has to be annotated with a recognizable name in the microarray files; and (iii) monoallelic parent-of-origin gene expression must be determined based on convincing published data. For that sake, we collected our information about the imprinted genes from two available resources including the “Catalogue of Imprinted Genes”1 and the “WAMIDEX: a Web Atlas of Murine Genomic Imprinting and Differential Expression”,60 in addition to at least two original publications per gene. Thus, 59 imprinted genes were chosen for the analysis of the microarray data. Monoallelic expression across tissues was assumed for genes not known to be tissue-specific imprinted genes.
Microarray data analysis
Perfect Match (PM) and Mismatch (MM) signals of each probe/gene were combined into an expression index using the MAS5.0 algorithm, together with the Presence/Absence calls of Affymetrix. For the purpose of our analysis, the expression intensities were log (base 2) transformed, and only genes with at least one presence call were kept for further analysis. Genes with either absent or marginal calls across the two replications and all tissues were discarded. Descriptive statistics and visual inspection were first used as an exploratory analysis of the data. Data from the two replications of each tissue were averaged for each gene, and mean, median, standard deviation, and percentiles were calculated for each category of genes (imprinted = monoallelically-expressed genes and nonimprinted genes across tissues). The results were summarized using box-plots and histograms (data not shown). Average expression differences between imprinted (monoallelically-expressed) and nonimprinted genes within each tissue were compared using a t-test for two independent groups with heterogeneous variances, and a significance level of 5%. The comparison of the relative expression imprinted:nonimprinted genes across tissues was performed by constructing 95% confidence intervals for each tissue. Non-overlapping intervals were considered statistically different at α = 0.05. The analyses were performed using the procedures PROC TTEST and PROC CAPABILITY of SAS v9.1 (SAS Institute, Inc., Cary, NC).
Quantitative real-time PCR
Mouse tissues and RNA preparation
All animals were maintained and used under protocols approved by the Institutional Animal Care and Use Committee of the University of Wisconsin-Madison. F2 conceptuses of 10.5 dpc and 14.5 dpc (where 0.5 dpc is the day of conception) were derived from matings between (C57Bl/6 × CBA) F1 mice (Jackson Laboratory, Bar Harbor, ME). Conceptuses were divested of decidual association, the parietal yolk sac was reflected and whole embryo, visceral yolk sac, umbilical cord, and trophectoderm derivatives (which included some reflected parietal endoderm) were harvested from 10.5-dpc conceptuses. Brain, liver, heart, gonads, umbilical cord and chorionic disk (which included the umbilical vessels of the labyrinth and a minor contribution of reflected parietal endoderm) were harvested from 14.5-dpc conceptuses. Tissues were kept in RNAlater reagent (Qiagen, Valencia, CA) at -20°C until RNA extraction. Only embryos of 14.5 dpc could be sexed by gross gonadal morphology in the microscope. Two litters from each developmental stage were collected. Total RNA was isolated using the RNAqueous-Micro kit (Ambion, Austin, TX) for all specimens except trophectoderm and whole embryo for which RNA was extracted using the RNeasy micro kit (Qiagen), in accordance with standard protocols. To eliminate DNA contamination, the eluted RNA was treated with Ambion RNase-free DNase I according to the instructions of the manufacturer, except for the incubation time, which was extended to 1 hour. First-strand cDNA was synthesized in accordance with the manufacturer's instructions using iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). The cDNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). A pilot experiment was conducted to investigate the validity of cDNA pooling for relative gene expression purposes. The average C(t) value from the individual specimens was similar to that obtained from their pool (data not shown). Tissue-specific pools of cDNA were prepared from the available tissues of 10.5 and 14.5 dpc mouse conceptuses. For 10.5-dpc conceptuses, tissue-specific-cDNA pools of eight specimens (samples of the same tissue type) were constructed for whole embryo, yolk sac, umbilical cord and trophectoderm. For male and female 14.5 dpc conceptuses, sex-and-tissue-specific-cDNA pools of four specimens (samples of the same tissue type) were constructed for brain, liver, heart, umbilical cord, placenta and gonads. The same amount of cDNA was taken from each sample used in constructing different pools. For the purpose of qPCR analysis, imprinted genes were selected based on the following criteria: (1) genes should be imprinted in mice, (2) genes are from different chromosomal locations/clusters, (3) paternally- and maternally-expressed genes are represented equally, (4) both protein-coding and noncoding imprinted transcripts are represented in the study, and (5) genes must be expressed in both 10.5 dpc and 14.5 dpc conceptuses. Thus, a total of 24 imprinted genes were selected for further analyses: 12 maternally-expressed genes, namely Asb4, Cd81, cDKn1c, Dcn, Gtl2, H19, Igf2r, Mash2, Osbpl5, Ppp1r9a, Tssc3 and Zim1, and 12 paternally-expressed genes, namely Air, Dio3, Igf2, Impact, Nap1l5, Ndn, Nnat, Peg1, Peg10, Peg3, Slc38A4 and Zac1. In addition, two control genes (AU042671 and Cry2) were chosen based on microarray data analysis (mentioned herein). Primers for Air, Ndn, Nnat and Peg1,61 Dcn, Tssc3, Impact and Zac1,62 were used as described in the original references. All other primers were designed using the Beacon Designer software (Premier Biosoft International, Palo Alto, CA) (Suppl. Table 2). Quantitative real-time PCR (qPCR) was run on a DNA Engine-Opticon 2 Detection System (MJ Research, Watertown, MA) using SYBR Green chemistry, specifically the iQ SYBR Green Supermix kit (Bio-Rad). A total of 70 ng of cDNA pools served as a template for qPCR. A calibrator pool was constructed from different tissue pools and was run in all qPCRs. The cycling conditions were as the followings: 95°C for 10 min and 40 cycles of 94°C for 30 s, 57°C for 30 s and 72°C for 45 s, followed by 72°C for 10 min. Gene expression of imprinted and control genes was normalized to Gapdh levels. Ratios of gene expression of each imprinted gene relative to both AU042671 and Cry2 levels were done separately, and quantification was calculated using the ΔΔ Ct method.63 All reactions were run in triplicate. Finally, the log2 ratios of each imprinted gene relative to the two control genes were averaged.
Choosing internal control genes and qPCR data analyses
The microarray data discussed above was used also to select two control genes to be used for the RT-PCR assays. The strategy was to use a data-based criterion to rank genes according to how stable (or constant) and close to the average their expression levels were across tissues. A score was calculated for each gene, using the expression:
where Sg is the score relative to gene g, ygt is the expression intensity of gene g in tissue t (t = 1, 2,..., T), T is the total number of tissues, and ȳ is the overall (across genes and across tissues) mean expression index. The score defined above penalizes genes that present expression levels systematically below or above the overall mean ȳ, as well as genes with variable expression. A subset of seven genes (Epha6, Sox5, Cdkn1b, Dll4, Mkks, AU042671 and Cry2) representing those with the most constant expression across tissues and close to the overall mean were selected for further analysis. Another round of filtering was conducted using qPCR to test the expression consistency of the selected seven genes across all tissue types of 10.5-dpc and 14.5-dpc conceptuses. The most consistent two genes (AU042671 and Cry2) were chosen to serve as internal controls to represent the average of global gene expression of all nonimprinted genes in the mouse, in order to investigate upregulation of imprinted genes using qPCR. The expression signals of the 24 imprinted genes relative to each of the two internal control genes (AU042671 and Cry2) were calculated across all tissues of 10.5-dpc and 14.5-dpc conceptuses. The agreement between signals from these two genes was assessed visually by scatter plots and by calculating the coefficient of correlation. Given the high correlation between the two control genes, the results of qPCR were averaged before further analysis. Averaged expression signals were then analyzed using ANOVA to study the effects of the expressed allele (paternally or maternally), sex, and their interaction within each tissue. The analyses were performed using the procedures PROC CORR and PROC GLM of SAS v9.1 (SAS Institute, Inc., Cary, NC). The analysis was performed by calculating the ratios of the log base 2 gene expression of the autosomal imprinted genes, one at the time, relative to the mean log base 2 of the whole genome expression (except known imprinted genes) across different tissue types. All the analyses were on log base 2 scale to meet normality assumptions, so values mentioned herein are of log base 2 unless otherwise indicated. For imprinted genes with no upregulation, the ratio of imprinted:nonimprinted gene expression would be -1 (equivalent to 0.5 on the normal scale) or lower. In contrast, for upregulated imprinted genes, the ratio of imprinted:nonimprinted gene expression would be of any value larger than -1, and a ‘zero’ value would be for genes with twofold upregulation.
