Abstract
The Saccharomyces cerevisiae origin recognition complex (ORC) is bound to origins of DNA replication throughout the cell cycle and directs the assembly of higher-order protein–DNA complexes during G1. To examine the fate of ORC when origin DNA is unwound during replication initiation, we determined the effect of single-stranded DNA (ssDNA) on ORC. We show that ORC can bind ssDNA and that ORC bound to ssDNA is distinct from that bound to double-stranded origin DNA. ssDNA stimulated ORC ATPase activity, whereas double-stranded origin DNA inhibited the same activity. Electron microscopy studies revealed two alternative conformations of ORC: an extended conformation stabilized by origin DNA and a bent conformation stabilized by ssDNA. Therefore, ORC appears to exist in two distinct states with respect to its conformation and ATPase activity. Interestingly, the effect of ssDNA on these properties of ORC is correlated with ssDNA length. Since double-stranded origin DNA and ssDNA differentially stabilize these two forms of ORC, we propose that origin unwinding triggers a transition between these alternative states.
Keywords: ATPase/conformation/electron microscopy/ORC/ssDNA
Introduction
Origins of DNA replication are sites at which multiple proteins must be assembled in a highly regulated manner and subsequently disassembled to allow formation of a mobile DNA replication machine. Studies of chromosomal replication in a number of organisms, including bacteria, phage and eukaryotic viruses, have demonstrated that a protein called the initiator participates in multiple early steps in this process (reviewed in Baker and Bell, 1998). Initiator proteins bind origins of replication and thereby select the site at which replication is to begin. These proteins also frequently facilitate the unwinding of origins required to generate the single-stranded DNA (ssDNA) template for polymerase action. Finally, initiator proteins recruit other replication proteins required for the assembly of replication forks at the origin.
In eukaryotic cells, the strongest candidate for an initiator protein is the origin recognition complex (ORC; Bell and Stillman, 1992; reviewed in Dutta and Bell, 1997). ORC was first identified in the yeast Saccharo myces cerevisiae and consists of six polypeptides (Orc1p–Orc6p) that are each essential for yeast viability (Bell et al., 1993, 1995; Li and Herskowitz, 1993; Micklem et al., 1993; Loo et al., 1995). The coordinate action of five of the six ORC subunits is required for origin binding as only the Orc6p subunit is dispensable for this activity (Lee and Bell, 1997). Two groups of subunits that contact distinct regions of the DNA mediate ORC–DNA binding, and ORC contacts more residues on one strand of the double-stranded DNA (dsDNA) than the other. Sequence analogs of ORC subunits have been identified in a number of organisms. In Xenopus laevis and Drosophila melanogaster, they have been shown to form a six-protein complex similar to that seen in yeast (reviewed in Dutta and Bell, 1997). Both in vivo and in vitro studies of replication strongly suggest that ORC is required for DNA replication in all eukaryotic species (Landis et al., 1997; Chesnokov et al., 1999; reviewed in Dutta and Bell, 1997).
In yeast, ORC binds to origins of replication throughout the cell cycle and directs the assembly of higher-order complexes in preparation for the initiation of DNA replication. In vivo DNase I protection assays and chromatin immunoprecipitation experiments suggest that ORC is present at the origin during the G2 and M phases to form the post-replicative complex (post-RC; Diffley et al., 1994; Aparicio et al., 1997; Tanaka et al., 1997). In G1, ORC is required to recruit additional replication proteins to origins, including Cdc6p and the Mcm2–7p (MCM proteins), to form the pre-replicative complex (pre-RC). Finally, Cdc45p and the replicative polymerases are recruited to origins in a manner correlated with the time of replication initiation (Aparicio et al., 1997, 1999; Zou and Stillman, 1998, 2000). During S phase, Cdc6p is degraded (Piatti et al., 1995; Drury et al., 1997), and MCM proteins and Cdc45p appear to be released from origins and move with the DNA polymerases as part of the replication fork (Aparicio et al., 1997; Labib et al., 2000; Tercero et al., 2000). ORC remains at the origin to repeat the process in the following cell cycle. The requirement for an ORC-dependent assembly of replication proteins on DNA is likely to be conserved throughout evolution, as replication in Xenopus extracts requires chromatin association of ORC, Cdc6p and MCM proteins, with the same dependence as in yeast (reviewed in Tye, 1999).
In addition to recognizing origin DNA, yeast ORC binds and hydrolyzes ATP, and these three activities are interrelated (Klemm et al., 1997). ATP binding to the Orc1p subunit is required for the complex to bind origin DNA. Interaction with origin DNA inhibits the Orc1p-dependent ATP hydrolysis activity. ATPase repression requires origin-containing dsDNA as non-origin dsDNA has no effect on the rate of ATP hydrolysis. Thus, ATP binding and sequence-specific DNA binding are coordinate processes that together regulate ATP hydrolysis by ORC. The inhibition of the ORC ATPase by origin DNA suggests that ATP hydrolysis by ORC will not occur throughout most of the cell cycle.
Origin DNA is generally double stranded and therefore inhibitory to the ORC ATPase, but once in each cell cycle the origin must become unwound to allow access of the enzymatic machinery to the DNA substrate. The consequence of this DNA unwinding event for ORC function as well as other proteins assembled at the origin is not understood. In experiments designed to determine how origin melting altered the ORC–origin complex, we found that ORC bound ssDNA. Furthermore, ssDNA altered ORC conformation (as determined by electron microscopy) and stimulated the ORC ATPase. For each activity, ssDNA had the opposite effect of double-stranded origin DNA, suggesting that ssDNA binding alters ORC function during the initiation of replication.
Results
ORC binds ssDNA in an ATP-independent manner
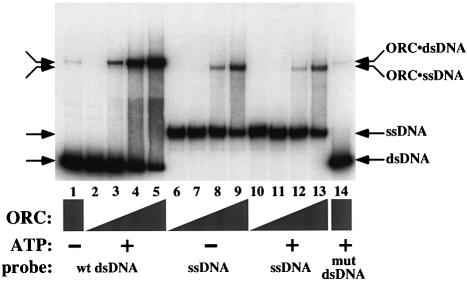
To determine whether ORC was capable of continued association with origin DNA after unwinding, we asked if ORC could bind ssDNA. We performed electrophoretic mobility shift assays (EMSAs) using radiolabeled ssDNA and dsDNA, each containing the wild-type ARS1 origin, as well as labeled dsDNA containing a mutated origin (Figure 1). As had been previously described (Bell and Stillman, 1992), the binding of purified ORC to dsDNA was both sequence specific (compare lanes 5 and 14) and dependent on the presence of ATP (compare lanes 1 and 5). We found that when ORC was incubated with labeled ssDNA, the mobility of the ssDNA was altered. Multiple monoclonal ORC antibodies could supershift this complex, indicating that this species included ORC (data not shown). Thus, ORC is able to bind ssDNA.
