Abstract
The multidomain homotetrameric tumor suppressor p53 has two modes of binding dsDNA that are thought to be responsible for scanning and recognizing specific response elements (REs). The C termini bind nonspecifically to dsDNA. The four DNA-binding domains (DBDs) bind REs that have two symmetric 10 base-pair sequences. p53 bound to a 20-bp RE has the DBDs enveloping the DNA, which is in the center of the molecule surrounded by linker sequences to the tetramerization domain (Tet). We investigated by electron microscopy structures of p53 bound to DNA sequences consisting of a 20-bp RE with either 12 or 20 bp nonspecific extensions on either end. We found a variety of structures that give clues to recognition and scanning mechanisms. The 44- and 60-bp sequences gave rise to three and four classes of structures, respectively. One was similar to the known 20-bp structure, but the DBDs in the other classes were loosely arranged and incompatible with specific DNA recognition. Some of the complexes had density consistent with the C termini extending from Tet to the DNA, adjacent to the DBDs. Single-molecule fluorescence resonance energy transfer experiments detected the approach of the C termini towards the DBDs on addition of DNA. The structural data are consistent with p53 sliding along DNA via its C termini and the DNA-binding domains hopping on and off during searches for REs. The loose structures and posttranslational modifications account for the affinity of nonspecific DNA for p53 and point to a mechanism of enhancement of specificity by its binding to effector proteins.
Keywords: protein, recognition, specificity
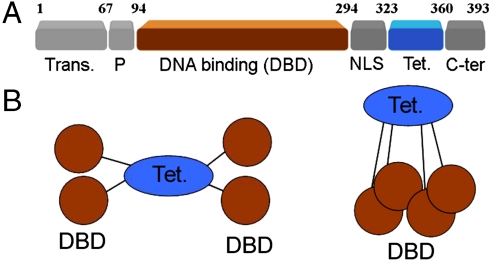
The tumor suppressor p53 is a transcription factor with a central role in genome integrity maintenance (1). Different types of cellular stresses activate the p53 pathway, whose transcriptional regulation of numerous proteins and microRNAs can promote cell cycle arrest, DNA repair, or apoptosis, mechanisms that explain the ability of p53 to inhibit tumor development (2). For its role as transcription factor, p53 tetramers recognize consensus sequences containing two half-sites of the decanucleotide 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′, half-sites that can appear separated by 0–13 base pairs (3). The protein is organized in two stably folded domains, the tetramerization (Tet) and DNA-binding domains (DBD) that are linked and flanked by intrinsically disordered segments (Fig. 1A). Two of these domains can bind DNA: the DBD displays sequence-specific activity in binding to the consensus motif (4); and a highly basic C-terminal domain (C-ter) binds DNA nonspecifically (5). The C-ter is a regulatory domain that modulates the p53 binding to specific sequences in DNA, but the mechanism and the biological role of this regulation are controversial (6). Blocking the C-ter DNA-binding properties enhances p53 binding to short specific DNA probes in the presence of nonspecific competitor DNA sequences (7–10). However, the C-ter does not show negative regulation on p53 binding to specific promoters within long DNA fragments (11). This behavior seems to relate to the capacity of p53 to slide along DNA while in continuous contact with the nucleic acid (12), a motion that requires the presence of the C-ter domain (13). It is thought that one-dimensional sliding might favor searching for specific sequences during initial response of the p53 pathway (14) and/or enhance dissociation of promoter-bound p53 to reduce gene transcription (15). Accordingly, posttranslational modifications of C-ter by kinases or acetylases might control these abilities by reducing the positive charge of the C-ter and its electrostatic interaction with nonspecific DNA.
Fig. 1.
Organization of p53 domains and isolation of p53 tetramers bound to DNA (A) Schematic domain structure of p53. (B) Cartoons depicting the overall quaternary structure of p53 tetramers. Stably folded domains are highlighted in brown (DBD) and blue (Tet.).
Structures of individual domains of p53 have been solved at high resolution. The Tet domain forms a dimer of dimers (16–18). Isolated DBDs self assemble on DNA to form tetramers that have been described bound to two half-site DNA duplexes (19), in oligomers stabilized by crosslinking between DBD and cognate DNA (20), and bound to full consensus sites without chemical modification (21, 22). These structures show varying dimer-dimer contacts and the overall assembly of the core domains on DNA is radically different from the one described in the first DBD·DNA complex solved (23). Taken together, the structural studies suggest that DBD domains can assemble in alternative arrangements for functional purposes.
