Abstract
Heterocyst differentiation in the filamentous cyanobacterium Anabaena PCC 7120 requires a functional hetR gene. Increased expression of the hetR gene is seen in developing and mature heterocysts in response to fixed nitrogen limitation. We mapped four likely transcriptional start sites for hetR and identified a specific transcript that is positively autoregulated. By using the copper-responsive petE promoter from Anabaena PCC 7120 to drive hetR expression, we show that ectopic expression of hetR increases heterocyst frequency and induces heterocyst differentiation under fully repressing conditions. Coexpression of a reporter gene shows that expression from the petE promoter is smoothly induced depending on the amount of copper supplied. In the heterocyst pattern mutant PatA, where terminally positioned heterocysts are formed almost exclusively, expression of the petE∷hetR fusion does not result in the formation of intercalary heterocysts. These results suggest that although the intracellular concentration of HetR has to be elevated for the differentiation decision, PatA plays a role as well. This role may be in the form of posttranslational modification of HetR, because PatA is a member of the response regulator family of proteins.
The cyanobacterium Anabaena grows in filaments of several hundred cells, each carrying out oxygen-evolving photosynthesis when there is an adequate supply of fixed nitrogen, such as nitrate or ammonia, in the environment. Deprived of fixed nitrogen, the filaments differentiate specialized cells, called heterocysts, spaced at nearly regular intervals. In the strain we study, Anabaena PCC 7120, heterocysts form at the ends of filaments and at approximately every 10th cell along their length, although the spacing can vary somewhat. Heterocysts are specialized nondividing anaerobic cells in which nitrogen fixation occurs, anaerobiosis being required for the activity of nitrogenase. Differentiation includes the inactivation of the oxygen-evolving photosystem II and the synthesis of a glycolipid layer external to the cell wall, which limits diffusion of gases into the heterocyst. These cells communicate with their neighboring vegetative cells through slender cytoplasmic bridges, providing amino acids and receiving carbohydrates. The differentiation process begins within a few hours of nitrogen step-down (removal of fixed nitrogen), and requires approximately 24 h to complete.
The formation and spacing of the heterocyst pattern is thought to depend not only on the physiological state of the individual cells along the filament, but also on the specific concentration of a postulated inhibitor of differentiation produced by developing and mature heterocysts. This inhibitor is thought to be broken down in vegetative cells, setting up a concentration gradient along the filament. This gradient would be responsible not only for the establishment of the initial pattern, but also its maintenance during diazotrophic growth. One potential candidate for this inhibitor is a small peptide produced from the patS gene, which apparently acts early in the differentiation process to prevent the formation of multiple contiguous heterocysts (1).
We have previously characterized several genes whose products are required for the normal heterocyst spacing pattern in Anabaena PCC 7120. Mutation of the hetR gene blocks the earliest steps in differentiation, without affecting vegetative growth. When a plasmid-borne hetR gene is introduced into wild-type Anabaena PCC 7120, the filaments are severely perturbed on nitrate or nitrogen-free media, differentiating supernumerary heterocysts in strings of two, three, or more. These results were interpreted to mean that HetR protein is titrated by the inhibitor postulated above and that increasing the level of HetR modifies the inhibitory threshold for differentiation (2). A second gene, patA, encodes a protein that appears to interact with HetR (3). A mutation in PatA leads to filaments in which differentiation is confined mostly to cells at the ends of the filaments. Although these heterocysts are capable of reducing acetylene, and therefore have active nitrogenase, they cannot provide fixed nitrogen along the filament to sustain vegetative growth. When the plasmid-borne hetR gene is introduced into the PatA strain, the phenotype remains that of PatA. Thus, the hetR and patA genes are in the same regulatory circuit.
To further examine the relationships between these genes and their products, we wished to establish a system for the independent and controlled expression of each one. Such a system would have other obvious advantages for the exploration of regulatory circuits. The system chosen is based on the observation of Straus and coworkers that transcription of the gene encoding the copper protein plastocyanin in the cyanobacterium Synechococcus PCC 7942 is regulated by copper (4). We describe here the use of the copper-regulated petE promoter from Anabaena PCC 7120 to drive transcription of the hetR gene in both wild-type and a PatA mutant of Anabaena PCC 7120.
Materials and Methods
Plasmid Constructions.
