Abstract
The yeast transcription factor Yap1 activates expression of antioxidant genes in response to oxidative stress. Yap1 regulation involves nuclear accumulation, but the mechanism sensing the oxidative stress signal remains unknown. We provide biochemical and genetic evidence that upon H2O2 treatment, Yap1 is activated by oxidation and deactivated by enzymatic reduction with Yap1-controlled thioredoxins, thus providing a mechanism for autoregulation. Two cysteines essential for Yap1 oxidation are also essential for its activation by H2O2. The data are consistent with a model in which oxidation of Yap1 leads to disulfide bond formation with the resulting change of conformation masking recognition of the nuclear export signal by Crm1/Xpo1, thereby promoting nuclear accumulation of the protein. In sharp contrast to H2O2, diamide does not lead to the same Yap1 oxidized form and still activates mutants lacking cysteines essential for H2O2 activation, providing a molecular basis for differential activation of Yap1 by these oxidants. This is the first example of an H2O2-sensing mechanism in a eukaryote that exploits the oxidation of cysteines in order to respond rapidly to stress conditions.
Keywords: oxidation/oxidative stress/sensing/thiol/Yap1
Introduction
Living cells have adaptive responses to oxidative stress, indicating that they sense increased levels of reactive oxygen species and respond to the signal by increasing the expression of defence activities. The mechanisms by which cells sense reactive oxygen species are relatively well understood in prokaryotes (Zheng and Storz, 2000), but not in eukaryotes.
The yeast transcription factor Yap1, a bZIP DNA-binding protein of the AP-1 family (Moye-Rowley et al., 1989), is an essential regulator of the H2O2 adaptive response (Schnell et al., 1992; Kuge and Jones, 1994) and controls many proteins of the H2O2 stimulon (Lee et al., 1999a). These include most cellular antioxidants and enzymes of the glutathione and pentose phosphate pathways.
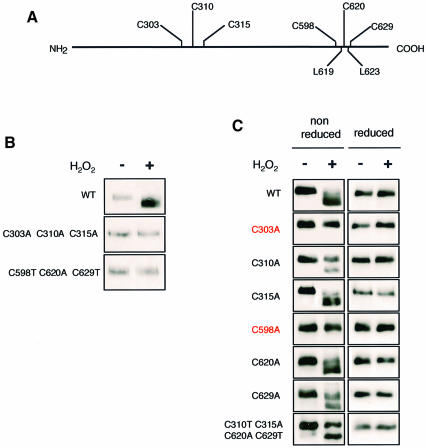
Yap1 is activated upon exposure to peroxides or to thiol-modifying drugs such as diamide (Kuge and Jones, 1994) by a mechanism acting at the level of subcellular protein localization (Kuge et al., 1997). Yap1 is restricted to the cytoplasm by virtue of rapid nuclear export by the export receptor Crm1/Xpo1 (Kuge et al., 1998; Yan et al., 1998). Upon exposure to oxidants, Yap1 accumulates in the nucleus due to loss of the Yap1–Crm1 interaction. Yap1 contains a short C-terminal domain (the CRD) carrying a non-canonical leucine rich nuclear export signal (NES) embedded in three repeated cysteine-containing motifs (see Figure 3A), which is important for the interaction with Crm1 (Kuge et al., 1997, 1998; Yan et al., 1998). Oxidation of these cysteines might modify the NES in a way that prevents recognition by Crm1, as suggested by (i) the constitutive cytoplasmic phenotype of a Yap1 allele bearing substitutions of all three C-terminal CRD cysteines (Kuge et al., 1998), (ii) the dithiothreitol (DTT) enhancement of the co-precipitation of Crm1 with Yap1 (Yan et al., 1998) and (iii) the sensitivity of the Yap1–Crm1 two-hybrid interaction to diamide (Yan et al., 1998). Yap1 has another cluster of three cysteines (C303, C310, C315), but these appear not to be involved in Yap1–Crm1 interaction (Yan et al., 1998). However, deletion mutagenesis has shown that a mutant protein lacking an internal segment between amino acids 225 and 335 was still inducible by diamide, but not by H2O2 (Wemmie et al., 1997). This suggested the intriguing possibility that different mechanisms are used to activate Yap1 by these two oxidants, and that Yap1 N-terminal cysteines might be important for activation by H2O2 but not by diamide.
Fig. 3. Identification of Yap1 redox-active cysteines. (A) Schematics of Yap1. (B) Both N- and C-terminal cysteines are required to oxidize Yap1. Δyap1 strain cultures expressing Myc-Yap1, Yap1C303A,C310A,C315A or Yap1C598T,C620A,C629T before and 5 min after treatment with 400 µM H2O2 were processed as in Figure 2A (lanes 1 and 2) and immunoblotted after non-reducing SDS–PAGE. (C) Cysteines C303 and C598 are the redox-active Yap1 cysteines. Δyap1 strain cultures expressing Myc-Yap1, Yap1C303A, Yap1C310A, Yap1C315A, Yap1C598A, Yap1C620A or Yap1C629A, and Yap1C310T,C315A,C620A,C629T before and 5 min after H2O2 treatment, as in Figure 2B and immunoblotted after non-reducing or reducing SDS–PAGE, as indicated.
We sought to understand how H2O2 activates Yap1. We provide biochemical and genetic evidence that, in response to H2O2, Yap1 is activated by oxidation and deactivated by enzymatic reduction with Yap1-controlled thioredoxins, thus providing a mechanism for autoregulation. Two cysteines essential for Yap1 activation by H2O2 are likely to form a disulfide bond upon oxidation. In sharp contrast, diamide does not lead to the same oxidized form of Yap1 and still activates mutants lacking cysteines essential for the H2O2 response, thus providing a molecular basis for the differential activation of Yap1 by these oxidants. This is the first example of an H2O2-sensing mechanism in a eukaryote that exploits the oxidation of cysteines in order to respond rapidly to stress.