High-resolution structural studies with full-length p53 are hindered by its flexibility and instability (24). Constructs of a p53 mutant with a superstable, structurally conserved DBD (25) were studied by a combination of SAXS, NMR and EM (26). Low resolution models for p53 tetramers describe a quaternary structure where subunits are linked by Tet domains, and while free p53 tetramers form open cross-shaped structures, the oligomers close around DNA targets via DBD·DNA interactions (Fig. 1B) (26). EM data show that free p53 tetramers also form closed structures, and an equilibrium between open and closed arrangements is postulated. The closed tetramers show Tet domains with D2 (dihedral) symmetry, whereas the remaining domains break this pattern resulting in overall C2 (cyclic) symmetry, an architecture common to other tetrameric DNA-binding proteins, such as the lac repressor (27).
We have investigated the quaternary structure of p53 tetramers while bound to DNA probes with a specific binding site flanked by nonspecific segments. When the nucleic acid has longer nonspecific segments, p53·DNA complexes show large variability in their quaternary structure, mostly in the arrangement of DBD, and the p53 tetramer is seen in equilibrium between tight and loose binding to DNA. Reduction of the nonspecific segments within the DNA probe clearly decreases the structural variability of p53 tetramers, and the DBDs form compact tetramers with extensive contacts between subunits. Also, p53 tetramers seem to bind DNA in different modes that might relate to specific and nonspecific recognition of the duplex. The results provide snapshots of dynamic p53 tetramers and support a two-binding site model for this transcription factor.
Results and Discussion
EM of Isolated p53 Tetramers on DNA.
Bacterially produced p53 binds with high affinity to specific sequences on DNA (28). Full-length p53 (flp53) was allowed to bind DNA sequences containing the GADD45 response element, a specific target for p53. A pure population of flp53 tetramers bound to DNA was isolated and crosslinked by a glycerol-glutaraldehyde combined gradient, a method known as GraFix (29). The procedure facilitates structural studies of fragile complexes and does not generally promote significant structural differences or artifacts in tested samples. Fractions of the gradient were analyzed by EMSA and SDS-PAGE and selected based on the presence of p53·DNA complexes compatible with a tetrameric organization of the protein (Fig. S1). The resulting p53·DNA complexes were visualized by EM in negative staining showing uniformly stained fields with particles homogenous in size (Fig. S1). We used previous structural data as initial reference or calculated new models by common-lines procedures (30). Both methods yielded similar results. The three-dimensional map (Fig. 2A and Fig. S1) is consistent with previous EM results for flp53·DNA, where the p53 tetramer adopts a closed conformation with the Tet region at the top, and the four DBDs at the bottom (26, 31). Two densities linking top and bottom are attributable to the four flexible regions that connect Tet and DBD, suggesting that the p53 tetramer is arranged as a dimer of dimers. There are missing parts that are not seen in the three-dimensional map. There are no signals for N- and C-terminal flexible regions, both of which seem to vanish in the three-dimensional averaging. Furthermore, as before (26, 31), the map does not display density for the nucleic acid, a typical outcome from negatively stained samples.
Fig. 2.
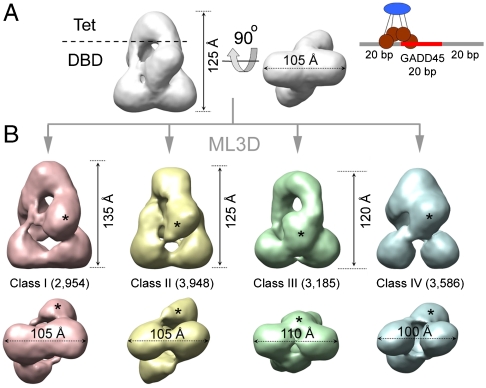
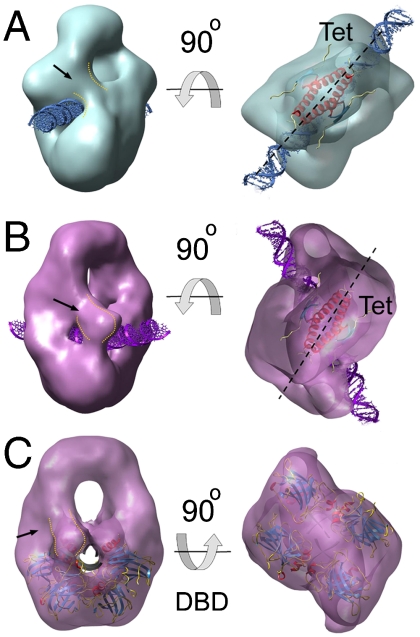
Three-dimensional classification of EM data for p53 tetramers on a 60 bp DNA probe (A) Three-dimensional EM map for the total set of images collected for p53 tetramers bound to a 60-mer DNA containing the GADD45 response element. The renderings show side (left) and bottom (right) views. The cartoon on the right illustrates the overall organization of a p53 tetramer on this DNA probe. (B) Three-dimensional EM maps for the four classes (in different colors) segregated from the p53·DNA sample after ML3D classification. The number of images contributing to each class is indicated. The asterisks label density attributed to the DBD domain that changes its position in the different classes.
P53 Tetramers on a 60 bp DNA Probe.