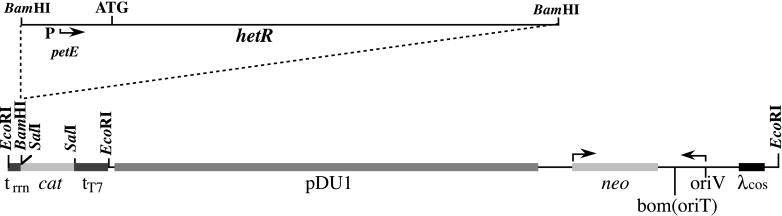
The petE promoter was amplified by PCR using the following two primers: 5′-GGATC CCAGT ACTCA GAATT TTTTG CT-3′ and 5′-GAATT CCATG GCGTT CTCCT AACCT G-3′. The resulting 372-bp fragment was blunt-end cloned into the HincII site of pUC19 to generate pPet1. The 5′-terminal region of the hetR gene was amplified by PCR using the following two primers: 5′-GGAAT TCACT CTGGG TGCTT AAT-3′ and 5′-GGAAT TCCAA TGAGT AACGA CATCG-3′. The resulting fragment was blunt-end cloned into the HincII site of pUC19 to generate pHet1. This fragment was fused to the petE promoter in pPet1 by digesting pHet1 with BspHI and XbaI and cloning it into pPet1 digested with NcoI and XbaI to produce pPetHet-prime. The complete hetR gene was reconstructed by subcloning an XbaI fragment of the wild-type gene into the XbaI site of pPetHet-prime to form pPetHetpuc. The final construction was made by removing the PpetE∷hetR fusion from pPetHetpuc using BamHI and cloning it into the BamHI site of pJL3 to generate pPetHetR (Fig. 3). It should be noted that pPetHetR forms a transcriptional fusion of the hetR gene to the cat gene.
Figure 3.
Structure of the plasmid pPetHetR, containing the Anabaena hetR gene, whose expression is driven by the petE promoter. Note that a cat gene reporter is located downstream of the hetR gene.
Cell Growth and Induction.
Standard growth conditions for Anabaena PCC 7120 in BG-11 liquid and on plates have been described (5). When Anabaena strains containing the petE promoter were being constructed, we used a modified BG-11 medium without copper sulfate and acid-washed, dry-heat-sterilized glassware, or disposable plasticware.
Cells were induced with copper by washing exponentially growing cells with fresh BG-11 medium, BG-11 containing no fixed nitrogen (BG-110), or BG-11 with 1 mM ammonium sulfate. Specified concentrations of total copper were attained by adding dissolved copper sulfate as needed. For liquid cultures, cells were grown in flasks with shaking at 150 rpm under continuous illumination at 32°C for 2 days. For slides, 10 μl of a dense cell suspension was placed in the center of a 300-μl 1% (wt/vol) agarose slab containing the appropriate medium, copper, and 10 mM potassium bicarbonate, and covered with a coverslip. Slides were incubated in a clear humid chamber under the same light conditions as the liquid cultures.
RNA Isolation.
Total RNA was isolated at specified times from 250-ml aliquots of cells induced in 2 liters of BG-110 medium. The cells were harvested by using vacuum filtration over 5-μm nylon filters and were flash frozen in liquid nitrogen. Frozen cells were broken, and total RNA was isolated by using glass beads and phenol as described ref. 2.
Northern Gel Analysis and Primer Extension.
A total of 5 μg of RNA from each sample was loaded on a 1.2% (wt/vol) agarose gel as described (2). After electrophoresis and transfer to a GeneScreen Plus membrane, the RNA was fixed by drying. An internal HincII fragment from hetR was random-labeled with 32P and used as probe.
Two primers, hetR-PEX2 (5′-CAAGATGCTCATTCCTCCGT-3′) and hetR-PEX3 (5′-TGCTCTTATGGCAGTGTAGG-3′) were end-labeled with 32P and used as probes for primer extension with 18-h RNA from induced cultures as substrate as described (6).
Nitrogen Fixation Assays.
Cells in liquid culture were induced for a period of 42 h after washing and transfer to BG-110 medium. Then, 2 ml of cells at roughly the same density were placed in 5-ml glass vials and stoppered with rubber injection caps. Next, 0.5 ml of air was withdrawn from each vial and replaced with an equal volume of acetylene, and the vials were incubated for 2 h under standard growth conditions as described above. Samples of gas from each vial were withdrawn and injected into a gas chromatograph to determine the amount of ethylene produced (7).
