Abstract
The influenza A virus pandemic of 1918–1919 resulted in an estimated 20–40 million deaths worldwide. The hemagglutinin and neuraminidase sequences of the 1918 virus were previously determined. We here report the sequence of the A/Brevig Mission/1/18 (H1N1) virus nonstructural (NS) segment encoding two proteins, NS1 and nuclear export protein. Phylogenetically, these genes appear to be close to the common ancestor of subsequent human and classical swine strain NS genes. Recently, the influenza A virus NS1 protein was shown to be a type I IFN antagonist that plays an important role in viral pathogenesis. By using the recently developed technique of generating influenza A viruses entirely from cloned cDNAs, the hypothesis that the 1918 virus NS1 gene played a role in virulence was tested in a mouse model. In a BSL3+ laboratory, viruses were generated that possessed either the 1918 NS1 gene alone or the entire 1918 NS segment in a background of influenza A/WSN/33 (H1N1), a mouse-adapted virus derived from a human influenza strain first isolated in 1933. These 1918 NS viruses replicated well in tissue culture but were attenuated in mice as compared with the isogenic control viruses. This attenuation in mice may be related to the human origin of the 1918 NS1 gene. These results suggest that interaction of the NS1 protein with host-cell factors plays a significant role in viral pathogenesis.
The influenza pandemic of 1918–1919 was uniquely severe, causing an estimated 20–40 million deaths worldwide (1, 2). Also unique was the age distribution of its victims: the death rate for young, previously healthy adults, who rarely suffer fatal complications from influenza, was exceptionally high (1, 3). Because the first human influenza virus was not isolated until 1933, samples of the 1918 influenza virus have not been available. The discipline of paleomicrobiology has begun by using PCR-based methods to identify the microbial pathogens that caused historic epidemics, including the 1918 influenza pandemic (4–8). Molecular analysis of the 1918 virus became possible on development of techniques for the reverse transcription–PCR (RT-PCR) amplification of viral RNA sequences from formalin-fixed and frozen tissue samples (2, 8, 9). It is hoped that the molecular characterization of the 1918 virus will shed light on both the reasons for its extraordinary virulence and its evolutionary origin.
The reasons for the 1918 virus' virulence remain obscure. A highly virulent phenotype could theoretically be encoded by a single or by multiple viral gene(s). The possibility of a second cocirculating pathogen that cooperated with the 1918 influenza virus to increase death rates has also been suggested (10, 11). To fully understand the pathogenesis of the 1918 virus and to pinpoint the genetic basis of virulence, the function of its proteins must be studied physiologically. Systems that permit the generation of influenza viruses from cloned cDNAs allow recombinant influenza viruses bearing genes of the 1918 pandemic virus to be constructed. The resulting viruses can be studied in vitro or in animal models (12, 13).
The full-length sequences of the two predominant surface proteins of the 1918 virus, hemagglutinin (HA) and neuraminidase (NA), have recently been described (2, 9). These sequences were determined first because influenza pandemics are thought to result from the emergence of influenza virus strains with novel surface antigens. Phylogenetic analyses of the 1918 HA and NA grouped them with human- and swine-adapted strains. However, the surface antigens of the 1918 virus are more closely related to avian influenza virus strains than those of any other mammalian virus and were probably derived from an avian source (2, 9).
Specific amino acid motifs within the HA and NA proteins of some influenza A viruses are associated with expanded tissue tropism and increased virulence. Analysis of the 1918 HA and NA sequences demonstrated that such sequence determinants were absent in the 1918 strain (2, 9). The 1918 HA lacked the multiple basic amino acid residues at the HA cleavage site characteristic of H5 and H7 proteins of some highly virulent avian viruses (2, 9, 14–17), including the avian H5N1 viruses that killed 6 of 18 infected patients in Hong Kong in 1997 (18). Likewise, the 1918 NA lacked an amino acid substitution analogous to that which confers on the mouse-adapted influenza A/WSN/33 (H1N1) virus, the properties of trypsin-independent growth in tissue culture, pantropism in newborn mice, and neurovirulence (19, 20).
It has recently been shown that the influenza A virus NS1 protein functions as a type I IFN-antagonist (21–23) and is required for influenza A virus virulence (23, 24). The NS1 protein prevents both type I IFN production, by inhibiting activation of the latent transcription factors IRF-3 (22), and NF-κB (21), and prevents activation of IFN-induced antiviral proteins, including PKR (25–27) and 2′,5′-oligoadenylate synthetase (unpublished observation). These functions may be mediated, at least in part, by the ability of NS1 to bind RNA (25, 26, 28).