Supplementary Material
Acknowledgements
We are grateful to J.K. Harting and M.L. Epstein for their support and encouragement. We thank B.W. Kirkpatrick and A.Z. Ansari for helpful discussions. We thank E.S. Kim for help with the statistical analysis. This study was supported by USDA Hatch grant no. WIS04895 from the University of Wisconsin-Madison to H.K. and RO1 HD042706 from the National Institutes of Health (K.M.D.).
Footnotes
Supplementary materials can be found at: www.landesbioscience.com/supplement/ZaitounEPI5-2-Sup.pdf
References
- 1.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC, Wood MD, Tunster SJ, Barton SC, Surani MA, John RM. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev Biol. 2007;7:53. doi: 10.1186/1471-213X-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.da Rocha ST, Charalambous M, Lin SP, Gutteridge I, Ito Y, Dean W, et al. Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: implications for the evolution of imprinting. PLoS Genet. 2009;5:1000392. doi: 10.1371/journal.pgen.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swaney WT, Curley JP, Champagne FA, Keverne EB. Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. Proc Natl Acad Sci USA. 2007;104:6084–9. doi: 10.1073/pnas.0609471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurrieri F, Accadia M. Genetic imprinting: the paradigm of Prader-Willi and Angelman syndromes. Endocr Dev. 2009;14:20–8. doi: 10.1159/000207473. [DOI] [PubMed] [Google Scholar]
- 6.Riccio A, Sparago A, Verde G, De Crescenzo A, Citro V, Cubellis MV, et al. Inherited and Sporadic Epimutations at the IGF2-H19 locus in Beckwith-Wiedemann syndrome and Wilms’ tumor. Endocr Dev. 2009;14:1–9. doi: 10.1159/000207461. [DOI] [PubMed] [Google Scholar]
- 7.Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.106. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce JA, Schofield PN. Genomic imprinting and cancer. Mol Pathol. 1998;51:185–90. doi: 10.1136/mp.51.4.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kainz B, Shehata M, Bilban M, Kienle D, Heintel D, Krömer-Holzinger E, et al. Overexpression of the paternally expressed gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in high-risk Bcell chronic lymphocytic leukemia. Int J Cancer. 2007;121:1984–93. doi: 10.1002/ijc.22929. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Zhang X, Zhang M, Zhu JD, Zhang YL, Lin Y, et al. Upregulation of DLK1 as an imprinted gene could contribute to human hepatocellular carcinoma. Carcinogenesis. 2007;28:1094–103. doi: 10.1093/carcin/bgl215. [DOI] [PubMed] [Google Scholar]
- 11.Lee JT. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr Biol. 2003;13:242–54. doi: 10.1016/s0960-9822(03)00162-3. [DOI] [PubMed] [Google Scholar]
- 12.Probst FJ, Hedera P, Sclafani AM, Pomponi MG, Neri G, Tyson J, et al. Skewed X-inactivation in carriers establishes linkage in an X-linked deafness-mental retardation syndrome. Am J Med Genet A. 2004;131:209–12. doi: 10.1002/ajmg.a.30308. [DOI] [PubMed] [Google Scholar]
- 13.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Lose F, Duffy DL, Kay GF, Kedda MA, Spurdle AB, Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer et al. Skewed X chromosome inactivation and breast and ovarian cancer status: evidence for X-linked modifiers of BRCA1. J Natl Cancer Inst. 2008;100:1519–29. doi: 10.1093/jnci/djn345. [DOI] [PubMed] [Google Scholar]
- 15.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–95. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19:1374–83. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verona RI, Thorvaldsen JL, Reese KJ, Bartolomei MS. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol Cell Biol. 2008;28:71–82. doi: 10.1128/MCB.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rougeulle C, Navarro P, Avner P. Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum Mol Genet. 2003;12:3343–8. doi: 10.1093/hmg/ddg351. [DOI] [PubMed] [Google Scholar]
- 19.Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 20.Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–5. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 21.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, et al. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 23.Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 24.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–7. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- 26.Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA. 2006;103:6623–8. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goedbloed JF. The embryonic and postnatal growth of rat and mouse I. The embryonic and early postnatal growth of the whole embryo A model with exponential growth and sudden changes in growth rate. Acta Anat (Basel) 1972;82:305–6. [PubMed] [Google Scholar]