Fig. 1. ORC binds ssDNA in an ATP-independent manner. ORC EMSAs were performed using a radiolabeled 244 bp dsDNA containing a wild-type ARS1 origin (wt dsDNA, lanes 1–5), a labeled 295 nt ssDNA containing wild-type ARS1 (ssDNA, lanes 6–13) and a labeled 244 bp dsDNA containing ARS1 with a mutation in the ORC-binding site (mut dsDNA, lane 14). ATP (50 µM final) was added to reactions in lanes 2–5 and 10–14. Two-fold titrations of ORC resulted in 2.5 ng (lanes 2, 6 and 10), 5 ng (lanes 3, 7 and 11), 10 ng (lanes 4, 8 and 12) or 20 ng of ORC per reaction (lanes 1, 5, 9, 13 and 14). The binding reactions were electrophoresed on a native polyacrylamide gel to separate bound and unbound DNA.
Unlike binding of ORC to dsDNA, this new DNA-binding activity of ORC did not require specific yeast origin sequences or the presence of ATP. Although initial experiments used ssDNA containing the ARS1 sequence, no preference has been observed for origin sequences over non-origin sequences (Supplementary Figure A, and data not shown). (Supplementary data are available at The EMBO Journal Online.) We have also seen that ORC can bind every naturally occurring ssDNA that we have tested, including ssDNA sequences derived from yeast, bacteriophage M13, bacteriophage lambda and humans (data not shown). Furthermore, ORC–ssDNA binding occurred in the absence of ATP (Figure 1, lanes 6–9). To demonstrate further that ATP was not required for ORC–ssDNA binding, two mutated ORC complexes that lack the ability to bind ATP (ORC-1A and ORC-5A; Klemm et al., 1997; see Materials and methods) were also tested. Neither of these mutant complexes showed any defect in ssDNA binding (data not shown). The affinity of ORC for ssDNA (see below) was also unaffected by addition of excess ATP, ADP or the non-hydrolyzable analog of ATP, ATP-γS (50 µM each, data not shown), indicating that nucleotide binding does not inhibit ssDNA binding. Thus, ssDNA binding occurs equally well whether ORC is bound to ATP or free of nucleotide.
The relative affinities of ORC for ssDNA or dsDNA depended on the presence or absence of ATP. The EMSA data suggested that, in the presence of ATP, the affinity of ORC was highest for specific dsDNA, weaker for ssDNA, and weaker still for non-specific dsDNA (Figure 1, lanes 5, 13 and 14, respectively). However, in the absence of ATP, ORC bound ssDNA more tightly than specific dsDNA (compare lane 9 with lane 1). We used the EMSA to determine the apparent association constants of ORC for a specific 244 bp dsDNA (in the presence of ATP) and for a corresponding 244 nucleotide (nt) ssDNA (see Materials and methods). The affinity of ORC for dsDNA in the presence of ATP (apparent KA = 1.4 × 109 M–1) was slightly higher than that of ORC for ssDNA (apparent KA = 0.9 × 109 M–1). The apparent association constant for ORC and ssDNA was also compared with that of the S.cerevisiae ssDNA-binding protein, ScRPA, and was found to be within 3-fold (apparent KA for ScRPA and ssDNA = 3.0 × 109 M–1, consistent with published results for ScRPA; reviewed in Wold, 1997). Therefore, ORC’s affinity for ssDNA is comparable to that of an established yeast ssDNA-binding protein.
ORC–ssDNA binding and ORC–dsDNA binding are mutually exclusive
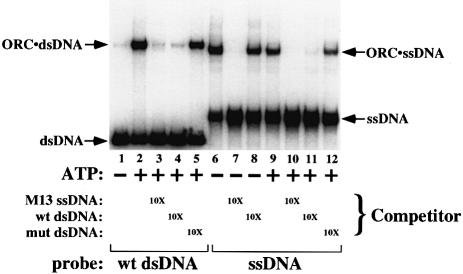
To examine whether ssDNA and dsDNA binding by ORC use overlapping or distinct binding sites, competition experiments were carried out (Figure 2). Binding of ORC to radiolabeled substrates (origin dsDNA and ssDNA) was assayed in the presence or absence of ATP, and in the presence or absence of one of three unlabeled competitor DNAs (present at a 10-fold molar excess): an M13 ssDNA circle, wild-type dsDNA and dsDNA containing a mutation in the ORC-binding site (mut dsDNA). Again, ORC–dsDNA binding was dependent on origin sequences and on ATP. The binding of ORC to labeled wild-type dsDNA (in the presence of ATP, lane 2) was efficiently competed by unlabeled wild-type dsDNA (lane 4) but not by the mutant dsDNA (lane 5). M13 ssDNA was also an effective competitor of ORC–dsDNA binding, resulting in a loss of the ORC–dsDNA signal without the appearance of a less mobile species indicative of ORC simultaneously binding dsDNA and ssDNA (lane 3). Binding of ORC to radiolabeled ssDNA (lanes 6 and 9) was efficiently competed by dsDNA, but only when the wild-type sequence was used as the competitor (compare lanes 11 and 12) and ATP was present (compare lanes 11 and 8). Thus, binding of ORC to ssDNA and binding to dsDNA are mutually exclusive, arguing that the binding sites on ORC for the two forms of DNA are at least partially overlapping or that ORC undergoes a conformational change such that one DNA molecule allosterically inhibits binding to the other (see below). Consistent with similar binding sites being used for both types of DNA, we have found that removal of the Orc1p subunit but not the Orc6p subunit from ORC eliminates ssDNA binding (data not shown). The same dependence is seen for dsDNA binding (Lee and Bell, 1997).
Fig. 2. ORC–ssDNA binding and ORC–dsDNA binding are mutually exclusive. The ssDNA and dsDNA EMSA was repeated in the presence or absence of one of three unlabeled competitor DNAs: an ssDNA M13 circle (lanes 3, 7 and 10), wild-type dsDNA (lanes 4, 8 and 11) and mutant dsDNA (lanes 5 and 12). Competitor DNAs were present at a 10-fold molar excess over the labeled DNA, and ATP (50 µM) was included as indicated.