One of the distinctive clinical characteristics of the 1918 influenza virus was its ability to rapidly produce extensive damage to the respiratory epithelium (29). Such a clinical course suggests a virus that replicated to a high titer and spread quickly from cell to cell. An NS1 protein that was especially effective at blocking the type I IFN system might have contributed to the exceptional virulence of the 1918 strain (23, 30). In this study, we report the complete sequence and a phylogenetic analysis of the smallest influenza gene segment, which codes for the NS1 protein and the nuclear export protein (NEP, previously NS2). To begin to understand the role NS gene products, NS1 and NEP, may have played in virulence, the 1918 virus NS1 gene and the entire 1918 virus NS segment were reconstructed, and transfectant influenza viruses bearing either the 1918 NS1 gene or the entire 1918 NS segment were generated. The virulence of these viruses in mice was then determined.
Materials and Methods
Virus Strains and Cells.
The transfectant influenza viruses, influenza A/WSN/33 (H1N1) virus (WSN) and influenza A/PR/8/34 (H1N1) virus (PR8), were propagated on Madin–Darby canine kidney (MDCK) cells (maintained in MEM, 10% FBS). 293T cells were maintained in DMEM, 10% FBS.
RNA Extraction, RT-PCR, and DNA Sequencing of Frozen Tissue Samples.
The 1918 case and the viral strain, A/Brevig Mission/1/18 (H1N1), used for this study were as described previously (2, 9). RNA was isolated from the frozen lung tissue by using RNAzol (Tel-Test, Friendswood, TX), following the manufacturer's instructions.
RT was carried out at 37°C for 45 min in 20 μl by using 300 units Maloney murine leukemia virus-reverse transcriptase/1× RT buffer (Life Technologies, Grand Island, NY)/5 μM random hexamers/200 nM dNTPs/10 mM DTT. RT reaction (2 μl) was added to an 18-μl PCR reaction mixture containing 50 mM KCl, 10 mM Tris-HCl, 2.5 mM MgCl2, 1 μM each primer, 100 nM dNTPs, 1 unit Amplitaq Gold (Perkin–Elmer), and 2 μCi (1 Ci = 37 GBq) 32P dATP (3,000 Ci/mmol). The entire NS coding sequence of A/Brevig Mission/1/18 (H1N1) (838 nucleotides) was amplified in 13 overlapping fragments, such that the sequences corresponding to primers could be confirmed. The primers were designed as degenerate NS consensus primers by using alignments of human, swine, and avian NS sequences, or as 1918-specific primers once partial sequence was available. Primer sequences used are available on request.
Phylogenetic Analyses.
Phylogenetic analyses of the NS genes used parsimony [Phylogenetic Analysis Using Parsimony (paup, Version 3.1.1)] (31) and neighbor joining (NJ) [Molecular Evolutionary Genetics Analysis (mega, Version 1.1)] (32). The optimization method used in paup was ACCTRAN. NJ analyses routinely used the proportion of differences between the sequences as the distance measure (p distance). All NJ analyses were bootstrapped 100 replications.
Plasmids
1918 NS1 ORF and 1918 NS Segment Constructs.
PCR products generated for sequencing were used for reconstruction. Products that had been generated by using degenerate primers were reamplified with 1918 sequence-specific primers. Overlapping products were combined and amplified by PCR by using only the extreme 5′ and 3′ primers. Accuracy of the full-length product was confirmed by sequencing. The 1918 NS1 ORF and NS segment products were cloned into pCR-TOPO (Invitrogen), and the constructs are subsequently referred to as pTopo-1918 NS1 and pTopo-1918 NS, respectively.
NS1–2A-NEP Clones.
A mutation at the splice acceptor site and a silent SmaI site were introduced into the 1918 NS1 ORF by site-directed mutagenesis. The NS sequence 525-CCAG↓GA-530 (the arrow shows the intron/exon junction) was changed to 525-CCCGGG-530 without changing any amino acids, creating the plasmid pTopo-1918 NS1 SAM (splice acceptor mutant). The 1918 NS1 SAM or a PR8 NS1 SAM ORF were then cloned in-frame with a foot-and-mouth disease virus 2A-protease-WSN NEP ORF, as detailed below, and cloned into pPolI-SapI-RT (33) creating the plasmids pPolI-1918 NS1–2A-NEP and pPolI-PR8 NS1–2A-NEP. These plasmids, in primate cells, produce negative-sense RNAs that possess the PR8 NS segment 3′ and 5′ noncoding regions flanking the 1918NS1–2A-NEP ORF or the PR8NS1–2A-NEP ORF.