Results
Kinetics of the activation of Yap1 by H2O2
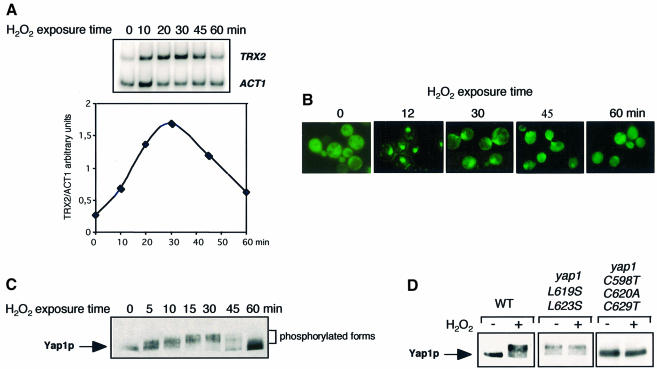
In response to 0.3 mM H2O2, the Yap1 target gene TRX2 is expressed rapidly and transiently, with expression peaking at ∼30 min and returning to baseline after 60 min (Figure 1A). The nuclear accumulation of a green fluorescent protein (GFP)–Yap1 fusion protein and its subsequent cytoplasmic redistribution paralleled the expression of TRX2 (Figure 1B). Nuclear GFP–Yap1 staining was maximal 5 min after H2O2 treatment and was sustained for 30 min. After 45 min, nuclear staining was half-maximal; after 60 min, it was indistinguishable from that of untreated cells. Yap1 activation also correlated with an altered migration of the protein on SDS–polyacrylamide gels. Within 5 min of H2O2 treatment, Myc-tagged Yap1 was shifted to more slowly migrating bands and this migration shift persisted for 1 h, being maximal at 30 min (Figure 1C). This migration shift must have been caused by protein phosphorylation, because it disappeared when extracts were treated with calf intestinal phosphatase (not shown). Yap1 phosphorylation could occur in the nucleus as suggested by (i) its kinetics, (ii) the constitutive phosphorylation of a constitutively nuclear mutant Yap1L619S,L623S and (iii) its absence in the constitutively cytoplasmic mutant Yap1C598T,C620A,C629T (Figure 1D). Therefore, although its role is not elucidated, phosphorylation of Yap1 correlates with its presence in the nucleus.
Fig. 1. Kinetics of Yap1 activation. (A) Expression of TRX2. Total RNA was isolated from a wild-type strain, grown to an OD600 of 0.3 in selective minimal medium, before and 10, 20, 30, 45 and 60 min after treatment with 300 µM H2O2, and TRX2 and ACT1 (control gene) mRNAs were quantified by RT–PCR. An autoradiogram of the separated 32P-labelled PCR products is shown in the upper panel and a plot of the TRX2/ACT1 signal ratio quantified on a PhosphorImager in the lower panel. (B) Nuclear redistribution of Yap1. A Δyap1 strain expressing GFP-Yap1 at an OD600 of 0.3, treated with 300 µM H2O2 for the time indicated and analysed for GFP staining. (C) Yap1 phosphorylation. Extracts of a Δyap1 strain expressing Myc-tagged Yap1 (yMyc-Yap1), before and 5, 10, 15, 30, 45 and 60 min after treatment with 300 µM H2O2 were immunoblotted with the anti-Myc monoclonal antibody after SDS–PAGE. (D) Yap1 phosphorylation correlates with Yap1 nuclear localization. Δyap1 strain cultures expressing Myc-Yap1, Yap1L619S,L623S or Yap1C598T,C620A,C629T, before and 20 min after H2O2 treatment, were processed as in (C).
Yap1 becomes oxidized upon exposure to H2O2
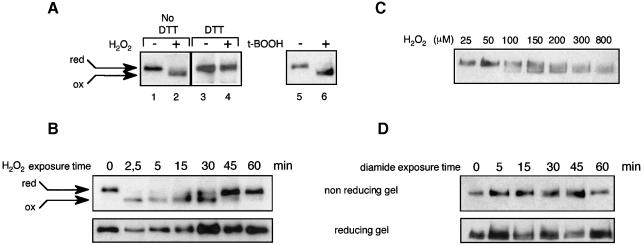
The non-inducible nature of the cytoplasmic Yap1C598T,C620A,C629T mutant demonstrated the importance of the C-terminal cysteines in Yap1 control and further suggested that Yap1 could be activated by direct oxidation. To test this hypothesis, we established a method to analyse directly the in vivo redox state of Yap1. If oxidation leads to covalent modification of Yap1, a change in conformation might be detectable by electrophoresis under non-reducing conditions, as previously shown for OxyR (Tao, 1999). After H2O2 treatment, yeast cells were lysed with trichloroacetic acid (TCA) to protonate all free thiols and the precipitated proteins were re-solubilized with excess iodoacetamide to alkylate free thiols irreversibly and prevent their subsequent oxidation. As suspected, Yap1 from H2O2-treated cells migrated more quickly than the protein from untreated cells under non-reducing conditions (Figure 2A, lanes 1 and 2). However, this mobility shift was not seen when the extract was treated with 200 mM DTT prior to iodoacetamide (Figure 2A, lanes 3 and 4) or after electrophoresis under reducing conditions (Figure 2B). These data demonstrate that the more quickly migrating species observed under non-reducing conditions is an oxidized form of Yap1. Cellular treatment with t-butyl hydroperoxide, an organic peroxide, resulted in a similar oxidation-dependent electrophoretic migration shift (Figure 2A, lanes 5 and 6). We next performed a time-course analysis of the Yap1 redox status after H2O2 treatment. In this experiment, Yap1 was dephosphorylated in vitro to prevent an offset of the redox-dependent migration by the shift induced by phosphorylation. The oxidized form was observed as early as 2.5 min after H2O2 treatment and disappeared at 60 min (Figure 2B). The minimum concentration of H2O2 required to detect oxidized Yap1 was 100 µM and the amount of the oxidized form increased with increasing H2O2 concentration (Figure 2C). Therefore, the appearance of the oxidized form of Yap1 was well correlated with the kinetics of Yap1 activation.
