Abstract
The ability to detect DNA damage within the context of the surrounding sequence is an important goal in medical diagnosis and therapies, but there are no satisfactory methods available to detect a damaged base while providing sequence information. One of the most common base lesions is 8-oxo-7,8-dihydroguanine that occurs during oxidation of guanine. In the work presented here, we demonstrate the detection of a single oxidative damage site using ion channel nanopore methods employing α-hemolysin. Hydantoin lesions produced from further oxidation of 8-oxo-7,8-dihydroguanine, as well as spirocyclic adducts produced from covalently attaching a primary amine to the spiroiminodihydantoin lesion, were detected by tethering the damaged DNA to streptavidin via a biotin linkage, and capturing the DNA inside an α-hemolysin ion channel. Spirocyclic adducts, in both homo- and hetero-polymer background single-stranded DNA sequences, produced current blockage levels differing by almost 10% from those of native base current blockage levels. These preliminary studies show the applicability of ion channel recordings not only for DNA sequencing, which has recently received much attention, but also to detecting DNA damage, which will be an important component to any sequencing efforts.
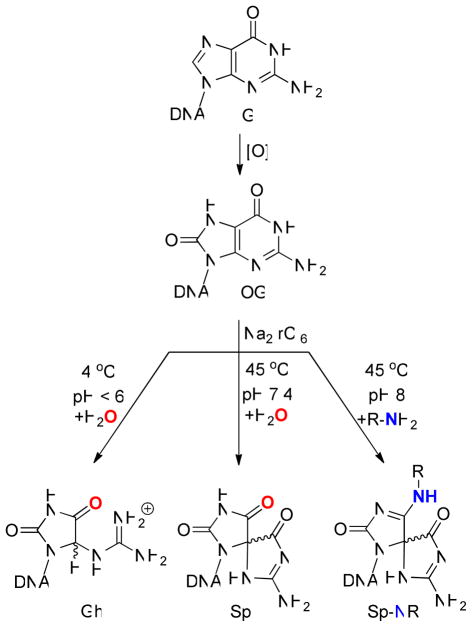
Oxidative stress in the cell underlies multiple age-related disorders including cancer, heart and neurological diseases.1 Reactive oxygen species (ROS) arising from metabolism, inflammation and environmental exposure to redox-active compounds lead to oxidation of many cellular components; those reactions occurring on DNA bases are of particular concern for their mutagenic potential.2,3 Chief among these DNA base lesions is 8-oxo-7,8-dihydroguanine (OG, Figure 1), an oxidized base that exists at the level of ~1 in 106 bases under normal cellular conditions,4 but at much higher levels under conditions of stress or in certain disease states.5 Present methods for detection of OG most commonly involve (1) the comet assay,6 which can be performed on a single cell although the lesion specificity of the assay is not high, and (2) HPLC-MS/MS methods which provide a more accurate count of specific lesions such as OG, but require complete enzymatic digestion to nucleosides before analysis.7 Neither of these methods yield sequence information,8 nor do they provide data on the occurrence of multiple lesions per strand, a phenomenon recognized as highly detrimental to proper DNA function.9
Figure 1.
Oxidation of the biomarker OG leads to the hydantoins Sp and Gh, depending on pH and base stacking context. The oxygen labeled “ ” is incorporated from H2O; inclusion of primary amines during oxidation leads to covalent adducts at this site.
” is incorporated from H2O; inclusion of primary amines during oxidation leads to covalent adducts at this site.
In contrast, single-molecule sequencing methods such as nanopore ion channel detection10 offer the potential to obtain both the identity and the sequence context of base damage sites on individual DNA strands as they translocate through the ion channel. Presently this method is focused on detection of the sequence of the native DNA bases (adenine (A), thymine (T), cytosine (C), and guanine (G)), in order to provide rapid genomic sequencing.11–17 However, sequencing methods based on translocation of DNA through ion channels, as well as solid-state pores, have proved problematic due to insufficient current resolution between the native bases.11,13–15,18 It has previously been shown that single-stranded DNA (ssDNA) molecules can be captured and immobilized within an α-hemolysin (α–HL) ion channel by linking a biotin (Btn) molecule directly to the DNA 3′ terminus and binding the DNA conjugate to streptavidin (Strep), as the streptavidin is too large to pass the α–HL channel.19–23 As a result of this immobilization technique, the longer residence time of the DNA molecule within the α-HL channel circumvents many of the issues involved with ssDNA translocation making it possible to distinguish not only the orientation of DNA entering the pore,19 but a single change in the native base sequence.22,23,24,25
The method described above should also be applicable to single-molecule sequencing of DNA damage, for which no method currently exists, but has great importance in medicinal diagnostics and in understanding the origins of diseases. Herein, we describe the detection of a single oxidative base lesion within a background of surrounding native bases by immobilizing single-stranded DNA in an α-HL ion channel.