The 1918 NS1 in the plasmid pTopo-1918 NS was PCR amplified and cloned into pPolI-SapI-RT (33). The resulting plasmid produces, in primate cells, an exact replica of the 1918 virus NS virion RNA (vRNA).
Generation of Transfectant Influenza Viruses.
To generate transfectant influenza viruses, 1 μg each of 12 plasmids was transfected in suspension into 293T cells, following the same method used to transfect MDCK cells at high efficiency (34). Each transfection contained vRNA expression plasmids for the WSN PA, PB1, PB2, NP, HA, NA, and M segments (13), the appropriate vRNA expression plasmid for the NS segment, and the protein expression plasmids pCAGGS-PA, -PB1, -PB2 and -NP. The latter were derived by transferring the ORFs of the WSN PA, PB1, PB2, and NP into the expression plasmid pCAGGS kindly provided by J. Miyazaki (Osaka University, Osaka, Japan) (35) and Y. Kawaoka (University of Wisconsin, Madison, WI). After transfection (16–20 h), the culture medium was replaced with DMEM/0.1% FBS/0.3% BSA. After transfection (48 h), supernatants were harvested, and plaque assays were performed. Transfectant virus plaques were picked and amplified on MDCK cells.
Growth of Transfectant Viruses in Tissue Culture.
All growth of virus in cell culture was performed in biological safety hoods under BSL-3+ Ag containment. MDCK cells were infected with the indicated virus at a multiplicity of infection of 0.001. Virus was grown in MEM/0.3% BSA/3 μg/ml trypsin. Titers were determined at the indicated time points by plaque assay on MDCK cells.
Mouse Infections.
Mice were anesthetized with ketamine–xylazine and inoculated intranasally with the indicated virus dose. Programmable transponders (IPTT-100, Electronic Laboratory Animal Monitoring Systems, BioMedic Data Systems, Seaford, DE) were implanted s.c. to identify individual mice. Mice were housed in cages inside hepa-filtered Horsfall units. All animal work was performed in specially separated negative-pressured hepa-filtered rooms within the larger BL-3+ Ag building. All personnel wore half-body Rocal hoods with back-pack hepa-filtered air supplies in the animal room and showered before entering the main BSL-3+ Ag building.
Results
Sequence Analysis.
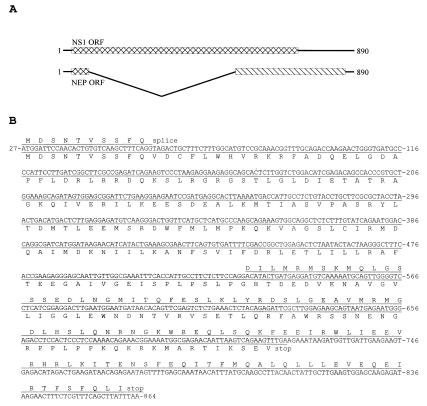
The entire NS coding sequence (838 nucleotides) from the shared start codon at nucleotide 27 of the NS segment to the stop codon of the NEP ORF at nucleotide 864 was determined from the frozen sample obtained from Brevig Mission, AK [A/Brevig Mission/1/18 (H1N1)]. The sequence and a map of the two overlapping reading frames are shown (Fig. 1). The theoretical translation of the two ORFs, NS1 and NEP, is indicated (Fig. 1B).
Figure 1.
Sequence of the 1918 NS segment. (A) Diagram of the NS gene segment showing the positions of the two overlapping ORFs. Nucleotides 1–26 and 862–890 are noncoding. (B) The sequence of the Brevig Mission/1/18 NS gene segment. The coding sequences start at nucleotide 27, and both NS1 and NEP share the initial 10 codons. The NEP translation product is shown above the line of nucleotides, whereas the NS1 product is below the line. The numbering of the gene is aligned to A/WSN/33 (GenBank no. M12597) and refers to the sequence in the sense (mRNA) orientation.
Several functional domains have been mapped to the NS1 protein. The theoretical translation of the 1918 NS1 sequence predicts that the RNA-binding domain (residues 19–38), the effector domain (residues 134–161), and the nuclear localization signals (residues 34–38 and 216–221) match the consensus sequences of influenza A NS1 proteins (of the A allele) (36–38).
Phylogenetic Analysis of the 1918 NS Segment.