Fig. 2. In vivo oxidation of Yap1. (A) yMyc-Yap1 cultures (grown to an OD600 of 0.4) before and after 2.5 min treatment with 400 µM H2O2 (lanes 1 and 2) or 1 mM t-butyl hydroperoxide (lanes 5 and 6) were lysed using TCA, treated with iodoacetamide and immunoblotted after non-reducing SDS–PAGE. Lanes 3 and 4 are the same as lanes 1 and 2 except that the TCA-precipitated protein pellet was dissolved in the presence of 200 mM DTT, before adding a 3.2 M excess of iodoacetamide. (B) Time course of Yap1 oxidation by H2O2. A yMyc-Yap1 culture before and 2.5, 5, 15, 30, 45 and 60 min after treatment with 400 µM H2O2, processed as in (A) (lanes 1 and 2), except that Yap1 was dephosphorylated and separated under non-reducing or reducing conditions as indicated. (C) The minimum H2O2 concentration required to oxidize Yap1 in vivo. A yMyc-Yap1 culture treated for 5 min with 25, 50, 100, 150, 200, 300 and 800 µM H2O2 and processed as in (A) (lanes 1 and 2). (D) Time-course analysis of Yap1 redox status in response to diamide. A yMyc-Yap1 culture before and 5, 15, 30, 45 and 60 min after treatment with 2 mM diamide, processed as in (A) (lanes 1 and 2) and separated under non-reducing or reducing conditions as indicated.
Since diamide can also activate Yap1-target gene expression and Yap1 nuclear redistribution (Kuge and Jones, 1994; Kuge et al., 1997), we monitored the in vivo redox status of Yap1 after treatment with this oxidant. Unexpectedly, ≤1 h after treatment with 2 mM diamide, a concentration able to activate Yap1 fully (not shown), we did not observe the redox-dependent migration shift of Yap1 observed on non-reducing PAGE after treatment with H2O2 (Figure 2D). The redox-dependent migration shift was not observed with higher concentrations of diamide (≤10 mM). These experiments indicate that treatment with H2O2 or t-butyl hydroperoxide, but not diamide, results in a distinctive oxidation-dependent change of conformation of Yap1 and suggest different modes of activation of Yap1 by these oxidants.
Cysteine residues important for the in vivo oxidation of Yap1
The residues important for the peroxide-induced redox-dependent mobility shift were identified by analysing Yap1 cysteine substitution mutants (Figure 3). The Yap1C598T,C620A,C629T constitutive cytoplasmic allele and a mutant allele, Yap1C303A,C310A,C315A, with alanine substitutions for all three N-terminal cysteine residues could not reproduce the redox-dependent mobility shift observed under non-reducing PAGE (Figure 3B), demonstrating that one or more cysteines of both the N- and C-terminal parts of the protein are required for the formation of the oxidized form of Yap1. Analysis of individual cysteine to alanine substitution mutants showed that of the six cysteines of Yap1, only C303 and C598 were required for the occurrence of the oxidized form (Figure 3C). These two residues were also sufficient for the occurrence of the oxidized form of Yap1 since a mutant protein with substitution of all four other cysteines, Yap1C310T,C315A,C620A,C629T, retained the redox-dependent mobility shift observed with the wild-type protein (Figure 3C). However, it is noteworthy that Yap1C310A and Yap1C310T,C315A,C620A,C629T, and, to a lesser degree, Yap1C629A, could not be fully oxidized, suggesting that C310 might play an accessory role in the oxidation of C303 and C598.
Cysteine C303 and C598 are essential for the regulation of Yap1 by H2O2
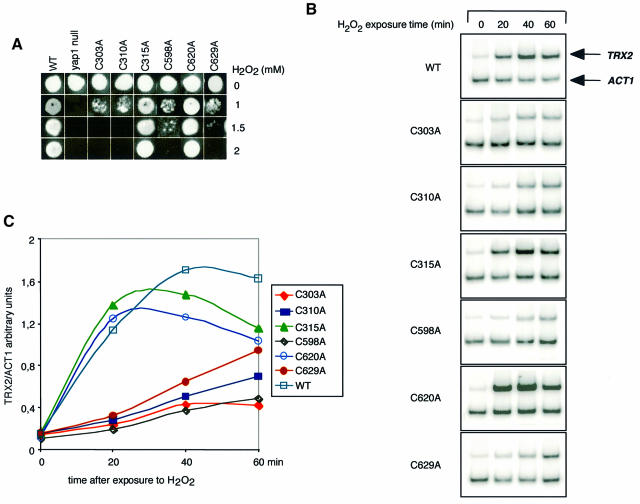
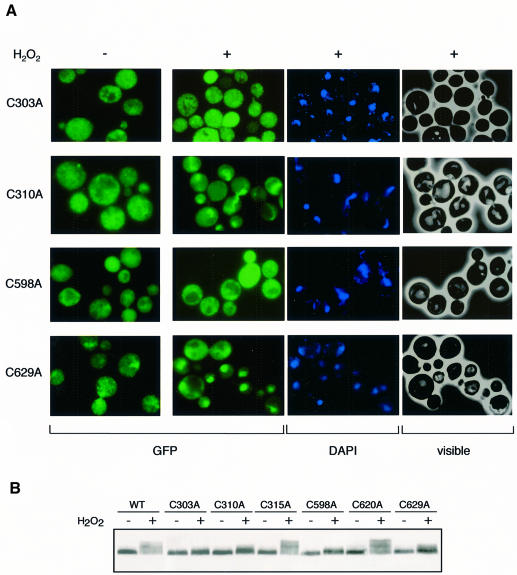
The importance of C303 and C598 for the in vivo oxidation of Yap1 by H2O2 suggested that these two amino acid residues are essential for the regulation of Yap1 by this oxidant. This suggestion was tested by investigating the capacity of cysteine substitution mutants to rescue the H2O2 hypersensitivity of the Δyap1 strain (Figure 4A). Yap1C315A and Yap1C620A had a wild-type H2O2 tolerance phenotype. All the other cysteine mutants were defective in this assay, with Yap1C303A and Yap1C310A displaying the highest sensitivity. The Yap1 cysteine mutants were also tested for their capacity to activate TRX2 expression (Figure 4B and C) and to redistribute into the nucleus in response to H2O2 (Figure 5A). Again, in both assays, Yap1C315A and Yap1C620A had a wild-type phenotype. In contrast, Yap1C303A and Yap1C598A were unable to activate TRX2 transcription and did not redistribute to the nucleus upon H2O2 treatment. Yap1C310A and Yap1C629A had a more complex phenotype. In these mutants, slight expression of TRX2 was seen 1 h after H2O2 treatment. Yap1C310A had an inefficient and delayed H2O2-induced redistribution to the nucleus and Yap1C629A had a partial constitutive nuclear phenotype but further concentrated in the nucleus after H2O2 treatment. The ability of cysteine mutants to redistribute to the nucleus was confirmed by their phosphorylation pattern (Figure 5B). Yap1C315A and Yap1C620A had a wild-type phosphorylation pattern whereas Yap1C303A and Yap1C598A did not undergo phosphorylation. Yap1C310A and Yap1C629A retained an inducible but lower phosphorylation level. Other Yap1 substitutions, C303S, C310S, C310T, C598S, C629S and C629T, were analysed and produced phenotypes similar to the corresponding C→A substitutions in all functional assays tested.