Cell Extracts and Total Protein Determination.
Cell extracts were prepared by vortexing harvested cells with 150 μl of glass beads (450–600 μm; Sigma) in a 1.5-ml plastic tube for 1.5 min in a buffer containing 10 mM Tris (pH 8.0), 1 mM EDTA, and 2 mM PMSF. Cell debris was removed by centrifugation at 16,000 × g for 4 min in a microfuge at 4°C and the supernatant was collected.
Protein assays were performed on cell extracts by using the Protein Assay kit from Bio-Rad, following the manufacturer's instructions for microassays.
Chloramphenicol Acetyltransferase (CAT) Assays.
Total CAT protein was detected with an Elisa kit from 5-Prime→3-Prime. Anabaena cells containing pPetHetR or pJL3 were grown to mid-log phase (approximately 2–4 μg/ml chlorophyll) in a modified BG-11 medium lacking copper and containing 35 μg/ml neomycin sulfate (Sigma), 1 mM (NH4)2S04, and 10 mM NaHCO3. Aliquots (5 ml) of these cells were placed into plastic tubes, and specific amounts of CuSO4 were added. After incubation for 48 h, heterocysts were viewed in a light microscope and CAT assays were performed on cell extracts. For certain cultures with high numbers of heterocysts, cells were induced by placement on agar plates containing copper, and heterocyst numbers were determined by light microscopy after 48 h.
Microscopy.
Cells on slides with agar slabs were observed under a ×40 dry objective on a Zeiss Axiophot microscope equipped with differential interference contrast microscopy and fluorescence optics. Photomicrographs were taken with Fujichrome ISO 64 daylight film.
Caution.
One comment regarding the methodology used for the copper induction follows: To reduce the background level of expression from the petE promoter, it was essential to acid-wash the glassware and to substitute plasticware wherever possible. Autoclaving glassware was avoided in preference to oven-baking to avoid steam deposition of trace copper. All solutions for growing the Anabaena strains were filter-sterilized.
Results
Identification of Transcriptional Start Sites.
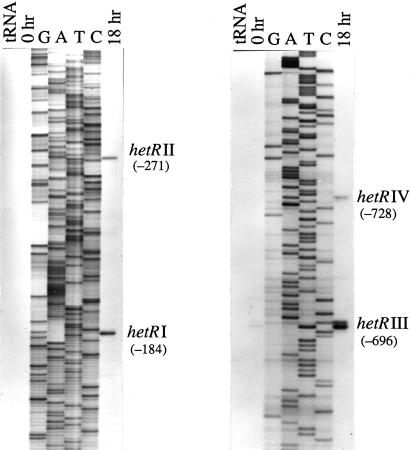
It seemed likely that the hetR promoter would show complex control, so as a first step in characterizing this control, we determined the likely transcriptional start sites. We used RNA isolated from cells grown in medium containing ammonia (zero time) and RNA isolated from cells 18 h after resuspension in a nitrogen-free medium in primer extension experiments. As shown in Fig. 1, there are four likely transcription start sites that clearly appear in the 18-h lanes, located at positions −184, −271, −696, and −728, relative to the start codon of HetR. Corresponding bands appear in the zero-hour lanes but are of much lower intensity.
Figure 1.
Identification of four potential transcription start sites of the hetR gene by primer extension. RNA isolated from cells before and 18 h after nitrogen starvation was used with specific labeled primers.
These results are in agreement with our previously reported results showing two major hetR transcripts of 1.9 and 1.4 kb on a Northern gel (2). The 1.4-kb band corresponds to the two smaller products, whereas the 1.9-kb band corresponds to the two larger. The 900-bp hetR coding region ends approximately 300 bp before the 3′ terminus of either message. There is a gap of nearly 800 bases after hetR before the start of the next ORF (an amino acid transferase; data not shown).
Early Expression of hetR.