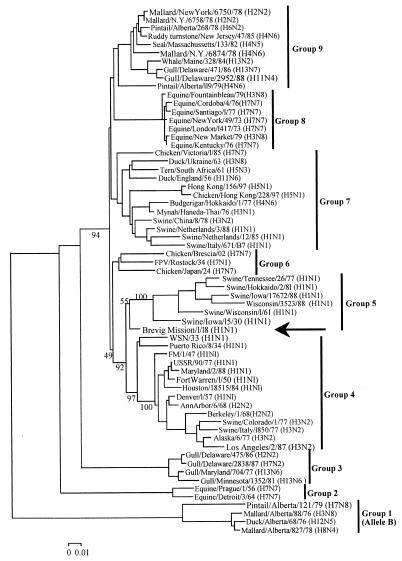
NJ analysis of 63 NS1 nucleotide sequences produced a tree with 9 clades: avian 1–5, equine 1 and 2, human, and swine (Fig. 2). NJ of 61 NEP nucleotide sequences produced a tree with the same 9 clades seen in the NS1 tree (not shown). The A/Brevig Mission/1/18 NS gene was within and near the root of the swine clade for NS1 and within and near the root of the human clade for NEP.
Figure 2.
Phylogenetic tree of influenza A NS1 genes. The tree is a NJ tree using p distance. Nine groups are indicated: groups 1, 3, 6, 7, and 9 are avian, groups 2 and 8 are equine, group 4 is human, and group 5 is swine. A/Brevig Mission/1/18 (arrow) is near the root of group 5. Bootstrap values are given for selected nodes, and a distance bar is shown under the tree.
A parsimony analysis of 37 NS1 protein sequences produced 6 trees of 386 steps (consistency index = 0.68), all of which were similar to that seen in Fig. 2. All placed the A/Brevig Mission/1/18 protein within and near the root of the human/swine family of sequences. Parsimony analysis of 35 NEP protein sequences produced 10 trees of 94 steps (consistency index = 0.69) that again placed the A/Brevig Mission/1/18 protein within and near the root of the human/swine family of sequences. Thus, our phylogenetic analyses produce trees similar to those of others (39, 40), and all place the 1918 pandemic virus within and near the root of the human/swine family of viruses.
Generating a Virus Bearing the NS1 ORF of the 1918 Virus.
To study the role of the NS gene in viral pathogenesis, transfectant influenza viruses were constructed containing either the 1918 NS1 ORF alone or the entire 1918 NS segment. All virus experiments were performed in a biosafety level 3+ facility at the U.S. Department of Agriculture Southeast Poultry Research Laboratory in Athens, GA, under an institutional Memorandum of Understanding and Agreement for Recombinant DNA Experiments.
To assess the individual role of the 1918 influenza virus, NS1 protein in viral pathogenesis, it was desirable to introduce only the 1918 NS1 gene into a transfectant influenza virus. The NS1 and NEP proteins are encoded, in wild-type viruses, by overlapping ORFs and are produced via alternative splicing of the NS gene. Therefore, a strategy was devised to separate the NS1 and NEP ORFs (Fig. 3). A single long ORF was generated such that the 1918 NS1 ORF, minus its stop codon, was fused in frame with the foot-and-mouth disease virus 2A protease, which exhibits autoproteolytic activity. The NS1–2A was further fused in frame to the entire NEP ORF. A control virus, identical to the 1918 NS1-containing virus, except that its NS1 gene was derived from the PR8 virus, was also generated. The NS1–2A-NEP constructs also contained a mutation at the splice acceptor site within the NS1 ORF to prevent formation of spliced products. These NS1–2A-NEP segments initially express a single polyprotein; however, because of the presence of the 2A autoprotease, these polyproteins are efficiently cleaved into two products (41). The first is an NS1 with 16 additional 2A-derived amino acids at its carboxy terminus. The second is an NEP with an additional proline at its amino terminus (41).
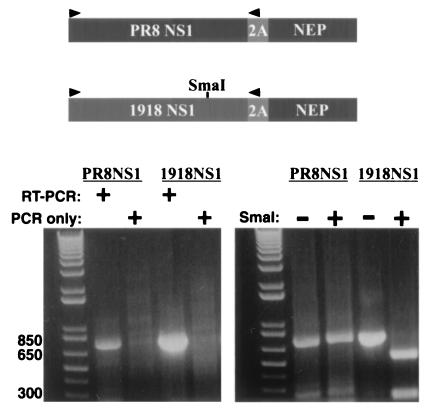
Figure 3.