Fig. 4. Activity of Yap1 cysteine substitution mutants. (A) Yap1 cysteine residues important for tolerance to H2O2. The Δyap1 strains not expressing or expressing Myc-Yap1, Yap1C303A, Yap1C310A, Yap1C315A, Yap1C598A, Yap1C620A or Yap1C629A were tested for their tolerance to increasing concentrations of H2O2 by the patch assay on solid medium. (B) The same strains as in (A) were used to measure the amount of TRX2 and ACT1 mRNAs by RT–PCR as in Figure 1A. Cells were treated with 300 µM H2O2 for the time indicated. (C) The TRX2/ACT1 signal ratios quantified from the RT–PCR shown in (B) on a PhosphorImager were calculated and plotted as a function of time of exposure to H2O2.
Fig. 5. Essential role of C303 and C598 for H2O2-induced Yap1 nuclear redistribution. (A) Analysis of the cellular distribution of GFP-tagged Yap1C303A, Yap1C310A, Yap1C598A and Yap1C629A. Cells taken before or 12 min after treatment with 300 µM H2O2 were analysed for GFP or DAPI staining or under visible light. (B) Phosphorylation of Yap1 cysteine mutants as an indicator of their subcellular localization. Δyap1 cultures expressing Myc-Yap1, Yap1C303A, Yap1C310A, Yap1C315A, Yap1C598A, Yap1C620A or Yap1C629A were taken before and after treatment with 300 µM H2O2 and processed as in Figure 1C.
The critical role of C303 and C598 is at the level of the Crm1–Yap1 interaction
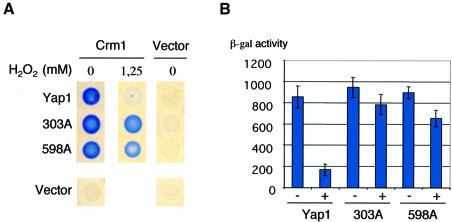
The data above demonstrate the critical role of the Yap1 N-terminal C303 and the C-terminal C598 in the activation of Yap1 by H2O2, probably due to their oxidation to a disulfide bond resulting in a change of conformation masking recognition of the Yap1 NES by Crm1. To provide further support for this model, we sought to evaluate whether the defective nuclear redistribution of Yap1C303A and Yap1C598A was due to their maintained interaction with Crm1/Xpo1 under oxidative stress conditions, using a two-hybrid assay. As shown by Kuge et al. (1997) and Yan et al. (1998), Crm1 and full-length Yap1 interact in the two-hybrid assay. This interaction has also been shown to be sensitive to diamide (Yan et al., 1998). We therefore evaluated the effect of H2O2 on the interaction between CRM1–LexABD and either full-length Yap1–B42AD, Yap1C303A–B42AD or Yap1C598A–B42AD (Figure 6). Whereas the interaction of Crm1 with wild-type protein was, as expected, sensitive to H2O2, interaction of Crm1 with both Yap1C303A and Yap1C598A was not modified by this treatment. These data demonstrate that the effect of these two mutations is exerted at the level of the interaction between Yap1 and Crm1.
Fig. 6. The critical role of C303 and C598 is at the level of Crm1–Yap1 interaction. β-galactosidase production from a lacZ reporter gene was assayed in strain EGY48 carrying pKW442 (Crm1–lexABD) or pEG202 (empty vector) and one of the following: pLDB439 (YAP1-B42AD or pYAP1C303A-B42AD), pYAP1C598A-B42AD or pJG4-5 (empty vector). (A) Assay performed on solid medium in the absence or presence of H2O2 at the concentration indicated. (B) Assay performed on liquid medium in the absence (–) or presence (+) of 500 µM H2O2. The results are the averages of four independent experiments.
Yap1 is deactivated (reduced) by the thioredoxin system
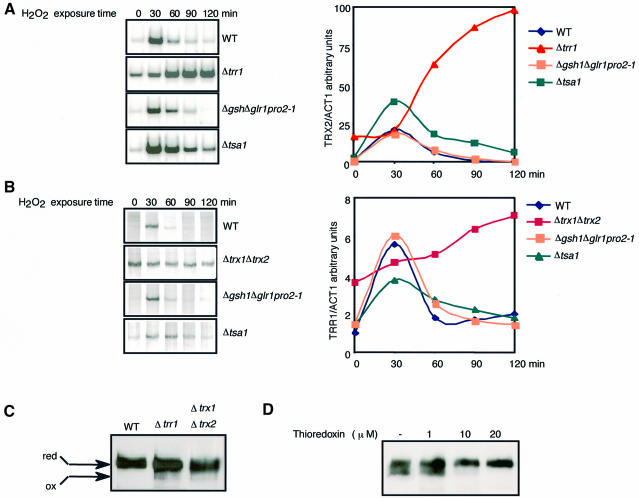
The data presented demonstrate that activation of Yap1 by H2O2 involves the oxidation of the protein to a probable disulfide bond, indicating that this regulator might directly sense the increased levels of H2O2. In yeast, the H2O2-sensing mechanism might be coupled to the systems that control the cell redox status, as in Escherichia coli (Zheng et al., 1998). To explore this possibility, Yap1 activation was monitored in strains carrying mutations of the two main cellular thiol disulfide reducing pathways (Figure 7). The GSH pathway was tested with strains deleted for the glutathione reductase gene GLR1 (Δglr1), or for both glutaredoxin genes GRX1 and GRX2 (Δgrx1Δgrx2), and with the Δgsh1pro2-1Δglr1 strain. The latter strain lacks the γ-glutamyl cysteine synthase gene (GSH1), encoding the rate-limiting enzyme of glutathione synthesis, and the glutathione reductase gene (GLR1), and harbours a third mutation in the proline biosynthesis enzyme gene PRO2, which restores the synthesis of GSH to ∼0.5% of the wild-type cellular content (our unpublished observations). The thioredoxin pathway was tested with strains lacking the thioredoxin reductase gene TRR1 (Δtrr1), or both thioredoxin genes TRX1 and TRX2 (Δtrx1Δtrx2). The time-course-induced expression of TRX2 and TRR1 in response to H2O2 was similar to that of the wild-type strain in Δglr1, Δgsh1pro2-1Δglr1 (Figure 7A and B) and Δgrx1Δgrx2 (not shown). By contrast, in strains inactivated in the thioredoxin pathway, Yap1 was constitutively partially active (∼20–50%), further activated after H2O2 treatment and remained maximally active after 2 h. This deregulated gene expression was specific to Yap1, since only the proteins of the Yap1 regulon were constitutively high in Δtrx1Δtrx2, as shown by two-dimensional gel electrophoresis (data not shown). We also monitored Yap1 activity in a strain deleted for TSA1 (Δtsa1), which encodes a thioredoxin-dependent thiol peroxidase (Chae et al., 1994). In this strain, Yap1 was not activated constitutively, but activation by H2O2 was prolonged (Figure 7A and B). However, Yap1 activity still decreased to baseline levels after ∼1.5 h, suggesting that the deregulation of Yap1 observed in the thioredoxin mutants is not solely related to a strain-intrinsic peroxide- scavenging