Our first experiment with flp53·DNA was carried out with a DNA probe enclosing the 20-mer specific sequence for the GADD45 gene, flanked by two 20-mer segments of unspecific random sequence (cartoon in Fig. 2A). The generated three-dimensional map is presented in Fig. 2A. Recently developed classification tools based on maximum-likelihood algorithms, ML3D (32), allow to explore the conformational variability in a population with no need of knowledge about the nature of the structural variability. Our aim was to search for possible distinct conformations of flp53 and localize flexible domains. Thus, we performed ML3D classification, initially sorting the dataset into four different groups (Fig. S2).
The EM maps for the four classes are rendered in Fig. 2B. At first sight, their overall design, i.e., the presence of the Tet region and four DBD domains, is the same as in the map for the total set of images (Fig. 2A). Thus, apparently, the classification does not reveal the regions of p53 that were absent in the initial map (i.e., the N- and C-terminal regions). In Fig. 2B we can see a gradual reduction (from left to right) in the overall dimensions of the maps, starting with the largest structure in class I, to more compact structures in classes III and IV. Interestingly, the four classes display large differences in the relative position of the DBDs (Fig. 1B). With the four maps aligned by the Tet and the two distal DBDs (distal to the Tet), densities corresponding to the proximal DBD change their position within the classes (one of the two symmetric DBDs labeled with asterisks in Fig. 2B). Comparing classes I (red map in Fig. 2B) and II (yellow map in the same), the mobile core domains change their position and make contacts with different densities at the bottom, indicating that DBDs can swap between dimers within the tetramer. The surfaces involved in these swapping interactions are relatively small, which could facilitate the ability of core domains to shift between alternative arrangements. In the other two classes, III and IV, we find the mobile DBDs between the pair at the bottom (class III, the green map in Fig. 2B) or close to the Tet region (class IV, blue map in the same Fig. 2), which describes an up-down motion. A bottom view of the four maps reveals that the interaction between distal DBDs ranges from tight (class I) to loose (class IV) contact (Fig. 2B). Overall, the four EM maps depict dynamic p53·DNA tetramers in a closed conformation where the DBDs are organized in different constellations. It is noteworthy that classification of this dataset yields four isopopulated classes with about 25% of the initial particles assigned to each group, and the conformational variability does not significantly increase using classification into a larger number of groups (up to twelve). The validity of the classification was further tested by cross-refinements between the classes, i.e., by refinement of particles from one of the classes using the three-dimensional EM map from another as initial reference. In all the cases the three-dimensional map for the particular group reemerged regardless of the initial reference used (Fig. S3). Thus, our classification succeeds in separating major conformational states. It is clear, however, that some variability still exists within the four classes, because flexible parts are still missing in the maps.
Differential DNA-Binding Modes of p53 Tetramers.
Because we did not detect a clear signal for the DNA in the two-dimensional images, poor contrast of the nucleic acid precludes direct localization within the EM maps. The location of the double helix, however, can be modeled within channels and grooves while avoiding clashes with densities attributable to p53. Fig. 3 displays some of the three-dimensional EM maps for the different classes in side views, revealing putative DNA trajectories. Classes II and IV showed larger cavities that allow DNA in different orientations, and no model is presented. For class I, we placed the double helix in a see-through channel as depicted in Fig. 3A. No other position is allowed without clashing with the protein. We modeled class III with the DNA in close contact with the DBD region (Fig. 3B), an interpretation consistent with known crystallographic data for DBD·DNA complexes (see below).
Fig. 3.
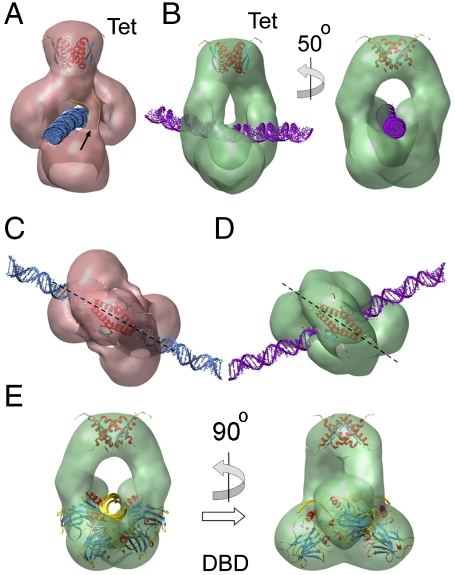
Different DNA-binding modes of p53 tetramers (A) Rendering of the three-dimensional EM map for class I showing a putative passage for the DNA (blue) and fitted coordinates for Tet domains [pdb code: 3SAK; (44)]. The fitted structure for Tet is depicted inside the EM maps in this figure. The arrow points to extra density in the vicinity of the DNA passage. (B) Two views of the EM map for class III showing DNA (purple). (C) and (D) Top views reveal that the relative orientations between DNA and fitted Tet region are different for class I (C) and class III (D). (E) Fitting of the atomic coordinates of a DBD tetramer bound to DNA [pdb code: 2ATA; (19)] within the EM map for class III. The DNA orientation (golden) in this rigid-body fitting is the same as in (B) and (D). The dashed lines in (C) and (D) run along the main symmetry axis of the Tet structure.