Construction of viruses that express either the 1918 NS1 protein or the control PR8 NS1 protein. Coding strategy and RT-PCR analysis of the 1918 NS1–2A-NEP and PR8 NS1–2A-NEP viruses. In the NS1–2A-NEP constructs, the NS1 coding sequences do not overlap the NEP coding sequences. The NS1 and NEP coding sequences are separated by the foot-and-mouth disease virus 2A autoprotease and are expressed as a single polyprotein. The polyprotein is cleaved into two peptides, an NS1 with 16 additional 2A-derived amino acids at its carboxy terminus and an NEP with a single 2A-derived proline at its amino terminus. RT-PCR was performed to amplify the NS1 ORFs. The primer-binding sites (sequences available on request) are indicated by arrows.
Viruses with NS segments derived from these constructs were generated by using a modified version of the cDNA-based rescue system of Fodor et al. (13). These viruses possessed seven viral segments from the WSN strain. The eighth (NS) segment of the two viruses differed only in the source (PR8 virus or 1918 virus) of the NS1 sequences. RT-PCR of the NS1 ORFs confirmed that the viruses possessed the correct NS segments. Products of the expected size were detected for each virus (Fig. 3). PCR performed without prior RT yielded no products, indicating that the products seen in the RT-PCR reactions were derived from viral RNA and not from contaminating plasmid DNA. The 1918 NS RT-PCR products were further shown to contain SmaI restriction enzyme sites, which had been engineered as a genetic tag (Fig. 3). The NS1–2A-NEP vRNAs from both viruses were also amplified in their entirety, sequenced, and shown to be error free.
The resulting viruses, plus a wild-type WSN virus also generated from cDNA, were plaque purified and amplified on MDCK cells. These viruses grew to comparable titers on MDCK cells. BALB/c mice were infected by intranasal inoculation with 1 × 104 plaque-forming units (pfu) of each virus. As expected, wild-type WSN virus killed all mice by 8 days after infection. This dose of PR8 NS1–2A-NEP virus also killed all mice, but with somewhat delayed kinetics compared with wild-type WSN-infected mice. In contrast, all mice infected with 1 × 104 pfu of 1918 NS1–2A-NEP virus survived infection (Fig. 4). Daily measurements of body weight showed that the 1918 NS1–2A-NEP viruses caused some illness with mice losing, on average, greater than 10% of their initial weight. However, the mice recovered and began to regain lost weight between 6 and 8 days after infection (data not shown).
Figure 4.
Percent survival of mice infected with PR8 NS1–2A-NEP or 1918 NS1–2A-NEP viruses. Mice were infected intranasally with 1 × 104 pfu of wild-type WSN virus (diamonds), PR8 NS1–2A-NEP virus (squares) or 1918 NS1–2A-NEP virus (triangles), or mock infected with PBS (not shown). Each group had six mice. The percentage of mice surviving on each day is indicated. All PBS-treated mice survived.
Viruses Bearing the 1918 NS Segment or the Control WSN NS Segment.
The results of the NS1–2A-NEP mouse infections might indicate that the 1918 NS1 protein does not function efficiently in mice and therefore attenuates the transfectant virus. Alternatively, the function of the 1918 NS1 protein might be affected by the addition of 16 carboxyl-terminal 2A-derived amino acids. Therefore, an alternate strategy was devised to study the role of the 1918 NS segment in viral pathogenesis. Isogenic viruses were generated that differed only in the presence of a wild-type WSN NS segment or the wild-type 1918 NS segment (they possessed seven common WSN segments). One wild-type WSN virus and two independently derived 1918 NS viruses were obtained. RT-PCR and restriction enzyme analysis confirmed the identity of the NS segments (published as supplemental data on the PNAS web site, www.pnas.org). Further, the NS vRNAs from both viruses were amplified by RT-PCR in their entirety, sequenced, and shown to be error free.
The growth on MDCK cells of the wild-type WSN virus and the two 1918 NS viruses was analyzed (Fig. 5A). Growth of the three viruses was identical after infection at low multiplicity of infection. Therefore, the 1918 NS1 and NEP proteins are compatible with the RNAs and proteins encoded by the 7 WSN-derived segments of the transfectant viruses.
Figure 5.
Replication of wild-type WSN and 1918 NS viruses in tissue culture and in mice. (A) Multicycle growth in MDCK cells of wild-type WSN virus (diamonds) or either of two independently isolated 1918 NS viruses (1918 A, squares or 1918 B, triangles). Cells were infected at a multiplicity of infection = 0.001. Viruses were titered by plaque assay on MDCK cells. (B) Survival of mice after infection with wild-type WSN virus or 1918 NS viruses. Mice were infected intranasally with 1 × 104 pfu. Eight mice were infected with each virus or were mock infected with PBS. The percent of mice surviving on each day is indicated. All PBS-treated mice survived. (C) Average lung titers of mice infected with wild-type WSN or with 1918 NS viruses on days 2, 4, and 6 after infection. The average titers on each day are reported. Three mice were used for each wild-type WSN time point and six mice were used for each 1918 NS virus time point. Viruses were titered by plaque assay on MDCK cells.