ORC preferentially binds longer ssDNA molecules
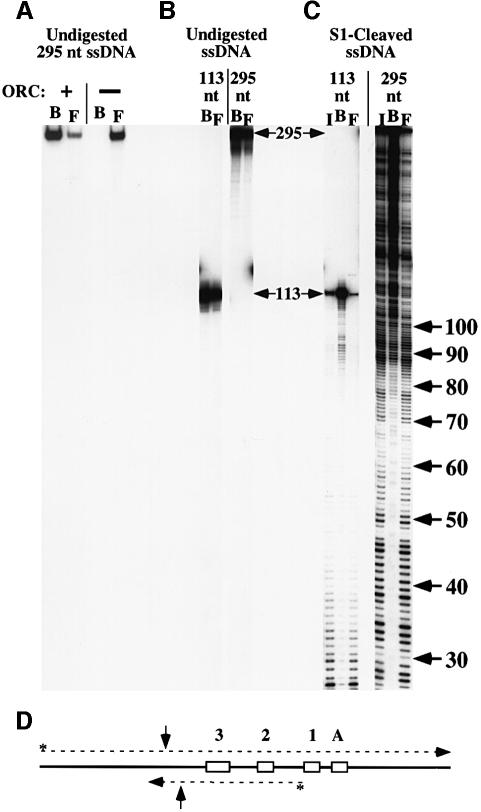
To determine the optimal length of ssDNA bound by ORC, we incubated ORC with ssDNA molecules of different lengths. Subsequent immunoprecipitation of ORC allowed us to compare the ssDNA bound by ORC (B) to the unbound or free ssDNA (F, Figure 3). In these experiments, the ability to immunoprecipitate labeled DNA was entirely dependent on the addition of ORC (Figure 3A) and ORC antibodies (data not shown). When ORC was incubated with end-labeled ssDNA molecules that were either 113 or 295 nt long, the two ssDNAs co-immunoprecipitated with ORC with similar efficiencies under these experimental conditions (Figure 3B). These ssDNA fragments were then treated with S1 nuclease to generate a population of differently sized molecules. Incubation of ORC with the cleaved ssDNA followed by immunoprecipitation of ORC showed that the bound population (B) was greatly enriched for molecules ∼90 nt or longer and showed reduced retention of shorter molecules (Figure 3C). Because similar results were obtained with two DNAs of different sequence (Figure 3C), the observed preference for longer ssDNA molecules is unlikely to be due to the presence of a specific sequence. Instead, increased ssDNA length correlates more closely with preferential association of ORC with both ssDNA sequences.
Fig. 3. ORC preferentially binds longer ssDNAs. (A) A 295 nt end-labeled ssDNA was incubated with or without ORC. Anti-ORC polyclonal sera and beads coupled to protein G were used to immunoprecipitate ORC and the associated ssDNA. ‘B’ and ‘F’ represent the bound and free DNA, respectively. (B) ORC was incubated with end-labeled ssDNA that was 113 or 295 nt long, and was precipitated as described. ORC binds these two (full-length) molecules with similar efficiencies under these experimental conditions. (C) ssDNAs of 113 and 295 nt were cleaved with S1 nuclease to generate random populations of ssDNA molecules of different lengths (input DNA, I). The resulting ssDNA was then incubated with ORC and immunoprecipitated as before. Input, bound and free DNAs were electrophoresed on a denaturing polyacrylamide gel. ssDNA lengths (in nucleotides) are shown on the right. (D) Schematic representation of the 113 and 295 nt ssDNAs (dashed lines) with respect to the pARS1/WT sequence (Marahrens and Stillman, 1992). The ARS1 ACS, B1, B2 and B3 elements are indicated by boxes labeled A, 1, 2 and 3, respectively. The 295 nt ssDNA encodes the top (A-rich) strand of ARS1, whereas the 113 nt ssDNA encodes the bottom strand. The 3′ end of each ssDNA is indicated by an arrowhead, and the radiolabeled 5′ end is indicated with an asterisk. The 90 nt position is indicated on each oligonucleotide with vertical arrows.
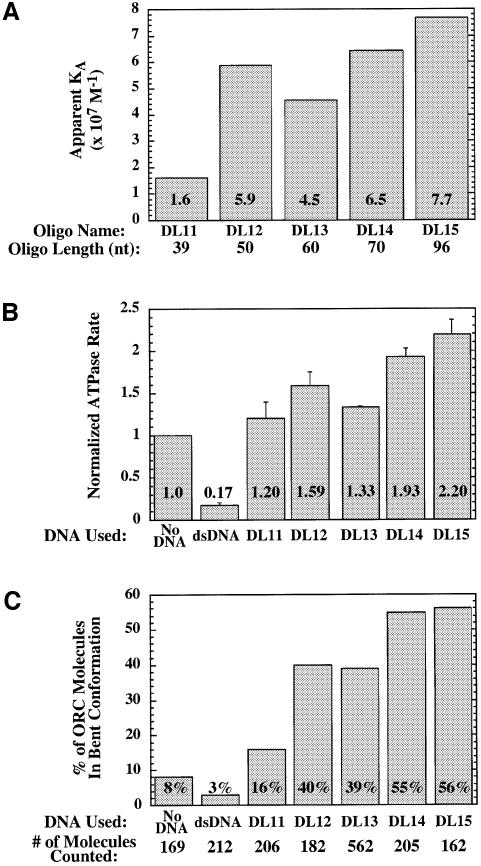
Because ORC was capable of binding ssDNA molecules <90 nt long, oligonucleotides of increasing lengths were tested directly using the EMSA to examine the length dependence of ssDNA binding. Binding of ORC was not detected with a molecule 20 nt long (data not shown), while weak binding was detected with a 30mer and a 39mer (DL11); in general, the affinity of ORC for ssDNA increased as the length of the ssDNA increased. Oligonucleotides for which apparent association constants could be measured are shown in Figure 4A, with affinities ranging from 1.6 × 107 M–1 (for the 39 nt DL11 oligonucleotide) to 7.7 × 107 M–1 (for the 96 nt DL15 oligonucleotide). The correlation between ssDNA length and affinity is not perfect. For example, the 50 nt DL12 oligonucleotide was bound more tightly by ORC than the 60 nt DL13 oligonucleotide. The different nucleotide sequences of these unrelated oligonucleotides are probably responsible for this discrepancy; nevertheless, overall length is a strong predictor of affinity (see below).