defect, but to a lack of a direct effect of thioredoxins on the protein redox status. Consistent with the deregulated expression of Yap1-target genes, Yap1 was constitutively partially oxidized in Δtrr1 and Δtrx1Δtrx2 (Figure 7C). Deregulation was also correlated with a constitutive nuclear localization of Yap1 in Δtrr1 and Δtrx1Δtrx2, but not in mutants of the GSH pathway or in the Δtsa1 strain (Izawa et al., 1999; data not shown). Because thioredoxins are known to catalyse protein disulfide bond reduction by NADPH-dependent thioredoxin reductase, they could catalyse the reduction (deactivation) of oxidized Yap1. To test this hypothesis, we incubated extracts containing oxidized Yap1 with increasing amounts of thioredoxin, thioredoxin reductase and NADPH. The oxidation-dependent shift of migration was completely eliminated within 30 min in the presence of 20 µM thioredoxin (Figure 7D).
Fig. 7. Deactivation (reduction) of Yap1 by the thioredoxin system. (A and B) Expression of TRX2 (A) and TRR1 (B) in wild-type, Δtrr1 (A), Δtrx1Δtrx2 (B), Δgsh1Δglr1pro2-1 and Δtsa1 strains. Cells were treated with 300 µM H2O2 for the indicated time and processed for RT–PCR as in Figure 1A. The ACT1 autoradiograms are not shown. (C) Yap1 is oxidized in mutants of the thioredoxin system. yMyc-Yap1, Δtrx1Δtrx2 and Δtrr1 expressing Myc-Yap1 (grown to an OD600 of 0.3) were processed as in Figure 2A and extracts were analysed by immunoblotting after non-reducing PAGE. (D) Reduction of oxidized Yap1 by the thioredoxin system. yMyc-Yap1 (grown to an OD600 of 0.4), treated with 400 µM H2O2, lysed with glass beads in 100 mM Tris–HCl pH 8, 0.1% SDS, 1 mM EDTA, complete protease inhibitors and PMSF. Twenty-five micrograms of extracts were incubated with 1, 10 or 20 µM thioredoxin (from Spirulina sp.), 1.3 µM E.coli thioredoxin reductase and 1 mM NADPH (Sigma) for 30 min at 37°C. Iodoacetamide (75 mM) was added and the samples were analysed by immunoblotting after non-reducing PAGE.
Discussion
Yap1 as the yeast H2O2 sensor
The data presented here establish that the Yap1 transcription factor is activated by oxidation within minutes of H2O2 treatment and therefore fulfils the function of a redox sensor. These data also provide evidence for an oxidation-induced change of conformation of Yap1, and the requirement of residues C303 and C598 in its formation. This requirement, and the lack of H2O2 reactivity of Yap1C303A and Yap1C598A, establish that C303 and C598 are part of the Yap1 redox centre. Although a glutathione mixed disulfide or a sulfenic acid are possible cysteine oxidation products, C303 and C598 probably oxidize to form an intramolecular disulfide bond. The prolonged activation of Yap1 in mutants of the thioredoxin pathway and the ability of thioredoxin to catalyse Yap1 reduction in vitro indicate that the thioredoxin system acts to deactivate (reduce) Yap1, which is consistent with the presence of a disulfide bond in its oxidized form. Since both TRX2 and TRR1 are targets of Yap1, activation of the pathway by H2O2 is autoregulated. These data substantiate the presence of a disulfide bond in oxidized Yap1, but the participation of a metal or another prosthetic group in its redox-active centre cannot be ruled out. Of the other cysteines of Yap1, C315, C620 and C629 do not appear essential for redox sensing. However, C620 and C629 might be important in Yap1 control by being close to or part of the NES (Kuge et al., 1998; Yan et al., 1998; Coleman et al., 1999). In the case of C620, an alanine substitution appears to be tolerated for NES function (this work), whereas a threonine substitution is not (Kuge et al., 1997). Similarly, an alanine substitution of C629 partially disrupts the NES function (Coleman et al., 1999; this work) while a threonine substitution has a wild-type phenotype (Kuge et al., 1997). The depressed activity of Yap1629A is similar to the previously observed, but not understood, decreased activity towards H2O2 of all nuclear constitutive Yap1 alleles tested (Kuge et al., 1997; Coleman et al., 1999; data not shown). The role of C310 is more complex. As the corresponding substitution mutant can reproduce the oxidation-dependent electrophoretic migration shift and is still capable of redistributing to the nucleus upon H2O2 treatment, this cysteine residue is dispensable for redox sensing. One remarkable phenotype of Yap1C310A is its inefficient oxidation and delayed nuclear redistribution, which suggests that C310 could have a role in the formation of the oxidized conformation, perhaps by modifying the vicinal electronic environment if this residue becomes partially oxidized to a sulfenate upon exposure to H2O2. However, the profoundly decreased activity of Yap1C310A may also indicate another undefined functional role for the domain containing this residue (Coleman et al., 1999). Our data also showed that Yap1 becomes phosphorylated upon exposure to H2O2. This modification occurs outside the C-terminal CRD since it was still observed in a Yap1 truncated version lacking this domain (not shown). As it did not occur in mutants unable to redistribute to the nucleus, and was constitutive in a constitutive nuclear mutant (Figure 1), this modification probably occurs in the nucleus, and therefore at a step after protein oxidation, which is the critical event triggering Yap1 nuclear redistribution. Although its role is not understood, phosphorylation could be important by providing a regulated interaction with other substrates or as a marker for protein degradation.