BALB/c mice were infected via the intranasal route with either wild-type WSN virus or with each of the two 1918 NS viruses. As expected, infection with 1 × 104 pfu of WSN virus killed all mice (Fig. 5B). However, mice infected with either 1918 NS virus did not die (Fig. 5B). Viral lung titers from infected mice were determined on days 2, 4, and 6 after infection. At each time point, the 1918 NS viruses replicated to lower titers than did wild-type WSN virus (Fig. 5C). On day 6 after infection, the average viral titer in wild-type WSN-infected mice was decreasing and was only about five times more than the virus titer for the 1918 NS virus-infected mice. This decrease likely reflected the advanced disease and imminent death of the WSN-infected mice.
Histopathology and immunohistochemistry of trachea, primary bronchi, and lung tissue mirrored the lung titers of the wild-type and the two 1918 NS viruses (see supplemental data).
Discussion
This paper presents the sequence of the 1918 NS segment and experiments testing the hypothesis that NS1 contributed to the virulence of the 1918 pandemic virus. Previous phylogenetic analyses of the 1918 HA and NA genes placed them within the mammalian clade, but both sequences are consistent with the hypothesis that they entered the mammalian population shortly before the pandemic (2, 9). Phylogenetic analyses place the 1918 NS1 and NEP genes within and near the root of the swine clade. These placements suggest that the 1918 NS is the ancestor of all subsequent swine and human NS genes. However, in the case of NS, the bootstrap values are low, and branch lengths are short (Fig. 2), suggesting that there are too few differences among the sequences to place them unambiguously. Because the NS1 and NEP sequences are more highly conserved than HA and NA (2, 9), their phylogenetic trees are less informative. This is because the HA and NA proteins are subjected to continual antigenic pressure in humans, whereas the NS-encoded proteins are not (42). The known functional domains of NS1 and NEP are highly conserved across species-adapted strains; no amino acid changes necessary for host adaptation have yet been identified, even though both proteins interact with cellular factors (43–52). Consequently, it is not possible to determine whether the 1918 NS segment derived from a novel (avian) source at about the same time as the 1918 HA and NA gene segments or was already part of a previously circulating human-adapted strain. In the absence of pre-1918 human influenza samples or contemporary avian influenza samples, this question will be difficult to address.
The role of NS1 in blocking the IFN response to influenza infection makes it an interesting candidate for increasing the virulence of the 1918 pandemic strain. A virus even marginally better at blocking IFN might be able to replicate to higher titers and spread to more cells in the lung, thus contributing to the severe lung pathology noted in many 1918 victims. We addressed this possibility by generating transfectant viruses possessing either the 1918 NS1 gene or the entire 1918 NS segment. The resulting viruses, although growing to comparable titers in MDCK cells, displayed reduced virulence in mice as compared with control viruses (Figs. 3 and 4). The attenuation of the virus expressing only the NS1 protein from the 1918 virus may reflect the inability of the 1918 NS1 to function efficiently in mouse cells. Alternatively, the 1918 NS1 might be more sensitive than the PR8 NS1 to the addition of extra carboxyl-terminal amino acids. Although significant growth differences between the control viruses and the 1918 NS1 and 1918 NS viruses were not seen in tissue culture, the presence of a highly efficient anti-IFN function should be more critical in vivo than in MDCK cells.
The inefficient function in mice of either the 1918 NS1 and/or the 1918 NEP could be caused by relatively poor protein stability in mouse cells or by the inefficient interaction of either protein with specific host factors. The ability of NS1 to affect the host IFN response may involve the binding of RNA by NS1 (21, 22, 25) or its interaction with specific proteins, including PKR (53). Possibly, the interaction of the 1918 NS1 protein with mouse PKR and/or other components of the type I IFN system is inefficient (53). Furthermore, the NS1 protein has been reported to interact with numerous other host-cell proteins (43–51). How the NS1 anti-IFN function is affected by interaction of NS1 with host-cell proteins is not clear.
The NEP also interacts with host-cell factors. The NEP promotes the nuclear export of viral ribonucleoprotein complexes late in the virus replication cycle through interaction with host-cell nuclear pore components including Rab/Rip1, Nup100 and Nup116 (52), and Crm1 (54). Inefficient interaction of the 1918 NEP with mouse nuclear pore components might also affect viral replication. However, because both the 1918 NS1 alone and the complete 1918 NS viruses were attenuated in mice, we favor the hypothesis that NS1 is the attenuating factor in both cases.