Comparing the density maps for classes I and III, we found distinct relative arrangements between the p53 tetramer and DNA. Keeping the structures aligned by the Tet and distal DBDs, there is a relative rotation of the double helix position of around 50° (Fig. 3 A–D). Furthermore, in class I, the symmetry axis of the Tet domain that divides the structure into two dimers, runs parallel to the axis of the dsDNA helix (Fig. 3C). However, due to the relative rotation of the DNA, class III breaks this arrangement (Fig. 3D), and the symmetry axis of the Tet region does not match with the orientation of the DNA. Overall, the results suggest that the arrangement of the p53 tetramer on DNA is of a different nature among the isolated groups.
While the map for class I depicts DBDs in loose contact (Fig. 2B and Fig. 3A), the same domains in class III are in tight contact (Fig. 2B and Fig. 3B). We fitted the atomic structures for DBD tetramers on DNA as rigid bodies inside our EM map for class III. While the structures on continuous DNA (20–22) are not compatible with the current architecture, the DBD tetramer bound to discontinuous DNA (19) shows a good agreement with our envelope (Fig. 3E). The DNA segments of the crystal structure follow the groove on the map and correspond to the orientation for the nucleic acid presented in Fig. 3B. In this atomic structure, however, DNA half-sites are out of register separated by a 2 bp spacer, and the dimers are rotated relative to each other. Our DNA probe is continuous, and to accommodate the crystallographic architecture some severe distortions at the level of the DNA are required. Several studies based on experimental data and simulations (33–35) have shown that the DNA suffers intense bending and twisting when bound by DBD or full-length p53. Some of the modeled DNA configurations would fit within our EM map for class III, but the lack of signal for the nucleic acid in the EM reconstruction precludes further insight. For classes I, II, and IV we have not found any previously described structure for specific DNA binding by DBD fitting into the EM maps, and the densities seem to be too far away from the putative DNA path to assume a tight specific interaction. Thus, class III that represents around 25% of the total population, depicts p53 tetramers compatible with a sequence-specific binding via DBDs, while in the rest of the groups the DBDs are in loose arrangements, not compatible with specific DNA recognition.
P53 Tetramers on a 44 bp DNA Probe.
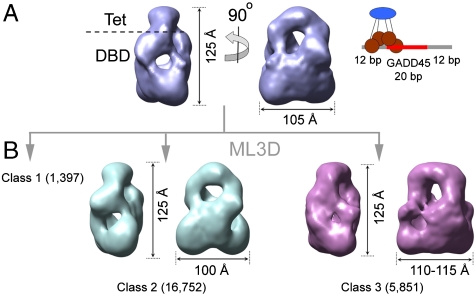
We performed additional structural characterization of flp53·DNA complexes on a DNA probe with shorter nonspecific flanking segments (cartoon in Fig. 4A). The rationale behind this experiment was to test whether the availability of noncognate sequences has any influence on the structural variability of p53 tetramers. In this new DNA probe, the flanking nonspecific DNA segments are still of 12 bp, in order to keep a length of the nucleic acid that overhangs a p53 tetramer, but short enough to avoid full nonspecific interactions. The three-dimensional map rendered in Fig. 4A again shows Tet (top) and DBD (bottom) regions in the same closed organization of the p53 tetramer around a see-through channel, the putative pathway for the DNA. As previously, ML3D classification was performed with this new set of images (Fig. S4). The first observation was that separation into three classes was sufficient to describe the structural variability within this population. The classification produced an asymmetric distribution of images, where around 70% of the images fall in one of the classes (class 2), while another group only recruits 5% of the data (class 1), with insufficient number of images to build up a three-dimensional map.
Fig. 4.
Three-dimensional EM and classification for p53 tetramers on a 44 bp DNA probe (A) Three-dimensional EM map for the total set of images collected for p53 tetramers bound to a 44-mer DNA containing the GADD45 response element. The renderings show two side views. The cartoon on the right depicts the organization of a p53 tetramer on this DNA probe. (B) Three-dimensional EM maps for two classes (in different colors) segregated from the p53·DNA sample after ML3D classification. No map for class 1 is shown since the limited number of images in this group precludes a three-dimensional reconstruction. The number of images contributing to each class is indicated.