Fig. 4. ssDNA stimulates ORC ATPase activity and induces a conformational change in ORC. (A) Apparent association constants of ORC for ssDNA oligonucleotides of indicated lengths were determined using EMSA. The KA was calculated by taking the inverse of the concentration of free ORC at half-maximal binding. Lengths in nucleotides are indicated below each oligonucleotide. (B) The rate of ATP hydrolysis by ORC was measured in the absence of DNA, in the presence of origin-containing dsDNA (the 244 bp dsDNA, see Materials and methods) or in the presence of oligonucleotides of various lengths. ATPase rates were normalized to the rate of hydrolysis seen in the absence of DNA. The averages and standard deviations for three experiments are shown. (C) Proportion of ORC molecules in the bent conformation as determined by electron microscopy (Figure 5). ORC was examined in the absence of DNA, in the presence of dsDNA or in the presence of ssDNA of various lengths. The total number of ORC molecules counted in at least two experiments is shown below each bar.
ssDNA and dsDNA have opposite effects on ORC ATPase activity
Previous studies of the ORC ATPase indicated that origin dsDNA (but not non-origin dsDNA) strongly inhibits this activity (Klemm et al., 1997). To determine if ssDNA affected the ORC ATPase, we measured the rate of ORC ATP hydrolysis in the absence of DNA, in the presence of origin-containing dsDNA and in the presence of ssDNA (Figure 4B). We found that ssDNA and origin DNA had opposite effects: in contrast to the ∼6-fold inhibition by origin DNA, ssDNA stimulated ATPase activity up to 2-fold. Like all previously detected ORC ATPase activity, the increased ATP hydrolysis that occurred in the presence of ssDNA was dependent on the activity of the Orc1p subunit but not the Orc5p subunit (as determined using the ORC-1A and ORC-5A mutant complexes; data not shown). To determine how different oligonucleotides stimulated ORC ATPase activity, we tested five oligonucleotides of increasing length in this assay (Figure 4B). Similar to the affinity of ORC for different ssDNAs, we found that there was a general correlation between the length of the oligonucleotide and the level of ATPase activity induced. As in the measurement of ORC affinity for these same oligonucleotides, we found that the DL12 and DL13 oligonucleotides were the exceptions to this general trend. Overall, these data support a hypothesis that ORC ATPase activity is induced by ssDNA as compared with origin DNA and that the affinity of an ssDNA molecule for ORC is predictive of its ability to induce the ORC ATPase.
ssDNA alters the conformation of ORC
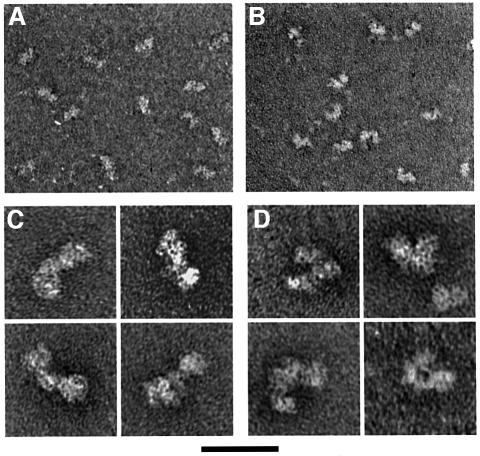
To examine the global structure of ORC, we have used electron microscopy. In the absence of DNA, ORC stained with uranyl acetate had a tri-lobed structure, with the smallest lobe in the middle and one of the outer lobes slightly larger than the other. A representative field of ORC molecules showed that the majority of complexes were straight or extended (Figure 5A and C), with an angle between the two ‘arms’ of ORC of 166 ± 11° (N = 100 straight molecules measured). A minority of ORC complexes (8%, N = 169) adopted a more bent or compact structure (Figure 4C). In this alternative conformation, the average angle between the two arms was reduced to 106 ± 13° (N = 100 bent molecules measured). Thus, ORC is present in at least two distinct conformations.
Fig. 5. Electron microscopy of ORC. Transmission electron microscopy of ORC was performed on samples negatively stained with uranyl acetate in the absence of DNA (A and C) or in the presence of the 96 nt DL15 ssDNA (B and D). Low magnification images show that, in the absence of DNA, ORC is primarily in a straight or extended conformation (A), whereas in the presence of ssDNA, many complexes adopt a bent or curved conformation (B). (C) High magnification images of straight ORC molecules in the absence of DNA. (D) High magnification images of bent ORC molecules in the presence of ssDNA. The scale bar represents 40 nm for (A) and (B) and 17 nm for (C) and (D).
To determine whether DNA binding influences the conformation of ORC, we performed electron microscopic analysis of ORC in the presence of saturating levels of origin DNA and ssDNA. When ORC was bound to double-stranded origin DNA (in the presence of ATP), the structure of ORC was similar to that of ORC in solution; however, a smaller fraction of the complexes (3%, N = 212) adopted the bent conformation (Figure 4C). In contrast, incubation of ORC with ssDNA resulted in a dramatic increase in the number of bent complexes (Figures 5B, D and 4C). When 100 molecules were chosen at random from the samples containing ORC alone (regardless of conformation), the average angle for this population of molecules was 151 ± 26°, consistent with the majority of complexes being in a straight conformation. In contrast, for ORC in the presence of the 96 nt DL15 ssDNA, the average angle for a population of molecules was 127 ± 31° (N = 100), consistent with roughly half of the complexes present being in the bent conformation. Similar to the effect of ssDNA on the ORC ATPase (Figure 4B), the fraction of ORC complexes in the bent conformation in the presence of ssDNA varied with the length of ssDNA added (Figure 4C). Addition of a 39 nt oligonucleotide (DL11) resulted in only 16% of the ORC molecules being in the bent conformation, whereas addition of a 96 nt oligonucleotide (DL15) resulted in 56% bent ORC molecules. Thus, the abilities of an ssDNA molecule to stimulate ATPase activity and stabilize the bent ORC conformation are closely related.