The attenuation in mice of the 1918 NS segment, sequenced directly from human tissue, demonstrates that NS1 is critical for the virulence in mice of WSN and PR8. The 1918 NS1 varies from that of WSN at 11 amino acids. Seven of these amino acid differences are shared between PR8 and WSN (amino acids 3, 22, 81, 114, 124, 224, and 227). The amino acid differences between the 1918, WSN, and PR8 NS segments may be important in the adaptation of the latter strains to mice and likely account for the observed differences in virulence in the experiment where only NS1 was changed.
Previous reassortment studies between avirulent and virulent viruses have identified a role for all viral segments, including the NS segment, in virulence and attenuation (55–59). Many of these studies also found virulence to be a multigenic trait, although at least one study demonstrated that the presence of an avian NS segment (of the B allele) in the background of a human influenza virus was attenuating in squirrel monkeys (36). The present study also points to the impact a single viral gene can have on virulence, and future experiments should identify the specific NS1 and/or NEP residue(s) that mediate virulence/attenuation in the mouse. The use of entirely cDNA-based reverse genetics systems to create specific reassortant and mutant viruses should greatly facilitate these and other future analyses of influenza virus virulence.
Supplementary Material
Acknowledgments
We thank Dr. Alan Hubbs and Ms. Ying Liu for their assistance in DNA sequencing. C.F.B. was supported by a National Institutes of Health National Research Service Award postdoctoral fellowship. A.G.-S. and P.P. were supported by grants from the National Institutes of Health. J.K.T. was supported by grants from the Department of Veterans Affairs and the American Registry of Pathology. This work was also supported in part by intramural funds of the Armed Forces Institute of Pathology. J.K.D. was supported by the U.S. Department of Agriculture Agricultural Research Service, CRIS research unit no. 6612–3200-22.
Abbreviations
- HA
hemagglutinin
- NA
neuraminidase
- NS
virus nonstructural
- NEP
nuclear export protein
- MDCK
Madin–Darby canine kidney
- RT-PCR
reverse transcription–PCR
- NJ
neighbor joining
- vRNA
virion RNA
- pfu
plaque-forming units
Footnotes
References
- 1.Crosby A. America's Forgotten Pandemic. Cambridge, U.K.: Cambridge Univ. Press; 1989. [Google Scholar]
- 2.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.United States Department of Commerce. Historical Statistics in the United States: Colonial Times to 1970. U.S. Govt. Printing Office; 1976. p. 58. [Google Scholar]
- 4.Raoult D, Aboudharam G, Crubezy E, Larrouy G, Ludes B, Drancourt M. Proc Natl Acad Sci USA. 2000;97:12800–12803. doi: 10.1073/pnas.220225197. . (First Published October 31, 2000; 10.1073/pnas.220225197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafi A, Spigelman M, Stanford J, Lemma E, Donoghue H, Zias J. Lancet. 1994;343:1360–1361. [PubMed] [Google Scholar]
- 6.Kolman C J, Centurion-Lara A, Lukehart S A, Owsley D W, Tuross N. J Infect Dis. 1999;180:2060–2063. doi: 10.1086/315151. [DOI] [PubMed] [Google Scholar]
- 7.Drancourt M, Aboudharam G, Signoli M, Dutour O, Raoult D. Proc Natl Acad Sci USA. 1998;95:12637–12640. doi: 10.1073/pnas.95.21.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 9.Reid A H, Fanning T G, Janczewski T A, Taubenberger J K. Proc Natl Acad Sci USA. 2000;97:6785–6790. doi: 10.1073/pnas.100140097. . (First Published May 23, 2000; 10.1073/pnas.100140097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilbourne E D. Influenza. New York: Plenum; 1987. [Google Scholar]
- 11.Noymer A, Garenne M. Popul Dev Rev. 2000;26:565–581. doi: 10.1111/j.1728-4457.2000.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez D R, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor E, Devenish L, Engelhardt O G, Palese P, Brownlee G G, Garcia-Sastre A. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klenk H D, Rott R, Orlich M, Blodorn J. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 15.Vey M, Orlich M, Adler S, Klenk H D, Rott R, Garten W. Virology. 1992;188:408–413. doi: 10.1016/0042-6822(92)90775-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perdue M L, Suarez D L. Vet Microbiol. 2000;74:77–86. doi: 10.1016/s0378-1135(00)00168-1. [DOI] [PubMed] [Google Scholar]