- 28.Marino R, Hegde A, Barnes KM, Schrier L, Emons JA, Nilsson O, et al. Catch-up growth after hypothyroidism is caused by delayed growth plate senescence. Endocrinology. 2008;149:1820–8. doi: 10.1210/en.2007-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkielstain GP, Forcinito P, Lui JC, Barnes KM, Marino R, Makaroun S, et al. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology. 2009;150:1791–800. doi: 10.1210/en.2008-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lui JC, Finkielstain GP, Barnes KM, Baron J. An imprinted gene network that controls mammalian somatic growth is downregulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol. 2008;295:189–96. doi: 10.1152/ajpregu.00182.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Oliver B. Dosage compensation goes global. Curr Opin Genet Dev. 2007;17:113–20. doi: 10.1016/j.gde.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O'Neill LP, et al. Dosage compensation in the mouse balances upregulation and silencing of X-linked genes. PLoS Biol. 2007;5:326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang M, Parker EA, Muller TJ, Haenen C, Mistry M, Finkielstain GP, et al. Changes in cell cycle kinetics responsible for limiting somatic growth in mice. Pediatr Res. 2008;64:240–5. doi: 10.1203/PDR.0b013e318180e47a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-ofwar. Trends Genet. 1991;7:45–9. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 35.Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–30. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res. 1996;92:91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 37.Allen ND, Logan K, Lally G, Drage DJ, Norris ML, Keverne EB. Distribution of parthenogenetic cells in the mouse brain and their influence on brain development and behavior. Proc Natl Acad Sc USA. 1995;92:10782–6. doi: 10.1073/pnas.92.23.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–8. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 39.Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci USA. 2002;99:327–32. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res. 2006;113:130–7. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- 41.Ke X, Thomas NS, Robinson DO, Collins A. The distinguishing sequence 17 characteristics of mouse imprinted genes. Mamm Genome. 2002;13:639–45. doi: 10.1007/s00335-002-3038-x. [DOI] [PubMed] [Google Scholar]
- 42.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–2. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 43.Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, et al. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet. 2002;31:216–20. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- 44.Richardson BJ, Czuppon AB, Sharman GB. Inheritance of glucose-6-phosphate dehydrogenase variation in kangaroos. Nat New Biol. 1971;230:154–5. doi: 10.1038/newbio230154a0. [DOI] [PubMed] [Google Scholar]
- 45.Sharman GB. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature. 1971;230:231–2. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- 46.Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 48.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 49.Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–7. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 50.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 52.Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 2007;39:945–9. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- 53.Yin S, Wang P, Deng W, Zheng H, Hu L, Hurst LD, et al. Dosage compensation on the active X chromosome minimizes transcriptional noise of X-linked genes in mammals. Genome Biol. 2009;10:74. doi: 10.1186/gb-2009-10-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243–8. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi K, Nakayama K, Nakayama K. Mice lacking a CDK inhibitor, p57Kip2, exhibit skeletal abnormalities and growth retardation. J Biochem. 2000;127:73–83. doi: 10.1093/oxfordjournals.jbchem.a022586. [DOI] [PubMed] [Google Scholar]
- 56.Sato N, Matsubayashi H, Abe T, Fukushima N, Goggins M. Epigenetic downregulation of CDKN1C/p57KIP2 in pancreatic ductal neoplasms identified by gene expression profiling. Clin Cancer Res. 2005;11:4681–8. doi: 10.1158/1078-0432.CCR-04-2471. [DOI] [PubMed] [Google Scholar]
- 57.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 58.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–58. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 60.Schulz R, Woodfine K, Menheniott TR, Bourc'his D, Bestor T, Oakey RJ. WAMIDEX: a web atlas of murine genomic imprinting and differential expression. Epigenetics. 2008;3:89–96. doi: 10.4161/epi.3.2.5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogawa H, Ono Y, Shimozawa N, Sotomaru Y, Katsuzawa Y, Hiura H, et al. Disruption of imprinting in cloned mouse fetuses from embryonic stem cells. Reproduction. 2003;126:549–57. doi: 10.1530/rep.0.1260549. [DOI] [PubMed] [Google Scholar]
- 62.Ogawa H, Wu Q, Komiyama J, Obata Y, Kono T. Disruption of parental-specific expression of imprinted genes in uniparental fetuses. FEBS Lett. 2006;580:5377–84. doi: 10.1016/j.febslet.2006.08.087. [DOI] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.