To determine whether ATP hydrolysis was necessary for ORC to assume the bent conformation, we tested the effect of the non-hydrolyzable ATP-γS on the proportion of bent complexes formed. Addition of ATP-γS did not alter the proportion of bent ORC complexes in the presence or absence of ssDNA (data not shown). We also tested a mutant ORC complex that lacked the Orc1p ATP-binding site and is incapable of hydrolyzing ATP (ORC-1A; Klemm et al., 1997). This mutant ORC achieved the same proportion of molecules in the bent conformation in the presence of ssDNA as did wild-type ORC (data not shown). Thus, consistent with the ATP independence of ssDNA binding, ATP binding or hydrolysis by ORC is not required for it to adopt the bent conformation.
ssDNA length influences ORC–ssDNA interactions
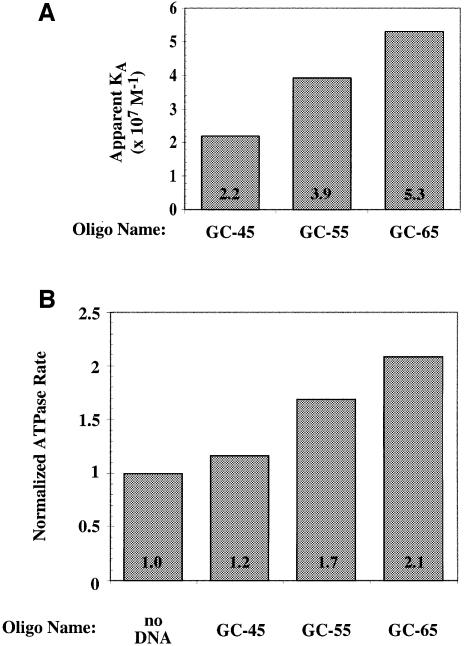
The data in Figure 4 show that ORC–ssDNA binding, ssDNA stimulation of ORC ATPase activity, and ssDNA-mediated conformational changes in ORC are correlated, and all three properties of ORC are more strongly affected by longer ssDNAs. The correlation between length and the extent to which ORC activities are altered is not perfect (the 50 nt DL12 oligonucleotide has a more pronounced effect than the 60 nt DL13 oligonucleotide), suggesting that DNA sequence also influences the strength of ORC–ssDNA interactions. To examine the effects of ssDNA length in the absence of sequence differences, we tested the ORC–ssDNA affinity and the ORC ATPase activity in the presence of three oligonucleotides containing increasing copies of a non-origin sequence, with overall lengths of 45, 55 and 65 nt (Figure 6). Both binding affinity (Figure 6A) and ATPase activity (Figure 6B) increased with ssDNA length. Thus, unlike the situation in Figure 4, in which ssDNA molecules of different length were unrelated in sequence, when length is increased by adding to the sequence of a shorter ssDNA molecule, we observe a consistent increase in affinity for ORC and activation of the ORC ATPase.
Fig. 6. ssDNA length is correlated with the strength of ORC–ssDNA interactions. Oligonucleotides made up of multiple copies of a non-origin DNA (50% GC bases) were used to determine the apparent association constant with ORC (A) or the effect on the rate of ORC ATP hydrolysis (B) as described in the legend to Figure 4. The GC-45, GC-55 and GC-65 oligonucleotides were 45, 55 and 65 nt long, respectively. The longer oligonucleotides each include all of the sequences of the shorter oligonucleotides with 10 nt of additional ssDNA.
Discussion
The studies presented here demonstrate two new activities of ORC. First, we show that ORC is capable of binding to ssDNA and that the affinity of ORC for ssDNA is correlated with ssDNA length. Secondly, we demonstrate that ORC can exist in two different conformations. By determining the effect of ssDNA and double-stranded origin DNA on the two ORC conformations and the intrinsic ATPase of ORC, we demonstrate that the type of DNA bound to ORC dramatically affects both its conformation and ATPase activity. These studies suggest that changes in the structure of origin DNA during initiation will have important effects on ORC function.
The conformation of ORC (straight or bent) correlates with the rate of ATP hydrolysis by ORC. Both ORC bending and ATPase activity are lowest when ORC is bound to dsDNA and greatest when ORC is bound to long ssDNA (compare Figure 4B and C). This correlation suggests that these two ORC activities are functionally related. Our studies indicate that ATP hydrolysis is not required for the observed conformational change. Instead, we hypothesize that the ssDNA-induced conformational change is required for ATPase activity, although our current data cannot prove that a causal relationship exists. If this is the case, then ORC ATPase activity is unlikely to participate directly in the melting of the origin. Instead, ORC is poised to detect melting of the origin and change its function in response to that initiation event.
Interestingly, the ability of ssDNA to bind to ORC is correlated with the length of ssDNA added. When ssDNA molecules of overlapping sequence but different lengths are compared, we consistently observe that increased ssDNA length results in increased affinity for ORC (Figures 3 and 6) and increased stimulation of the ORC ATPase (Figure 6). In the context of origin DNA, this type of ssDNA comparison is particularly relevant, as ongoing origin unwinding would produce single-stranded regions of increasing length but overlapping sequence. It is clear that ssDNA length is not the only determinant of the ability of an ssDNA to interact with ORC. For example, the 50 nt DL12 oligonucleotide bound ORC with higher affinity than the 60 nt DL13 oligonucleotide of non-overlapping sequence (Figure 4). Nevertheless, specific origin elements are not a prerequisite for ORC–ssDNA binding since non-origin ssDNAs are bound equally well as origin sequences (Supplementary Figure A). Indeed, no particular sequence is apparently required for ORC recognition of ssDNA, as both ssDNAs derived from multiple natural sources and synthetic poly(dC) or poly(dT) can be bound by ORC (data not shown). Thus, although DNA sequence influences the affinity of ORC for ssDNA in the context of any particular stretch of DNA sequence, length of ssDNA is the most robust predictor of affinity for ORC.
There are two possible explanations for the different rates of ORC ATPase and proportions of ORC molecules in the bent conformation induced by longer ssDNAs. First, because the affinity of ORC for ssDNA generally correlates with increased ssDNA length (see above and Figures 3, 4 and 6), the greater stimulation with longer ssDNAs could be due to a higher proportion of ORC molecules bound to ssDNA. This is unlikely, however, because both the ATPase and electron microscopy studies were performed at saturating concentrations of oligonucleotide, based on measurements of ORC affinity for each ssDNA. Moreover, increased concentrations of ssDNA oligonucleotide failed to stimulate ORC ATPase activity further (data not shown). Instead, our findings argue that ORC can detect differences in the length and sequence of ssDNA bound and that, in general, longer ssDNAs stimulate the ORC ATPase more effectively than shorter ssDNAs. This suggests the interesting possibility that ORC could alter its activity in accordance with the extent of origin DNA unwinding (see below).
Does ORC interact with ssDNA in vivo?