The data presented here establish that activation of Yap1 by H2O2 involves the critical oxidation of the N-terminal CRD C303 and the C-terminal CRD C598 probably to a disulfide bond, with the resulting change of conformation masking recognition of the Yap1 NES by Crm1/Xpo1, thereby promoting accumulation of Yap1 into the nucleus. This model would provide a new example of a protein regulated by reversible disulfide bond formation, as a way to respond rapidly to oxidative stress (Åslund and Beckwith, 1999).
The mechanisms by which H2O2 and diamide activate Yap1 are different
The model proposed above seems to contradict studies that analysed the activation of Yap1 by diamide (Kuge et al., 1997, 1998; Yan et al., 1998). In particular, Kuge et al. (1997) showed that the Yap1 C-terminal CRD by itself could confer regulated nuclear redistribution to a reporter protein in response to diamide, while Yan et al. (1998) showed that any one of the C-terminal cysteines was required for the diamide regulation of Yap1. These published data clearly rule out involvement of disulfide bond formation between N- and C-terminal cysteine in the activation of Yap1 by diamide. This apparent contradiction could indicate, as pointed out by Wemmie et al. (1997), that there is a difference in the mechanism by which H2O2 and diamide activate Yap1. This is now clearly demonstrated by lack of the H2O2-induced redox-dependent migration shift upon diamide treatment (Figure 2), indicating that this oxidant does not generate the H2O2 oxidation product of C303 and C598. In addition, Yap1C303 and Yap1C598 still redistributed to the nucleus in response to diamide (data not shown), which demonstrates the specificity of their defective H2O2 response, and is consistent with the notion that any one cysteine can fulfil the required regulatory function needed in this process (Yan et al., 1998). In conclusion, we propose that diamide may activate Yap1 by direct modification of one or more C-terminal cysteines, perhaps by creation of a stable sulfenylhydrazine thereby modifying the vicinal NES and, hence, the recognition of Yap1 by Crm1.
Conservation of the H2O2-sensing mechanism between eukaryotes and prokaryotes
The two best characterized oxidative stress sensors are the E.coli transcriptional regulators OxyR and SoxR. SoxR is activated by reversible one-electron oxidation of its iron–sulfur centres in response to superoxide-generating compounds or nitric oxide (Gaudu et al., 1997; Hidalgo et al., 1997). OxyR is directly activated by H2O2 through formation of an intramolecular disulfide bond, and is deactivated by glutaredoxins (Zheng et al., 1998). Therefore, Yap1 and OxyR appear functionally homologous. Both activators control activities important for cellular thiol redox control and for scavenging reactive oxygen species (Lee et al., 1999a; Zheng and Storz, 2000) and share the function of sensing increased H2O2 levels. The quite different molecular control of these regulators, which is exerted at the level of the subcellular localization of the protein in the case of Yap1 (Kuge and Jones, 1997) and at the level of promoter selection and DNA binding in the case of OxyR (Toledano et al., 1994), reflects the greater cellular complexity in eukaryotes compared with prokaryotes. However, the biochemical modifications that reversibly activate these regulators, involving disulfide bond formation and deactivation by a thioltransferase system, appear to be conserved. Furthermore, although OxyR is preferentially deactivated by the glutathione pathway and Yap1 by the thioredoxin pathway, in both cases the whole system is autoregulated since each factor is deactivated by its target genes. It will still be important to demonstrate directly the presence of a disulfide bond in oxidized Yap1, and whether Yap1, like OxyR (Åslund et al., 1999), is directly oxidized by H2O2. We indeed do not rule out the possibility that another molecule acts to transduce the H2O2 redox potential to Yap1, in the same way as protein disulfide isomerase transmits the reducing equivalent generated during photosynthesis to the chloroplast polyadenylate binding protein, cPABP (Kim and Mayfield, 1997).
Yap1 does not sense GSH/GSSG status
Apart from their conserved features, significant differences could exist between the prokaryotic and eukaryotic redox sensing systems. OxyR can sense the cellular thiol–disulfide status, in addition to sensing H2O2. This disulfide stress sensing is based on OxyR constitutive activation in E.coli mutants simultaneously compromised in both thioredoxin and GSH pathways, and was proposed to explain OxyR activation by diamide (Åslund et al., 1999). However, the data presented here do not support that conclusion for Yap1. We observed that Yap1 is only constitutive in mutants of the thioredoxin pathway, but not in mutants of the GSH pathway. This could be interpreted as meaning that, in yeast, the drop in the cellular redox status needed to activate Yap1 is achieved after inactivation of the thioredoxin but not the glutathione pathway. However, this possibility can be ruled out by the status of the thiol–disulfide balance, which is more oxidized in mutants of the GSH pathway. Indeed, the percentage of oxidized GSH (GSSG), which is thought to be the best index of the cellular thiol redox balance, was 54% in Δglr1 compared with 22% in Δtrx1Δtrx2; in wild type the proportion of GSSG was <5% (not shown), as shown previously (Muller, 1996). Alternatively, the constitutive activation of Yap1 in mutants of the thioredoxin pathway could be interpreted as the sole consequence of the defective enzymatic deactivation of oxidized protein, which accumulates as a result of endogenous production of reactive oxygen species, and not as re-equilibration of Yap1 with an oxidizing cellular redox environment. This hypothesis would be consistent with the idea that diamide, although dramatically shifting the GSH/GSSG ratio towards its oxidized form (Kosower and Kosower, 1995; data not shown), activates Yap1 not through this perturbation, as proposed for OxyR (Åslund et al., 1999), but directly, as discussed above. We conclude that Yap1 has evolved to sense increased peroxide levels but not the GSH redox balance.