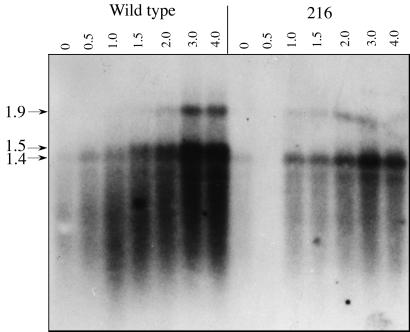
To help characterize how hetR is expressed immediately after nitrogen step-down, RNA was isolated from induced cells at short intervals and run on a moderately high percentage agarose gel in an attempt to separate the specific transcripts. For comparison, RNA was isolated in parallel from the hetR point mutant 216, which cannot initiate differentiation. As shown in Fig. 2, an internal hetR fragment probe distinguishes three separate transcripts of 1.4, 1.5, and 1.9 kb that quickly increase in the wild type. In contrast, the hetR mutant 216 shows a reduced induction of the 1.9- and 1.4-kb transcripts, and virtually no induction of the 1.5-kb transcript. This transcript corresponds to the −271 transcription start site seen in Fig. 1, which provides a clear indication of autoregulation of hetR.
Figure 2.
Northern gel analysis of the early expression of hetR after nitrogen starvation in wild type and the hetR point mutant 216. The lane labels refer to hours postinduction. The 0.5-h 216 lane is blank, likely to be a result of sample degradation, and the 1.9-kb band in the 216 3.0- and 4.0-h lanes is distorted because of an air bubble during transfer of the RNA.
This result suggests that high, sustained expression of HetR is necessary to induce differentiation of heterocysts. It was already known that complementation of the hetR mutant 216 with a multicopy plasmid carrying hetR showed high heterocyst frequencies (2). To determine whether hetR expression is sufficient to induce differentiation, we expressed hetR from a promoter that could be controlled ectopically.
Vector Construction.
The promoter of the petE gene, encoding the protein plastocyanin, was shown to respond to copper added to the medium in which the cyanobacterium Anabaena PCC 7120 was growing (4). Based on the DNA sequence, we PCR-amplified the promoter region of petE from Anabaena PCC 7120, fused it to the native hetR gene, and inserted it into pJL3 (8) to form pPetHetR (Fig. 3). Features of this plasmid include transcription terminators upstream of the petE promoter and downstream of the cat gene, a gene encoding neomycin resistance, replicons for both Escherichia coli and Anabaena, and a mobilization origin for transfer from E. coli to Anabaena by conjugation. This plasmid was transferred by conjugation to wild-type Anabaena PCC 7120, selecting for resistance to neomycin.
CAT Expression from the petE Promoter.
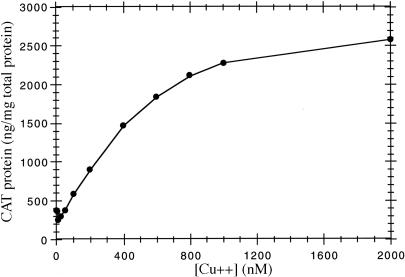
The control of expression from this construct can be monitored by determining the level of CAT antigen, whose coding region is downstream of both the petE promoter and the hetR gene. Fig. 4 shows the induction of CAT antigen as a function of added copper concentration in a culture of Anabaena grown with added ammonia. There is a smooth increase in CAT antigen over the range of added copper up to 2 μM. The latter concentration is close to the maximum tolerated by wild-type Anabaena PCC 7120 (9).
Figure 4.
Dependence of CAT antigen induction as a function of added copper ion in wild-type Anabaena 7120 containing pPetHetR. Cells were grown for 24 h in BG-11 medium containing 1 mM (NH4)2S04, and CAT antigen was assayed as described in Materials and Methods.
HetR Expression from the petE Promoter.
HetR expression induced by added copper has dramatic consequences. The frequency of heterocysts in wild-type Anabaena PCC 7120 cultures grown with ammonia or nitrate as nitrogen source is zero or 1%, respectively. At 24 h after nitrogen step-down, the heterocyst frequency is approximately 10%. The same is true for the pPetHetR-containing strain in the absence of added copper. However, when 2 μM copper is added to the latter strain, the heterocyst frequency becomes 21, 23, and 29%, respectively, in ammonia, nitrate, and nitrogen-free medium (n = 500, Fig. 5). Intermediate numbers of heterocysts are produced at correspondingly lower concentrations of copper (data not shown). The key result of this experiment is that all of the upstream controls of HetR expression can be bypassed; expression of HetR alone suffices to turn on the differentiation pathway.