Several lines of evidence support the hypothesis that ORC is exposed to ssDNA forms of the origin DNA during the initiation process. First, both chromatin immunoprecipitation and in vivo footprinting studies suggest that ORC is associated with origin DNA throughout the cell cycle (Diffley et al., 1994; Aparicio et al., 1997; Tanaka et al., 1997). Because origin DNA must be unwound for the initiation of replication to proceed, ORC is likely to be present for this change in origin DNA structure. Secondly, studies mapping the start site of DNA replication in vivo suggest that DNA unwinding initiates immediately adjacent to the ORC-binding site at ARS1 (Bielinski and Gerbi, 1999). Thirdly, although it is possible that the ssDNA-binding activity of ORC could be used at sites other than origins of replication, the relatively low abundance of ORC and the constitutive binding of ORC to both active and inactive origins argues that the majority of ORC will be bound to these sites in the cell (Santocanale and Diffley, 1996; Santocanale et al., 1999). Thus, the primary exposure of ORC to ssDNA is likely to be at origins of replication. The requirement of ATP for origin binding by ORC and the inhibitory nature of origin DNA on the ORC ATPase activity argue that ORC remains in an ATP-bound state when associated with origin DNA. Thus, exposure of ORC to ssDNA during origin melting represents the most likely opportunity for DNA-bound ORC to hydrolyze ATP.
The alterations in ORC induced by hydrolysis of ATP and the conformational change may play one or more roles during the initiation of replication. First, these changes may inactivate ORC when the origin is unwound. In the absence of ATP, ORC binds ssDNA more tightly than dsDNA (Figure 1) and it should not be able to re-bind origin DNA until it has re-bound ATP. Coupling the inactivation of ORC–origin recognition to origin unwinding would provide a simple mechanism to reset the replication process after each round of initiation. Secondly, the switch in conformation may remodel replication complexes at active origins. Proteins such as MCMs, Cdc45p and the DNA polymerases must first be assembled at origins in a stable fashion but are probably released to form a moving replication fork upon initiation (Aparicio et al., 1997). A change in ORC conformation may alter protein–protein contacts within this higher-order complex to couple pre-RC remodeling to the unwinding of origins. The stronger response of ORC to longer ssDNAs suggests that ORC could monitor the extent of origin unwinding, ensuring that these changes in ORC activity do not occur prematurely. Finally, an ssDNA-regulated conformational switch in ORC may also inactivate pre-RCs at passively replicated origins. Because many origins are active during only a small proportion of cell cycles (Yamashita et al., 1997), it is imperative that pre-RCs assembled at these non-firing origins (Santocanale and Diffley, 1996; Santocanale et al., 1999) are inactivated when the locus is passively replicated. The ssDNA generated by a moving replication fork (Park et al., 1998) could trigger the ORC conformational switch to aid in the inactivation of these pre-RCs. A rigorous test of these models will require an in vitro reconstitution of the pre-RC to examine the effects of ORC activities in the context of the other origin-associated proteins.
In addition to examining the effects of ssDNA on ORC function in the presence of pre-RC components, a better understanding of the in vivo relevance of this putative switch in ORC function will require the use of more physiologically relevant DNA substrates and mutants in ORC specifically defective for ssDNA binding. The likely ssDNA substrates in vivo are ssDNA bubbles rather than the oligonucleotides with free 5′ and 3′ ends used in these studies. Preliminary experiments using an ARS1 substrate containing a 63 bp ssDNA bubble spanning the B1 and B2 elements indicate that ORC is competent to bind such a structure in an ATP-independent manner (data not shown). Identification of mutants in ORC specific for ssDNA binding is complicated by the potential overlap between dsDNA- and ssDNA-binding sites in ORC (Figure 2) and a lack of understanding of the domains of the six ORC subunits required for DNA (ss or ds) binding. Mapping these sites more carefully in vitro will facilitate more directed approaches to identifying mutants that affect specific DNA-binding activities of ORC.
Interaction with ssDNA is observed for other known initiator proteins. The viral initiators Sendai virus 40 (SV40) T-antigen, bovine papilloma virus E1 protein and herpes simplex virus UL9 each possess ssDNA-binding activities associated with an intrinsic DNA helicase (Borowiec et al., 1990; Yang et al., 1993; Boehmer and Lehman, 1997). In addition, like ORC (Lee and Bell, 1997), modification interference assays with SV40 T-antigen showed a preferred interaction with one of the two strands of the origin prior to DNA unwinding (SenGupta and Borowiec, 1992). Initiators lacking an intrinsic DNA helicase activity also exhibit ssDNA-binding activity. Studies of the Escherichia coli initiator protein, dnaA, show that it preferentially binds one of the two strands of the origin after DNA melting (Hwang and Kornberg, 1992). In each case these interactions are critical for the function of the initiator, suggesting that an understanding of ssDNA binding by ORC and its function in the initiation process will be vital for understanding the events of eukaryotic DNA replication initiation.
Materials and methods
Wild-type and mutant ORC complexes
Wild-type ORC was expressed in insect cells and purified as described (Lee and Bell, 1997). Mutant ORC complexes containing point mutations in the ATP-binding domain of the Orc1p subunit (the ORC-1A complex, containing a K485E substitution in the Walker A motif of Orc1p) or the Orc5p subunit (the ORC-5A complex, containing a K43T substitution in the Walker A motif of Orc5p), were described previously (Klemm et al., 1997).
dsDNA and ssDNA
All DNAs were prepared and purified as described (Lee and Bell, 1997) unless otherwise noted:
244 bp dsDNA (wild-type and mutant). The wild-type and mutant versions were an EcoRI–HindIII fragment of pARS1/WT (Marahrens and Stillman, 1992) or pARS1/a–b2– (Lee and Bell, 1997), respectively. When used as radiolabeled probes, these DNAs were end-labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP at the HindIII site.
575 bp dsDNA. This was an ARS1-containing PCR product of pARS1/WT, prepared using oligonucleotides FSP24 and BUBLR1 as primers.
244 nt ssDNA. The end-labeled 244 bp dsDNA (wild-type) was resuspended in 51 µl of TE containing 9% dimethylsulfoxide (DMSO), heated to 95°C for 10 min to denature the DNA, and quickly chilled in ice water. An equal volume of ice-cold 0.1 M NaOH was added and the entire mixture was separated on a 5% acrylamide gel (50:1 acrylamide:bisacrylamide, 0.56× TBE) cooled to 4°C. Electrophoresis was performed for 6000 Vh at 4°C and the gel-purified labeled ssDNA was detected by exposing the wet gel to film. The HindIII-labeled ssDNA was separated from the other single strand and from duplex DNA (data not shown). Since this ssDNA was derived from the end-labeled dsDNA, the two DNAs had identical specific activities and were therefore used to compare the ORC association constants for ssDNA and dsDNA (see below).