The two obtained reconstructions for classes 2 and 3 (Fig. 4B) portray p53 tetramers with similar dimensions and with the DBD region forming a compact density. We did not detect a significant population of tetramers in a loose conformation, as described for the longer DNA probe. Furthermore, at this resolution (around 30 Å), the DBDs are arranged in a similar tetrameric conformation for both classes. Thus, by reducing the fraction of nonspecific DNA, p53 tetramers are seen structurally more uniform at the level of the DBD. The main difference between both maps is related to the position and orientation of channels that could accommodate the DNA (Fig. 5). The rendering of the map for class 2 (Fig. 5A) illustrates that the double helix would run parallel to the longitudinal symmetry axis of the Tet region. By contrast, in the map for class 3 (Fig. 5B), the DNA is tilted with respect to the Tet axis. In the map for class 3, the atomic structure of tetrameric DBD on discontinuous DNA (19) fits accurately within the boundaries of the p53 tetramer and the passage for the DNA (Fig. 5C). The two different configurations for p53·DNA complexes that we observe for classes 2 and 3 (Fig. 5), are similar to the ones described for the classes I and III with the previous DNA probe (Fig. 3). Noteworthy, in both classifications, there is a subset of images (classes III and 3) that displays a clear agreement with one of the crystal structures for DBD·DNA complexes (19) and represents about 25% of the total ensemble.
Fig. 5.
Binding modes of p53 tetramers on a 44-mer DNA (A) Side (left) and top (right) views of the EM map for class 2. In the top view coordinates for Tet domains [pdb code: 3SAK; (44)] are shown in the semitransparent depiction together with a representation of the DNA. (B) The map for class 3 is depicted in orientations similar to the ones shown in (A) for class 2. (C) Side (left) and bottom (right) views of a semitransparent rendering for class 3 together with fitted coordinates of a DBD tetramer bound to DNA [pdb code: 2ATA; (19)]. The arrow in each indicates densities coming down from the Tet region and running close to the DNA passage. The dashed lines in (A) and (B) run along the main symmetry axis of the Tet structure.
C-Terminal Domain in p53·DNA Complexes.
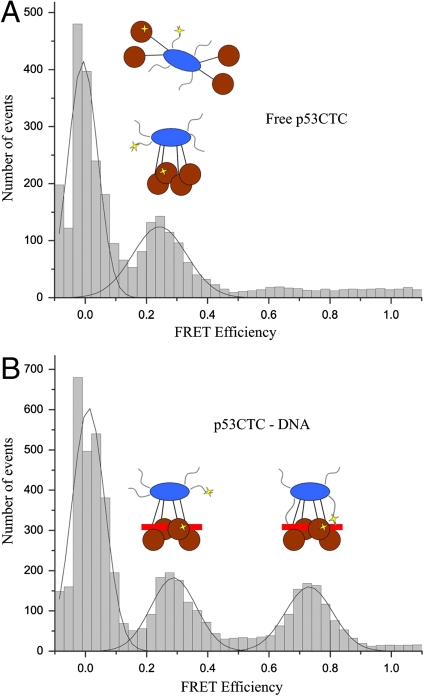
In some of the EM maps, in addition to the two main densities that connect Tet and DBD, we observed additional density, next to the DNA for several classes, namely in class I and in classes 2 and 3 (arrows in Fig. 2A and Fig. 5 A and B). This density is clearly absent in class III (Fig. 3B). This difference might be caused by reorganization of the linker that connects Tet and DBD, and/or by the presence of flexible regions that are stabilized in some of the classes. In all three cases, these bodies are projections coming from the Tet region that reach the vicinity of the DNA duplex. One interesting possibility is that the flexible C-ter, an extension of the Tet domain, is stabilized in some of the p53 conformations. This hypothesis is consistent with the observation that C-ter binds nonspecific DNA sequences (36). The limited resolution of the maps, however, cannot resolve this assignment. We therefore investigated the dynamics of the C-ter domain by single-molecule fluorescence resonance energy transfer (SM-FRET). All surface-exposed cysteines in p53CTC (p53 residues 94–393 containing DBD, Tet, and C-ter) were mutated to alanine, and two new cysteines, one in the DBD (residue 292) and the other in the C-ter (residue 371) were introduced and labeled in a random manner with Alexa fluor 546 and 647 as previously described (37). The mutant protein exhibited the same DNA-binding properties as the wild type.
In the FRET data, the histogram obtained for the free p53CTC has a peak with a low FRET efficiency of about 0.2 (Fig. 6A). In the DNA-bound form (Fig. 6B) a new peak emerged at a FRET efficiency of ∼0.7, an indication of a new ensemble with shorter distance between C-ter and DBD of p53. The FRET peak at 0.2 is unaffected even in the presence of excess DNA. The previous study on the quaternary structure of p53 using SAXS and EM (26) showed that free p53 tetramers can adopt open and closed conformations, while the DNA-bound p53 is closed around the DNA. The peak at 0.2 represents a conformation (or ensemble of conformations) in which C-ter and DBD are far apart. The FRET signal at 0.7 occurs when the C-ter of p53 makes nonspecific contacts with DNA, bringing C-ter and DBD closer together as a result. This conclusion is further supported by another observation. In the presence of a 15 bp DNA probe, the high FRET efficiency peak at 0.7 is not seen, while the peak at 0.2 remains unaffected (histogram as in Fig. 6A), indicating that the C termini are released from the DNA and point away from the DBD region.