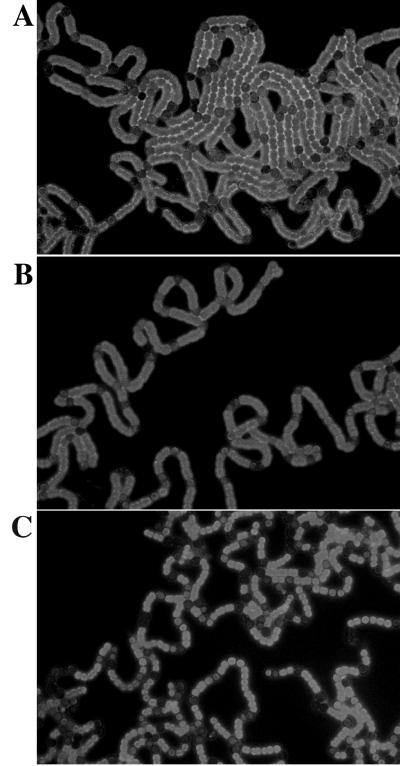
Figure 5.
Chlorophyll fluorescence in Anabaena cultures 2 days after the addition of 2 μM Cu2+ to induce expression of the hetR gene. In each case, the culture contained wild-type Anabaena carrying the plasmid pPetHetR, and the fluorescence was viewed with a wide bandpass filter. (A) The culture contained 1 mM (NH4)2S04. (B) The culture contained 17 mM nitrate. (C) The culture contained no fixed nitrogen. The heterocysts can be identified by the low fluorescence attributable to the loss of chlorophyll from photosystem II and the degradation of the phycobiliproteins.
Although all of the heterocysts appear similar in the fluorescence images shown in Fig. 5, they are not functionally the same. In the cultures containing ammonia or nitrate, there is no nitrogenase activity detectable by the acetylene reduction assay. In the nitrogen-free culture, the activity is roughly comparable to that of the same culture induced without added Cu2+ (data not shown).
In nitrogen-free media, the Cu2+-induced culture had 29% heterocysts and nitrogenase activity comparable to a culture induced by nitrogen step-down alone, in which the heterocyst frequency is 10%. In the former case, it can be supposed that the remaining vegetative cells cannot reduce carbon fast enough to keep up with the demand from heterocysts for reductant. Indeed, cultures with this many heterocysts grow poorly (2).
HetR Expression from the petE Promoter in a PatA Mutant.
The Anabaena PatA mutant makes heterocysts mainly at the ends of filaments (3). When a hetR-containing plasmid, which complements a HetR mutant and causes multiple contiguous heterocysts in wild-type Anabaena PCC 7120, is introduced into the PatA mutant, heterocysts differentiate only at the ends of the filaments (3). Thus, the PatA mutation suppresses the multiheterocyst phenotype normally produced by extra copies of the hetR gene. Further evidence for interaction of the hetR and patA gene products come from the pattern of expression from the hetR promoter studied with a hetR∷gfp fusion. When such a fusion is introduced into wild-type Anabaena PCC 7120, green fluorescence protein fluorescence appears in the developing heterocysts. The same fusion introduced into a PatA mutant results in fluorescence only in the differentiating cells at the ends of filaments (W.J.B., unpublished data). Thus, functional PatA is required for the increase in expression of the hetR gene in internal cells of the filament.
These experiments do not distinguish between two possibilities: PatA is required either for hetR transcription or for HetR modification. In the latter case, HetR modification would have to be involved in the increase in hetR transcription in heterocysts. In either case, the role of PatA could be indirect, involving other factors as well. With the petE∷hetR fusion it became possible to rule out the first possibility. The Cu2+-regulated hetR plasmid was introduced into a PatA mutant. The addition of Cu2+ that led to 29% heterocysts in the wild type did not increase the frequency of heterocysts in the PatA mutant. This result means that PatA is not involved in the transcription of the hetR gene directly, but rather is required for HetR modification or some other posttranscriptional step.
Discussion
Previously we found that extrachromosomal copies of the hetR gene cause heterocyst differentiation in nitrogen-replete media and the formation of multiple contiguous heterocysts in nitrogen-free media (2). By using the copper-responsive petE promoter fused to the hetR gene, we now show that the level of expression of hetR is correlated with the number of heterocysts produced along the filaments. The petE promoter shows a smooth induction of expression within a physiological range of copper concentrations, and will be a useful tool for the regulated expression of other genes in Anabaena 7120.