- 17.Horimoto T, Kawaoka Y. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 19.Schulman J L, Palese P. J Virol. 1977;24:170–176. doi: 10.1128/jvi.24.1.170-176.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Schulman J, Itamura S, Palese P. J Virol. 1993;67:6667–6673. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Li M, Zheng H, Muster T, Palese P, Beg A A, Garcia-Sastre A. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, Garcia-Sastre A. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 24.Talon J, Salvatore M, O'Neill R E, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. Proc Natl Acad Sci USA. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. . (First Published March 21, 2000; 10.1073/pnas.070525997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Wambach M, Katze M G, Krug R M. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 26.Hatada E, Saito S, Fukuda R. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida T, Shaw M W, Young J F, Compans R W. Virology. 1981;110:87–97. doi: 10.1016/0042-6822(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 29.Winternitz M C, Wason I M, McNamara F P. Pathology of Influenza. New Haven, CT: Yale Univ. Press; 1920. [Google Scholar]
- 30.Garcia-Sastre A, Durbin R K, Zheng H, Palese P, Gertner R, Levy D E, Durbin J E. J Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swofford D L. paup: Phylogenetic Analysis Using Parsimony, Version 3.1.1. Champaign, IL: Illinois Natural History Survey; 1991. [Google Scholar]
- 32.Kumar S, Tamura K, Nei M. molecular evolutionary genetics analysis, Version 1.01. University Park, PA: Pennsylvania State Univ.; 1993. [Google Scholar]
- 33.Pleschka S, Jaskunas R, Engelhardt O G, Zurcher T, Palese P, Garcia-Sastre A. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basler C F, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk H D, Garcia-Sastre A, Palese P. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. . (First Published October 10, 2000; 10.1073/pnas.220398297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 36.Treanor J J, Snyder M H, London W T, Murphy B R. Virology. 1989;171:1–9. doi: 10.1016/0042-6822(89)90504-7. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Yamakita Y, Krug R M. Proc Natl Acad Sci USA. 1998;95:4864–4869. doi: 10.1073/pnas.95.9.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenspan D, Palese P, Krystal M. J Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawaoka Y, Krauss S, Webster R G. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buonagurio D A, Nakada S, Parvin J D, Krystal M, Palese P, Fitch W M. Science. 1986;232:980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- 41.Percy N, Barclay W S, Garcia-Sastre A, Palese P. J Virol. 1994;68:4486–4492. doi: 10.1128/jvi.68.7.4486-4492.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jameson J, Cruz J, Ennis F A. J Virol. 1998;72:8682–8689. doi: 10.1128/jvi.72.11.8682-8689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, Li Y, Krug R M. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemeroff M E, Barabino S M, Li Y, Keller W, Krug R M. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 45.Qiu Y, Krug R M. J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu Y, Nemeroff M, Krug R M. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 47.Aragon T, de la Luna S, Novoa I, Carrasco L, Ortin J, Nieto A. Mol Cell Biol. 2000;20:6259–6268. doi: 10.1128/mcb.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marion R M, Zurcher T, de la Luna S, Ortin J. J Gen Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 49.Falcon A M, Fortes P, Marion R M, Beloso A, Ortin J. Nucleic Acids Res. 1999;27:2241–2247. doi: 10.1093/nar/27.11.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolff T, O'Neill R E, Palese P. J Virol. 1996;70:5363–5372. doi: 10.1128/jvi.70.8.5363-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff T, O'Neill R E, Palese P. J Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Neill R E, Talon J, Palese P. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan S L, Katze M G. J Interferon Cytokine Res. 1998;18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 54.Neumann G, Hughes M T, Kawaoka Y. EMBO J. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown E G. J Virol. 1990;64:4523–4533. doi: 10.1128/jvi.64.9.4523-4533.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown E G, Bailly J E. Virus Res. 1999;61:63–76. doi: 10.1016/s0168-1702(99)00027-1. [DOI] [PubMed] [Google Scholar]
- 57.Clements M L, Subbarao E K, Fries L F, Karron R A, London W T, Murphy B R. J Clin Microbiol. 1992;30:655–662. doi: 10.1128/jcm.30.3.655-662.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herlocher M L, Clavo A C, Maassab H F. Virus Res. 1996;42:11–25. doi: 10.1016/0168-1702(96)01292-0. [DOI] [PubMed] [Google Scholar]
- 59.Ward A C. J Neurovirulence. 1996;2:139–151. doi: 10.3109/13550289609146876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.