Fig. 6.
SM-FRET between labels in C-ter and DBD within a p53 construct. SM-FRET histogram for free p53CTC (A) and p53CTC bound to DNA (I). Cartoons for p53 tetramers in both represent the probable p53 populations centered at 0.24 and 0.73. The location of the FRET labels on p53 is highlighted with yellow stars in the cartoons.
Binding Affinities of C-ter and DBDs to Specific and Nonspecific DNA.
We examined the dissociation constants of p53 and a mutant, p53Cmut, that had all the positive charges in the C-ter removed by mutation. (Table 1, SI Text, and Fig. S5). Whereas both p53s bound to a 20-bp high affinity sequence (X1_20) with a dissociation constant, Kd, of ∼10 nM, neither bound detectably to a “random” 20-bp weak site (X2_20, Kd > 10-4 M). Addition of a further 80 bp of random DNA led to a Kd ∼ 150 nM for p53 at 122–172 mM ionic strength, but ∼2,200 nM for p53Cmut (Table 1). The poor binding of both to X2_20 supports a structural model where DBDs bind only very weakly to nonspecific DNA if they are constrained to the tight structure required to envelope a minimal 20 bp stretch. Further consistency with the models from EM is that long stretches of random DNA, which allow the more relaxed modes of binding, will accommodate the DBDs, as shown by the p53Cmut data, and the presence of unmutated C-ter enhances the binding a further 15 fold.
Table 1.
Dissociation constants for p53 and p53Cmut * binding to DNA from fluorescence anisotropy measurements †
| Protein |
p53 (log P50%/M) |
p53Cmut (log P50%/M) |
| X1_20 ‡ | -7.89 ± 0.01 | -8.30 ± 0.07 |
| X1_100 § | -8.09 ± 0.01 | -8.63 ± 0.01 |
| X2_20 ¶ | > -4 | > -4 |
| X2_100 ∥ | -6.86 ± 0.00 | -5.61 ± 0.01 |
*C terminus of full-length p53 mutated to remove positive charges.
†All experiments were performed at 20 °C in 5 mM DTT, 25 mM NaPi (pH 7.2), 10% glycerol and 100 mM NaCl—see SI Text. (log P50%/M) = logarithm of concentration of p53 for 50% binding.
‡X1_20= high-affinity binding site GGACATGTCCGGACATGTCC).
§X1_100=X1_20+random 80 base-pairs.
¶X2_20= low affinity binding site GAAGATCTCCCAAGATCTTG (the random DNA stretch).
∥X2_100=X2_20+80 further random base pairs. See SI Text for sequences.
Dynamic p53 Tetramers on DNA. Specific vs. Nonspecific DNA Recognition.
In recent years, numerous studies have uncovered the role of the p53 C terminus in the regulation of the DNA binding by p53. In vitro, it has been shown that part of this regulation is the result of competition between nonspecific (by C-ter) and specific (by DBD) binding to DNA (28). Our results show that p53 tetramers bound to DNA have a dynamic behavior. In the presence of the long DNA probe, p53 shows a highly variable architecture at the level of DBD packing (Fig. 2). Two of the DBDs (proximal DBDs) appear at radically different positions, and in most of the maps the DBDs only weakly interact and are in a loose arrangement unsuited for target recognition. This type of loose conformation for nonspecific binding has been described previously for other protein·DNA complexes (38). Only 25% of the population displays a compact DBD tetramer (class III) where dimers appear rotated relative to each other. This architecture is compatible with the atomic structures of DBD engaged in specific DNA recognition of discontinuous DNA (19, 22) but cannot accommodate structures of DBD bound to continuous half-sites that display dimers arranged in parallel fashion (20–22). Our DNA probe, however, contains continuous half-sites and should be overtwisted to accommodate the four DBD in specific binding. This type of DNA deformation has been observed for p53·DNA complexes reaching values as high as 70 ° of overtwisting for DNA in complex with flp53 (35). It is also possible that the compact configuration of DBD observed in class III represents an intermediate with partial sequence recognition. Nevertheless, the existence of p53 tetramers that can be engaged in specific or in nonspecific DNA interactions is supported by the two DNA-binding modes detected in the EM maps (Fig. 3), and by our SM-FRET data that monitor the motions of the C termini with respect to the DBD. We conclude that the flexible C termini embrace the DNA as in a “monorail” system where the DNA functions as the track, and thus p53 tetramers hold the DNA running parallel to the longitudinal symmetry axis of Tet. In this mode, the DBDs are also involved in nonspecific binding embracing the nucleic acid. The results have also implications regarding the assembly and interactions of p53 on DNA. Noteworthy, the two distal DBDs (with respect to Tet) keep a similar position in all the EM maps, facilitating closure of the tetramer around the DNA. What emerges is a model where one of the DBDs of the dimer (distal DBD) first binds nonspecifically to DNA until it reaches the specific sequence. Next, the other DBD within the dimer (proximal to the Tet region) is engaged in the specific binding following a cooperative process as proposed earlier (39). This model provides the structural basis for the linear diffusion of p53 along DNA (13) where the C-ter serves as anchor to scan DNA trails and the DBDs allow p53 linear diffusion with low friction and high stability in a loose arrangement (12). Recently, the sequence analysis of p53 response elements as full-site palindromes composed of four quarter-sites (each quarter being a pentanucleotide), revealed that coupling between quarter-sites 1 and 4 was correlated with higher p53 binding affinity (40). Those quarter sites correspond to the position of the distal DBD in our EM maps, which supports the hypothesis of an initial specific recognition via these sites that could modulate the symmetry of the first pair of DBD bound to DNA and the cooperativity for the tetrameric assembly.