There is further information on the relationship between HetR and PatA. Studies with the hetR∷gfp fusion, mentioned above, show that hetR is normally transcribed in developing heterocysts. Shortly after a nitrogen step-down, most cells show an increase in fluorescence. Then, as proheterocysts develop, fluorescence in the vegetative cells decreases whereas that in the proheterocysts increases. In the PatA mutant, the vegetative cells are not fluorescent, meaning that they do not transcribe the hetR gene. However, if the same hetR∷gfp fusion is introduced into a HetR mutant strain, all of the cells show an increase in fluorescence. There is no pattern of bright developing cells interspersed among dull vegetative cells.
These results indicate that hetR gene expression responds in two ways to HetR itself: in developing cells, HetR activates hetR expression; in vegetative cells under conditions allowing heterocyst differentiation, hetR expression is repressed. Only one of these functions requires PatA. Recall that the hetR∷gfp fusion placed in a PatA mutant results in a strain that shows fluorescence only in the terminal cells that differentiate. The hetR gene in vegetative cells is repressed as it is in the wild type. Therefore, PatA is required for the activation of HetR and the increased expression of hetR in developing cells, but it is not required for the repression of hetR in vegetative cells.
The lack of nitrogenase activity in the nitrogen-replete cultures induced to differentiate heterocysts using the petE promoter is not surprising. Recall the properties of the NtcA protein of Anabaena PCC 7120. NtcA is a transcription activator responsible for inducing genes encoding nitrate transport, nitrate reductase, nitrite reductase, glutamine synthetase, nitrogenase, and the excision enzyme required for nif gene rearrangement. The NtcA protein binds to the consensus sequence TGT- (N9–10)-ACA. NtcA-dependent promoters are active in nitrate-containing media and repressed in ammonia-containing media. In nitrogen-free media, NtcA has all these responsibilities in addition to activating hetR. The hetR gene promoters described here all lack a canonical NtcA-binding site, so this requirement for NtcA must be indirect (10).
In ammonia-containing media, NtcA is not active. Accordingly, the nif genes cannot be transcribed or rearranged and the observation of no nitrogenase activity is expected. In nitrate-containing medium, NtcA is active, so all of the genes it regulates are active, and ammonia is generated internally. Regardless of the state of expression of the nif and xis genes, the ammonia will inactivate nitrogenase itself. Thus the observation of no nitrogenase activity is expected in this case as well.
The PatA protein sequence indicates that it is the response regulator of a two-component environment-sensing system. PatA has the conserved Asp residue that one expects to be phosphorylated by a histidine kinase. The immediate neighborhood of the patA gene does not contain a kinase gene. Response regulators of the PatA class demonstrate one of three functions: they may phosphorylate other proteins, they may bind to DNA and activate transcription, or they may cause rotation of a flagellar motor (11).
The HetR protein sequence is not related to any other in GenBank and it has no recognizable domains with known function. However, the original HetR mutation in our mutant strain is a missense mutation that converts Ser→Asn. The Ser seems to be very important. Five revertants picked from the Ser→Asn strain were all found to have reverted precisely Asn→Ser. No second-site suppressors or revertants to other amino acids were found (W.J.B., unpublished results).
Why should the Ser be critical? One formal possibility is that HetR is a protease (2). A protease ought to act catalytically, whereas HetR acts stoichiometrically, based on the Cu2+-induction experiment. However, a great deal of evidence from the laboratory of J. Zhao in Beijing (College of Life Sciences, Peking University) indicates that HetR may indeed be a protease (12). First, purified recombinant HetR expressed in E. coli has autodigestion activity, inhibited by phenylmethane sulfonyl fluoride and dansyl fluoride. The latter compound derivatizes the HetR protein, determined by mass spectroscopy of isolated peptide containing the dansyl group. This peptide sequence is internal to HetR. Thus, HetR is either a totally new kind of serine protease or it is a different kind of protease requiring activation by phosphorylation on serine, yet is reactive with dansyl fluoride (12).