Materials and methods
Strains and culture conditions
Most of the experiments were performed with the wild-type strain YPH98 (Sikorski and Hieter, 1989) (Mata ura3-52 lys2-801amber ade2-101ochre trp1Δ1 leu2Δ1) and its isogenic derivatives. The Δyap1, Δglr1, Δtsa1, Δgsh1 and Δtrr1 strains were described previously (Lee et al., 1999b). Δtrx1Δtrx2 was prepared by a one-step amplification protocol that successively replaced the entire TRX1 and TRX2 open reading frames (ORFs) with the TRP1 and kanamycin genes, respectively. Δgsh1pro2-1Δglr1 is the Δgsh1 strain carrying a PRO2 mutation that rescues the strain’s GSH auxotrophy by allowing the synthesis of traces of GSH (0.5% of the wild-type GSH content as determined by a microbiological assay and by thin layer chromatography after [35S]methionine pulse labelling) and a deletion of the entire GLR1 ORF replaced by URA3. The pro2-1 suppressor mutation acts by diverting the γ-glutamate synthesized in the first step of the proline biosynthetic pathway into the synthesis of GSH (D.Spector and M.B.Toledano, unpublished observations). Given the GSH auxotrophy of the Δgsh1 mutation, the pro2-1 suppressor mutation ensures the presence of very low and constant levels of GSH on minimal medium. Δgrx1, Δgrx2 and the double deletion Δgrx1Δgrx2 are not isogenic with YPH98 and were a gift from Dr C.Grant (UMIST, Manchester) (Luikenhuis et al., 1998). EGY48 (Matα ura3-52 his3 LEXAop(X6)-LEU2 <pSH18-34 URA3 2µ>) (R.Brent) was used for the two-hybrid assay. The minimal medium (SD) was yeast nitrogen base (6.7 g/l) and dextrose (20 g/l) supplemented with the required amino acids. Dextrose can be replaced by 2% raffinose (S raffinose) or 2% galactose (S galactose). The H2O2 sensitivity assays were performed as described (Lee et al., 1999a).
Preparation of cell extracts and western blot analysis
Pelleted cells were washed in 20% TCA, frozen and lysed by vortexing after adding glass beads and 12.5% TCA. The lysate was centrifuged and the precipitated pellet dissolved in 2% SDS, 5% β-mercaptoethanol, 62.5 mM Tris–HCl pH 8.7, 10% glycerol, 0.01% bromophenol blue, boiled and loaded on a 10% SDS–polyacrylamide gel (30:0.4 acrylamide:bisacrylamide). To monitor the Yap1 redox state, the TCA-precipitated pellet was washed in acetone, air dried, dissolved in 75 mM iodoacetamide, 1% SDS, 100 mM Tris–HCl pH 8, 1 mM EDTA, complete protease inhibitors (Boehringer Mannheim) and incubated at 25°C for 15 min. Where indicated, Yap1 was dephosphorylated with 20 U calf intestinal phosphatase (New England Biolabs) for 45 min at 37°C, after dialysis of the resuspended iodoacetamide-treated protein pellet for 4 h against 10 mM Tris–HCl pH 8, 50 mM NaCl, 10 mM MgCl2,. Protein extracts were separated by non-reducing SDS–PAGE after adding 3× Laemmli sample buffer not containing β-mercaptoethanol and boiling. The separated proteins were blotted on to a nitrocellulose membrane and probed with the 9E10 anti-Myc monoclonal antibody; bound antibodies were detected using an enhanced chemiluminescence western blotting reagent kit (Amersham Pharmacia).
Analysis of cell GFP staining by immunofluorescence microscopy
Five minutes after addition of DAPI, cells were centrifuged, washed, resuspended in DABCO (75% glycerol, 0.25× phosphate-buffered saline, 200 mM diazabicyclooctane) and analysed with a Leica DMRXA fluorescent microscope equipped with a Roper Scientific Micro-Max cooled CCD camera and MetaMorph software (Universal Imaging, Inc.).
Two-hybrid assay
EGY48 carrying pSH18-34 (2µ plasmid carrying a lacZ reporter gene under the control of eight lexA operators) (Gyuris et al., 1993) was transformed with PKW442 [CRM1-LexABD (Stade et al., 1997)] or pEG202 (lexA(1-202)) and one of the following: pJG4-5 [B42AD haemagglutinin (HA) epitope tag TRP1], pLDB439 [YAP1-B42AD: full-length Yap1 cloned between the EcoRI and XhoI sites (Yan et al., 1998) of pJG4-5], pYAP1C303A-B42AD or pYAP1C598A-B42AD. The C303 and C598 mutations were introduced by cloning into YAP1-B42AD a mutagenized fragment generated by the two-step PCR method and inserted in pLDB439 between the BamHI and PacI sites (C303) or between the PacI and XhoI sites (C598). To test interaction in liquid medium, cells pre-grown in 2% S raffinose lacking uracil, histidine and tryptophan were re-inoculated in the same medium to an OD600 of 0.3–0.5 and were transferred to an equivalent volume of 2% S galactose for 30 min. Cells were then treated or not with H2O2 (500 µM) for 1 h. The β-galactosidase assay was performed as described (Miller et al., 1972). To test interaction on solid medium, cells grown in 2% S raffinose were patched at a density of 4.5 × 107 cells on solid 1% S raffinose/2% galactose medium and incubated for 2.5 h at 30°C. The assay was revealed by overlaying the cells with top agar containing X-gal (0.04%).