Crosslinking might induce small conformational changes by stapling nearby regions, but sample preparation for long and short DNAs was the same, and differences in the structure of p53 tetramers can therefore be attributed to the distinct nature of the interactions with the nucleic acid. It is clear that the loose architecture of p53 tetramers on DNA depends on the availability of nonspecific segments, since the p53·DNA complex on the 44 bp probe does not show this loose configuration of DBDs (Fig. 4). Within the DBD-DNA superdomain, the four DBD monomers assemble as a tetramer with extensive intra and interdimer contacts. The EM maps could suggest a specific recognition of the DNA, however, only one of the classes, class 3 (Fig. 5C), fits well with known atomic structures for DBD tetramers on DNA (19). The EM map for class 2 (Fig. 5A) fits better with a different p53 binding mode, where the nucleic acid runs parallel to the longitudinal Tet axis, and this suggests a nonspecific DNA recognition. In both maps, nevertheless, we detected extra densities attributable to C-ter next to the nucleic acid (Fig. 5). The structures point to mixtures of DNA interaction types: specific by DBD and nonspecific by C-ter for class 3; and simultaneous nonspecific binding by DBD and C-ter in class 2.
Remarkably, the group compatible with the specific recognition (class 3) represents approximately 25% of the total ensemble, the same ratio as for the long DNA probe (class III). This ratio would indicate that the equilibrium between p53 tetramers in specific and nonspecific DNA-binding mode is not shifted by the use of our distinct DNA targets. It is understood that progress to sequence-specific binding mode and subsequent activation of p53 occurs via posttranslational modifications. Among the positively charged amino acids of the C-ter, there are several lysines that can be acetylated by CBP/p300 (7). Such posttranslational modifications reduce the number of charges in the C-ter, which has a negative effect on p53 linear diffusion properties and will shift the interconverting p53·DNA complex in favor of the specific DNA-binding mode. The outcome of this regulation by C-ter would depend on the posttranslational modifications, the length of the DNA to be scanned, the competition between specific and nonspecific sequences, and on the distribution of single or multiple response elements in target genes. Thus, variable modification of C-ter might supply a fine-tuning device to select downstream genes at the level of DNA binding.
The observation that only the relaxed DBD conformations of p53 bind with significant affinity to nonspecific DNA suggests that there might be proteins that bind to the C termini of p53 (or elsewhere) that constrain it to the quaternary structure specific for binding to response elements. Possible candidates are the 14-3-3 ϵ and γ isoforms that require phosphorylation in the C terminus of p53 to bind to it and enhance binding to response elements, as well as preventing the C termini binding to DNA (41).
Materials and Methods
A variant of human p53 (1–393) carrying four mutations (M133L/V203A/N239Y/N268D) in the DNA-binding domain was expressed and purified as described previously (42). For isolation of p53 tetramers bound to DNA, solutions containing p53 at a concentration of 23 μM and the DNA probe at 30 μM were incubated for 30 min at room temperature, and then stored on ice. The reactions were performed in 25 mM sodium phosphate buffer, pH 7.2, 150 mM NaCl, 5 mM DTT, and 5% glycerol (vol/vol). The DNA probes were preannealed and contained the response element for gene GADD45 flanked by random nonspecific sequences. Tetramers of p53 bound to DNA were isolated using GraFix methodology (29). Selected fractions were adsorbed on carbon coated grids and stained with uranyl formate in double carbon sandwich. EM images were recorded on a 4 k × 4 k CCD resulting in a 2.1 Å final pixel size. Reference-based projection matching was performed in EMAN (43) using a previously obtained EM model for p53·DNA as a starting reference (26). Nonsupervised three-dimensional classification of the images for p53·DNA complexes was performed by the maximum-likelihood based method (32). Resolutions for the EM maps were calculated using cutoffs of 0.5 and in the Fourier Shell Correlation (FSC) and the estimated values were in the range of 24–30 Å. Atomic coordinates for DBD·DNA complex [pdb code: 2ATA; (19)] and Tet domains [pdb code: 3SAK; (44)] were fitted as rigid bodies in Chimera (45), which was also used to produce figures. SM-FRET experiments were carried out as described previously (37). In the gene encoding for the fragment p53CTC (94–393) of human p53 all the surface-exposed cysteines (124, 182, 229, 275, and 277) were mutated to alanine. Two cysteines were introduced at positions 292 and 371. Protein samples were labeled with Alexa fluor 546 and Alexa fluor 647 (Invitrogen, United Kingdom). 60-mer dsDNA, as used in EM studies, was used in single-molecule experiments.