113 nt ssDNA and 295 nt ssDNA. These ssDNAs were generated by combining 50 pmol of an end-labeled oligonucleotide with 8 µg of a plasmid cut with a restriction enzyme in a standard PCR mixture (Ausubel et al., 1994) and subjecting the reaction to 30 cycles of linear amplification with Taq polymerase. The reaction was precipitated with ethanol, resuspended in a formamide–dye mixture and electrophoresed on a 6% sequencing gel to purify the end-labeled ssDNA. For the 113 nt ssDNA, oligonucleotide DLX833-814 was used with XbaI-digested pARS1/WT, and for the 295 nt ssDNA, oligonucleotide RSP23 was used with EcoRI-digested pARS1/WT. One hundred and twenty counts per minute each of 113 and 295 nt ssDNA were digested with S1 nuclease (0.6 U, Boehringer Manheim) for 1 min at room temperature. The reaction was stopped by the addition of 200 ml of 1% sodium dodecyl sulfate (SDS)/20 mM EDTA, and the DNA was purified by phenol extraction and precipitation with ethanol.
The sequences for the oligonucleotides used in this study were FSP24: cgccagggttttcccagtcacgac; BUBLR1: cgttcttccttctgttcggag; DLX833-814: aaggcctgcaggcaagtgca; RSP23: agcggataacaatttcacacagg; DL11: gttaccatggcatcgagttckttcaacaagactacaatgg; DL12: acttgctcgagatgtccttacttgcttacaagcaaacagaagacttatac; DL13: attatttttttttggaagtgtttttcgacagaagttgcatcatcgatgaattcgagctcg; DL14: aggttctcgagatgtccccaaagaagaagaggaag gtcttagcggagtccaaagccatcctggcactat; DL15: acctgtcgtgccagctgcattaatgaa tcgcgaccccccattcaagaacagcaagcagcattgagaactttggaatccagtccctcttccacctgc; GC-45: gcagtagcacatggagcagtagcacatggagcagtagcacatgga; GC-55: gcagtagcacatggagcagtagcacatggagcagtagcacatggagcagtagcac; GC-65: gcagtagcacatggagcagtagcacatggagcagtagcacatggagcagtagcacatggagcagt.
EMSAs
ORC–DNA binding reactions were carried out in 10 µl of binding buffer [100 mM HEPES–KOH pH 7.6, 1 mM EDTA, 1 mM EGTA, 5 mM magnesium acetate and 0.15 M KCl, 10% (v/v) glycerol, 0.01% (w/v) NP-40, 1 mM dithiothreitol (DTT), 0.1 mg/ml poly(dG-C)]. ATP was included as indicated at 50 µM (unless otherwise noted). For gel shifts comparing dsDNA and ssDNA binding at various concentrations of ORC (Figure 1), 15 fmol of DNA was incubated with 2.5, 5, 10 or 20 ng of ORC (6, 12, 24 and 48 fmol of ORC, respectively). The radiolabeled DNAs corresponded to the 244 bp dsDNA (wild-type and mutant) and the 295 nt ssDNA. Competition experiments (Figure 2) were performed using 24 fmol of ORC with 15 fmol of labeled (wild-type) 244 bp dsDNA or 36 fmol of ORC with 15 fmol of labeled 295 nt ssDNA. Where indicated, 150 fmol of competitor DNA was used (i.e. a 10-fold molar excess of competitor to probe DNA). The unlabeled competitor DNAs were the 244 bp dsDNA (wild-type and mutant) and an M13 ssDNA circle. ORC was added to the DNA (a mixture of the labeled and unlabeled DNA in competition experiments), binding reactions were incubated at room temperature for 7 min and at 4°C for 7 min and EMSAs were performed as described (Lee and Bell, 1997).
Apparent association constants were determined from EMSAs by quantifying the proportion of the radiolabeled probe that was bound and using this value to calculate the concentration of bound ORC (assuming one molecule of ORC bound per molecule of DNA). Binding curves (fraction of DNA bound versus concentration of free ORC) were plotted to determine the free concentration of ORC at half-maximal binding, and the inverse of this value was calculated to determine KA. Comparisons of the affinities for dsDNA and ssDNA were performed using the 244 bp ARS1 dsDNA and the 244 nt ssDNA that was derived from it. The 244 nt ssDNA was also used to calculate the KA for ScRPA. Because the amount of ssDNA contacted by ORC is not yet known, the KAs were not corrected for the possibility of multiple ORC molecules binding a single ssDNA. This probably accounts for the significantly higher affinity of the 244 nt ssDNA compared with the shorter oligonucleotides.
DNA co-immunoprecipitation assays
Approximately 40 c.p.m. of ssDNA (undigested or digested with S1 nuclease as described above) was incubated with 1 µg of ORC as indicated in 50 µl of binding buffer lacking poly-d(G-C). Reactions were incubated at room temperature for 10 min and at 4°C for 10 min. The volume was increased to 200 µl with binding buffer, 1 µl of polyclonal anti-ORC antiserum was added, and each reaction was incubated at 4°C for 1 h with constant mixing. Twenty microliters of protein G-coupled Sepharose beads (Pharmacia) were added, followed by an additional incubation (with mixing) for 30 min. Beads were precipitated by centrifugation and the supernatant was removed. Unbound ssDNA in the supernatant was isolated by precipitating with ethanol. The beads were washed four times with 1 ml of binding buffer, and the bound ssDNA was eluted by adding SDS to 1%, removing the beads and proteins by phenol extraction and precipitating the ssDNA with ethanol. ssDNA samples were separated on an 8% DNA sequencing gel. For S1-cleaved samples (Figure 3B), equal amounts of radioactivity were loaded for the input, bound and free samples.
ATPase assays
ATP hydrolysis assays were performed as described (Klemm et al., 1997) in a 25 µl reaction containing 1 pmol of ORC, and 4 pmol of DNA where indicated. In each experiment, the rate of ATP hydrolyzed was normalized to the activity seen in the absence of ORC. The averages and standard deviations for three independent experiments are shown.