Construction of plasmids
Myc-Yap1 contains a Myc9 epitope tag inserted at the unique AccI site, three codons downstream from the ATG, and is expressed from the centromeric pRS316 plasmid (Lee et al., 1999b). A 1.17 kb fragment encoding the C303A, C310A and C315A substitutions, amplified from PldB583 (a gift from L.Davis, Brandeis University) (Yan et al., 1998), was recombined to Myc-Yap1 by gap repair, resulting in Yap1C303A,C310A,C315A. A 327 bp NdeI–PacI fragment of Pldb518 encoding the C598T, C620A and C629T substitutions (Yan et al. 1998) was subcloned into Myc-Yap1, resulting in Yap1C598T,C620A,C629T. Yap1L619S,L623S, Yap1C303A, Yap1C310A, Yap1C598A, Yap1C620A, Yap1C629A and Yap1C315A were prepared by a two-step PCR method and the mutagenized fragments were introduced into Myc-Yap1 by gap repair. GFP–Yap1 is a GFP N-terminal fusion expressed from vector pRS cp-GFP HA-YAP1 GFP (TRP1, CEN6-ARSH4) (a gift from S.Kuge, University of Tokyo) (Kuge et al., 1997). GFP–Yap1 fusions carrying individual C→A mutations were prepared by subcloning the corresponding mutagenized fragments into the BamHI and BstEII sites of pRS cp-GFP HA-YAP1. All the constructs used in this study were sequenced completely.
Quantitative RT–PCR assays
Total RNA was isolated as described by Lee et al. (1999b). RT–PCR assays were performed as described by Godon et al. (1998). cDNAs were synthesized by random hexanucleotide-primed reverse transcription from 1 µg of total RNA. Quantitative PCR was performed using primers specific for TRX2 (CGTCACTCAATTAAAATCCGCTTC, GGAAGC AATAGCTTGCTTGCTTGATAGC), TRR1 (CCAGGGCAGAAA TCAAGCCAATCC, ATAAACCGCTGACTGGCAAATCGG) and ACT1 (TTGGATTCCGGTGATGGTGTTACT, TGAAGAAGATTG AGCAGCGGTTTG). 32P-labelled PCR products were separated on a native polyacrylamide gel, dried, exposed to autoradiography film and quantified on a PhosphorImager.
Acknowledgments
Acknowledgements
We are indebted to Scott Moye-Rowley, Shusuke Kuge, Laura Davis and Karsten Weis for reagents, Jean Labarre for two-dimensional gel analysis and Dan Spector for measuring GSH redox balance. We especially thanks Françoise Stutz for her gifts of plasmids and Christophe Créminon for the 9E10 monoclonal antibody. Many thanks to Jean-Marie Buhler, Jean Labarre, Carl Mann, André Sentenac and Daniel Spector for critical review of the manuscript. This work was supported by grants from the French Ministère de l’Education Nationale and l’Association pour la Recherche contre le Cancer (no. 9575) to M.B.T.
References
- Åslund F. and Beckwith,J. (1999) Bridge over troubled waters: sensing stress by disulfide bond formation. Cell, 96, 751–753. [DOI] [PubMed] [Google Scholar]
- Åslund F., Zheng,M., Beckwith,J. and Storz,G. (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl Acad. Sci. USA, 96, 6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.Z., Chung,S.J. and Rhee,S.G. (1994) Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem., 269, 27670–27678. [PubMed] [Google Scholar]
- Coleman S.T., Epping,E.A., Steggerda,S.M. and Moye-Rowley,W.S. (1999) Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol., 19, 8302–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudu P., Moon,N. and Weiss,B. (1997) Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem., 272, 5082–5086. [DOI] [PubMed] [Google Scholar]
- Godon C., Lagniel,G., Lee,J., Buhler,J.-M., Kieffer,S., Perrot,M., Boucherie,H., Toledano,M.B. and Labarre,J. (1998) The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem., 273, 22480–22489. [DOI] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Ding,H. and Demple,B. (1997) Redox signal transduction: mutations shifting [2Fe–2S] centers of the SoxR sensor–regulator to the oxidized form. Cell, 88, 121–129. [DOI] [PubMed] [Google Scholar]
- Izawa S., Maeda,K., Sugiyama,K., Mano,J., Inoue,Y. and Kimura,A. (1999) Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem., 274, 28459–28465. [DOI] [PubMed] [Google Scholar]
- Kim J. and Mayfield,S.P. (1997) Protein disulfide isomerase as a regulator of chloroplast translation activation. Science, 278, 1954–1957. [DOI] [PubMed] [Google Scholar]
- Kosower N. and Kosower,E.M. (1995) Diamide: an oxidant probe for thiols. Methods Enzymol., 251, 123–133. [DOI] [PubMed] [Google Scholar]
- Kuge S. and Jones,N. (1994) YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J., 13, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Jones,N. and Nomoto,A. (1997) Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J., 16, 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Toda,T., Lizuka,N. and Nomoto,A. (1998) Crm1 (Xpo1) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells, 3, 521–532. [DOI] [PubMed] [Google Scholar]
- Lee J., Spector,D., Godon,C., Labarre,J. and Toledano,M.B. (1999a) A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem., 274, 4537–4544. [DOI] [PubMed] [Google Scholar]
- Lee J., Godon,C., Lagniel,G., Spector,D., Garin,J., Labarre,J. and Toledano,M.B. (1999b) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem., 274, 16040–16046. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S., Perrone,G., Dawes,I.W. and Grant,C.M. (1998) The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell, 9, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Moye-Rowley W.S., Harshma,K.D. and Parker,C.S. (1989) Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev., 3, 283–292. [DOI] [PubMed] [Google Scholar]
- Muller E.G.D. (1996) A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell, 7, 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N., Krems,B. and Entian,K.-D. (1992) The PAR1(VAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr. Genet., 21, 269–273. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Tao K. (1999) In vivo oxidation–reduction kinetics of OxyR, the transcriptional activator for an oxidative stress-inducible regulon in Escherichia coli. FEBS Lett., 457, 90–92. [DOI] [PubMed] [Google Scholar]
- Toledano M.B., Kullik,I., Trinh,F., Baird,P.T., Schneider,T.D. and Storz,G. (1994) Redox-dependent shift of OxyR–DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell, 78, 897–909. [DOI] [PubMed] [Google Scholar]
- Wemmie J.A., Steggerda,S.M. and Moye-Rowley,W.S. (1997) The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem., 272, 7809–7914. [DOI] [PubMed] [Google Scholar]
- Yan C., Lee,L.H. and Davis,L.I. (1998) Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J., 17, 7416–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M. and Storz,G. (2000) Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol., 59, 1–6. [DOI] [PubMed] [Google Scholar]
- Zheng M., Aslund,F. and Storz,G. (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation. Science, 279, 1718–1721. [DOI] [PubMed] [Google Scholar]