Supplementary Material
Acknowledgments.
We thank Dr. Fang Huang for help with single-molecule experiments. This work was supported by the Medical Research Council and by European Community FP6 funding. M.V. is supported by Ministerio de Ciencia e Innovación Grant (BFU2009-11682), by Etortek Research Program (The Department of Industry, Tourism and Trade of the Government of the Autonomous Community of the Basque Country) and from the Innovation Technology Department of the Bizkaia County. J.M.C. was supported by National Institute of Health (NIH) (HL70472), Ministerio de Ciencia e Innovación (BIO2007-67150-C03-01) and Comunidad Autónoma de Madrid Grants (S-GEN-0166/2006).
Footnotes
The authors declare no conflict of interest.
See companion article on page 563.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015520107/-/DCSupplemental.
References
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 4.Bargonetti J, Manfredi JJ, Chen X, Marshak DR, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 5.Bakalkin G, et al. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn J, Prives C. The C terminus of p53: the more you learn the less you know. Nat Struct Biol. 2001;8:730–732. doi: 10.1038/nsb0901-730. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 8.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 9.Hupp TR, Sparks A, Lane DP. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 10.Jayaraman J, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 11.Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 12.Tafvizi A, et al. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J. 2008;95:L01–03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Lagowski JP, Vanderbeek GE, Kulesz-Martin MF. Facilitated search for specific genomic targets by p53 C-terminal basic DNA binding domain. Cancer Biol Ther. 2004;3:1102–1108. doi: 10.4161/cbt.3.11.1189. [DOI] [PubMed] [Google Scholar]
- 15.Sauer M, et al. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 2008;36:1900–1912. doi: 10.1093/nar/gkn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clore GM, et al. Interhelical angles in the solution structure of the oligomerization domain of p53: correction. Science. 1995;267:1515–1516. doi: 10.1126/science.7878474. [DOI] [PubMed] [Google Scholar]
- 17.Jeffrey PD, Gorina S, Pavletich NP. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 18.Lee W, et al. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 19.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Malecka KA, Ho WC, Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2009;28:325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Dey R, Chen L. Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure. 2010;18:246–256. doi: 10.1016/j.str.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitayner M, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 24.Bell S, Klein C, Muller L, Hansen S, Buchner J. p53 contains large unstructured regions in its native state. J Mol Biol. 2002;322:917–927. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 25.Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem. 2004;1279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 26.Tidow H, et al. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc Natl Acad Sci USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman AM, Fischmann TO, Steitz TA. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. Regulation of DNA binding of p53 by its C-terminal domain. J Mol Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 29.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 30.Van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 31.Scheres SH, Melero R, Valle M, Carazo JM. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure. 2009;17:1563–1572. doi: 10.1016/j.str.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres SH, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 33.Balagurumoorthy P, et al. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc Natl Acad Sci USA. 1995;92:8591–8595. doi: 10.1073/pnas.92.19.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaich AK, Appella E, Harrington RE. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J Biol Chem. 1997;272:14842–14849. doi: 10.1074/jbc.272.23.14842. [DOI] [PubMed] [Google Scholar]
- 35.Nagaich AK, et al. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc Natl Acad Sci USA. 1999;96:1875–1880. doi: 10.1073/pnas.96.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang F, et al. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2009;106:20758–20763. doi: 10.1073/pnas.0909644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 39.McLure KG, Lee PW. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma B, Pan Y, Zheng J, Levine AJ, Nussinov R. Sequence analysis of p53 response-elements suggests multiple binding modes of the p53 tetramer to DNA targets. Nucleic Acids Res. 2007;35:2986–3001. doi: 10.1093/nar/gkm192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajagopalan S, Sade RS, Townsley FM, Fersht AR. Mechanistic differences in the transcriptional activation of p53 by 14-3-3 isoforms. Nucleic Acids Res. 2010;38:893–906. doi: 10.1093/nar/gkp1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ang HC, Joerger AC, Mayer S, Fersht AR. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J Biol Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 44.Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structure with a pseudopotential for the radius of gyration. J Am Chem Soc. 1999;121:2237–2338. [Google Scholar]
- 45.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.