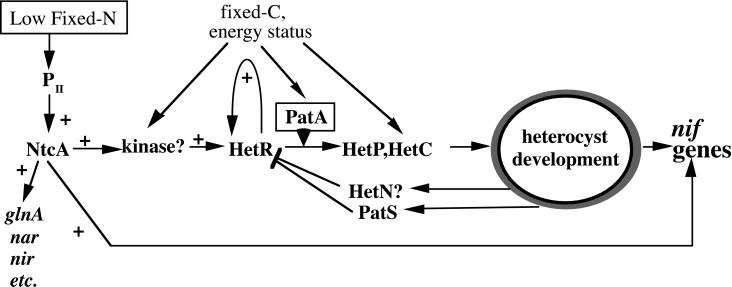
The second possibility is that HetR is a protein kinase or transcription activator that is itself phosphorylated on the critical Ser residue. If PatA is the kinase (direct or indirect) of HetR, the relationship between the two would be clear. In wild type, PatA would phosphorylate HetR only in developing cells, resulting in activation of hetR expression. In vegetative cells, hetR expression is repressed in both the wild type and the PatA mutant, because unphosphorylated HetR does not activate hetR expression. These relationships are included in Fig. 6.
Figure 6.
Model of the relationships among some of the gene products that control heterocyst differentiation in Anabaena. Normally, the environmental signal of low fixed nitrogen is perceived by the NtcA protein, as modified by the regulator PII. NtcA activates transcription of many genes, eventually including hetR. When modified by PatA, HetR activates expression of itself and other genes leading to heterocyst differentiation. HetR action is countered by both PatS and HetN.
Fig. 6 shows NtcA to be upstream of hetR. An NtcA mutant does not activate expression of hetR. However, the hetR promoters do not contain the canonical binding site for NtcA, so this activation is probably indirect. Presumably the missing components are used when the cells encounter severe nitrogen-starvation conditions. Candidates for such components might be two-component systems or eukaryotic-type serine/threonine kinases (13). These systems may be used to integrate diverse nutrition and environmental signals in addition to the nitrogen-starvation signal. Indeed, analysis of the Anabaena PCC 7120 genome released by Kazusa (Kazusa DNA Research Institute, http://www.kazusa.or.jp/cyano/) indicates the presence of over 100 two-component systems, and over 30 serine/threonine kinases, including a new class of 13 hybrid kinases that contain an N-terminal serine/threonine domain linked to a C-terminal histidine kinase domain (data not shown).
Acknowledgments
We thank Neil Straus (University of Toronto) for providing information about the petE promoter, and Sean Callahan and Kathryn Jones for critical reading of the manuscript. This work was supported by Research Grant GM-21823 from the National Institute of General Medical Sciences.
Abbreviation
- CAT
chloramphenicol acetyltransferase
References
- 1.Yoon H S, Golden J W. Science. 1998;282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 2.Buikema W J, Haselkorn R. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 3.Liang J, Scappino L, Haselkorn R. Proc Natl Acad Sci USA. 1992;89:5655–5659. doi: 10.1073/pnas.89.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghassemian M, Wong B, Ferreira F, Markley J L, Straus N A. Microbiology. 1994;140:1151–1159. doi: 10.1099/13500872-140-5-1151. [DOI] [PubMed] [Google Scholar]
- 5.Buikema W J, Haselkorn R. J Bacteriol. 1991;173:1879–1885. doi: 10.1128/jb.173.6.1879-1885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahamsha B, Haselkorn R. J Bacteriol. 1991;173:2442–2450. doi: 10.1128/jb.173.8.2442-2450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming H, Haselkorn R. Cell. 1974;3:159–170. doi: 10.1016/0092-8674(74)90121-4. [DOI] [PubMed] [Google Scholar]
- 8.Lang J D, Haselkorn R. J Bacteriol. 1991;173:2729–2731. doi: 10.1128/jb.173.8.2729-2731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phung L T, Ajlani G, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:9651–9654. doi: 10.1073/pnas.91.20.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frias J E, Flores E, Herrero A. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 11.Swanson R V, Alex L A, Simon M I. Trends Biochem Sci. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou R, Wei X, Jiang N, Li H, Dong Y, Hsi K-L, Zhao J. Proc Natl Acad Sci USA. 1998;95:4959–4963. doi: 10.1073/pnas.95.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C-C. Proc Natl Acad Sci USA. 1993;90:11840–11844. doi: 10.1073/pnas.90.24.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]