ORC electron microscopy
ORC–ssDNA complexes were formed in a 10 µl reaction containing 20 mM HEPES–KOH pH 7.6, 2 mM EDTA, 2 mM EGTA, 5 mM magnesium acetate, 0.15 M KCl, 725 fmol of ORC and 8 pmol of oligonucleotide ssDNA. ORC–dsDNA binding reactions were performed in 50 µl of the same buffer, with 530 fmol of ORC, 53 fmol of ARS1-containing dsDNA (575 bp PCR DNA) and 100 µM ATP. Binding reactions were incubated at room temperature for 10–17 min. Samples were either not diluted, diluted 5-fold or diluted 10-fold in the same buffer, mounted on glow-charged carbon supports and stained with a 2% aqueous solution of uranyl acetate as described (Makhov et al., 1996). A Philips CM12 transmission electron microscope was used to analyze the samples. The proportion of bent complexes was determined by counting complexes directly on the microscope screen or from negatives of images. These experiments were performed at least twice for each DNA. ORC–dsDNA complexes were counted only if dsDNA was detected in association with ORC. In other experiments, ORC–dsDNA samples were treated with 0.6% glutaraldehyde for 10 min at room temperature, purified on a 2 ml BioGel A5m column (Bio-Rad) prior to mounting on carbon supports, and analyzed by electron microscopy using uranyl acetate staining or rotary shadow-casting with tungsten as described (Griffith and Christiansen, 1978). Neither fixation and column purification nor rotary shadowing altered the proportion of bent ORC complexes when bound to dsDNA (data not shown).
The angle between the two arms of straight or bent ORC complexes was measured by analyzing ORC alone or ORC plus DL15 oligonucleotide samples respectively using NIH Image software. To determine the average angle for a population of molecules in each of two experimental conditions (ORC alone or ORC plus ssDNA), 100 molecules were randomly selected for measurements regardless of their conformation. Figures were prepared for publication by scanning the negatives using a Nikon Film Scanner LS-4500AF (Nikon Corporation, Japan) and Adobe Photoshop software was used to adjust the brightness and contrast.
Acknowledgments
Acknowledgements
We thank Mark Biggin for advice on native gel separation of ssDNA, Richard Austin for anti-ORC polyclonal sera, Anthony Schwacha for ScRPA purification, and Tania Baker and Carl Pabo for critical reading of the manuscript. This work was supported by a National Institutes of Health grant (GM52339) and the Rita Allen Foundation. D.G.L. was supported in part by a 1967 Science and Engineering Scholarship from the Natural Sciences and Engineering Research Council of Canada.
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Stout,A.M. and Bell,S.P. (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- Baker T.A. and Bell,S.P. (1998) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Kobayashi,R. and Stillman,B. (1993) Yeast origin recognition complex functions in transcription silencing and DNA replication. Science, 262, 1844–1849. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Mitchell,J., Leber,J., Kobayashi,R. and Stillman,B. (1995) The multi-domain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell, 83, 563–568. [DOI] [PubMed] [Google Scholar]
- Bielinsky A.-K. and Gerbi,S.A. (1999) Chromosomal ARS1 has a single leading strand start site. Mol. Cell, 3, 477–486. [DOI] [PubMed] [Google Scholar]
- Boehmer P.E. and Lehman,I.R. (1997) Herpes simplex virus DNA replication. Annu. Rev. Biochem., 66, 347–384. [DOI] [PubMed] [Google Scholar]
- Borowiec J.A., Dean,F.B., Bullock,P.A. and Hurwitz,J. (1990) Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell, 60, 181–184. [DOI] [PubMed] [Google Scholar]
- Chesnokov I., Gossen,M., Remus,D. and Botchan,M. (1999) Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev., 13, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Drury L.S., Perkins,G. and Diffley,J.F. (1997) The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J., 16, 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A. and Bell,S.P. (1997) Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol., 13, 293–332. [DOI] [PubMed] [Google Scholar]
- Griffith J.D. and Christiansen,G. (1978) Electron microscope visual ization of chromatin and other DNA–protein complexes. Annu. Rev. Biophys. Bioeng., 7, 19–35. [DOI] [PubMed] [Google Scholar]
- Hwang D.S. and Kornberg,A. (1992) Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J. Biol. Chem., 267, 23087–23091. [PubMed] [Google Scholar]
- Klemm R.D., Austin,R.J. and Bell,S.P. (1997) Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell, 88, 493–502. [DOI] [PubMed] [Google Scholar]
- Labib K., Tercero,J.A. and Diffley,J.F. (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Landis G., Kelley,R., Spradling,A.C. and Tower,J. (1997) The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc. Natl Acad. Sci. USA, 94, 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.G. and Bell,S.P. (1997) Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol., 17, 7159–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.J. and Herskowitz,I. (1993) Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science, 262, 1870–1874. [DOI] [PubMed] [Google Scholar]
- Loo S., Fox,C.A., Rine,J., Kobayashi,R., Stillman,B. and Bell,S.P. (1995) The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol. Biol. Cell, 6, 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhov A.M., Boehmer,P.E., Lehman,I.R. and Griffith,J.D. (1996) Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol., 258, 789–799. [DOI] [PubMed] [Google Scholar]
- Marahrens Y. and Stillman,B. (1992) A yeast chromosomal origin of replication defined by multiple functional elements. Science, 255, 817–823. [DOI] [PubMed] [Google Scholar]
- Micklem G., Rowley,A., Harwood,J., Nasmyth,K. and Diffley,J.F. (1993) Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature, 366, 87–89. [DOI] [PubMed] [Google Scholar]
- Park K., Debyser,Z., Tabor,S., Richardson,C.C. and Griffith,J.D. (1998) Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J. Biol. Chem., 273, 5260–5270. [DOI] [PubMed] [Google Scholar]
- Piatti S., Lengauer,C. and Nasmyth,K. (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J., 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Cocker,J.H., Harwood,J. and Diffley,J.F.X. (1995) Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J., 14, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F.X. (1996) ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J., 15, 6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Santocanale C., Sharma,K. and Diffley,J.F. (1999) Activation of dormant origins of DNA replication in budding yeast. Genes Dev., 13, 2360–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta D.J. and Borowiec,J.A. (1992) Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science, 256, 1656–1661. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tercero J.A., Labib,K. and Diffley,J.F. (2000) DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J., 19, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Hori,Y., Shinomiya,T., Obuse,C., Tsurimoto,T., Yoshikawa,H. and Shirahige,K. (1997) The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells, 2, 655–665. [DOI] [PubMed] [Google Scholar]
- Yang L., Mohr,I., Fouts,E., Lim,D.A., Nohaile,M. and Botchan,M. (1993) The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 90, 5086–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (1998) Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol., 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]