Because OG is similar in size and shape to the parent base G, we elected to magnify the difference between the two by taking advantage of the greatly reduced redox potential of OG compared to G (0.74 and 1.29 V. vs. NHE, respectively26), which results in a high reactivity of OG toward further oxidation. The mild, water-soluble oxidants ferricyanide or hexachloroiridate, oxidize OG selectively in a strand of DNA leading to hydantoin lesions, particularly spiroiminodihydantoin (Sp)27 when the oxidation of ssDNA is carried out at pH 8.28 When a primary amine, such as lysine or spermine (Spm), is present during oxidation, an oxidized intermediate is trapped by the nucleophilic amine generating a covalent adduct to OG as a spirocyclic adduct (Sp-NR) (Figure 1).29,30
Single oxidized base modifications, as outlined in Figure 1, were placed within a homopolymer poly-dC background sequence and in the K-ras heterosequence. Both sequences were examined for single base oxidative damage through ion channel recordings. The DNA immobilization technique, introduced by the Bayley and Schmidt laboratories,22,23 was applied to the experiments presented here, and carried out using a wild-type α-HL ion channel reconstituted within a lipid bilayer suspended across a glass nanopore membrane.31–33
In the present work, we demonstrate that a simple chemical derivatization of OG to form hydantoin adducts of various sizes can lead to a nearly 10% change in the ion channel blockage current, not only for homopolymer sequences, but heterosequences as well, providing an excellent entry into the single-molecule sequencing of this oxidized lesion. To our knowledge, this is the first presentation of the ability to detect and identify singe-point base lesions within homo- and hetero-polymer native base sequences through ion channel recordings.
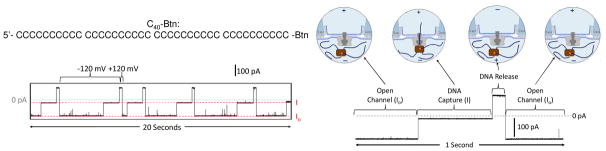
A single WT α-HL protein ion channel was reconstituted into a 1,2-diphytanoyl-sn-glycero-3-phosphocholine lipid bilayer suspended across the orifice of a glass nanopore membrane (GNM), with an orifice radius between 500 and 1000 nm. The GNM provides a robust bilayer support with low electrical noise.31–33 Because the translocation of ssDNA through α-HL is too fast for accurate single-base identification, we immobilized ssDNA oligomers within the ion channel via a tethered biotin-streptavidin (Strep-Btn) complex (Figure 2).22,23
Figure 2.
Strep-Btn-C40 structure, example i-t trace, and molecule capture/release schematic. A C40 ssDNA strand is tethered to a Btn molecule on the 3′ end, and for strand immobilization within the αHL channel, the Btn-DNA is bound to Strep. Schematic not drawn to scale
A voltage was applied across the α-HL ion channel to electrophoretically drive the Strep-Btn DNA complex into the α-HL ion channel, where it was held for 1 to 2 s to collect current blockage event data. It was then released by reversing the voltage bias, which drives the Strep-Btn DNA into the bulk solution, restoring the open channel current. Figure 2 shows a typical current-time (i-t) trace for the capture of Strep-Btn-C40 (hereinafter termed “C40”). When −120 mV was applied (cis vs. trans), an open channel current level, Io, and current blockage level, I, are observed when the channel is unblocked and blocked by C40, respectively. This capture and release cycle was repeated to collect a population of current blockage events (>100 events) for multiple and different DNA strands within the sample of interest. The schematic in Figure 2 illustrates the open channel, capture, release, and open channel cycle for the Strep-Btn immobilization experiment; brief current fluctuation in the open channel current are attributed to noise and unbound single-stranded DNA. All voltages were applied cis vs. trans with respect to the α-HL channel (corresponding to external vs. internal solutions with respect to the GNM).
Previous work has shown that the native base at position 14, relative to the 3′ terminus (ω14) in the DNA strand, has a particularly sensitive influence on the K+ and Cl− ion flux in the β-barrel sensing region of α-HL.22 The DNA lesions under study were initially incorporated into the ω14 position of a poly-dC background with a total length of 40 nucleotides. The ion channel current blockage level of the base lesions at this position were evaluated relative to the unmodified, homopolymer sequence C40.
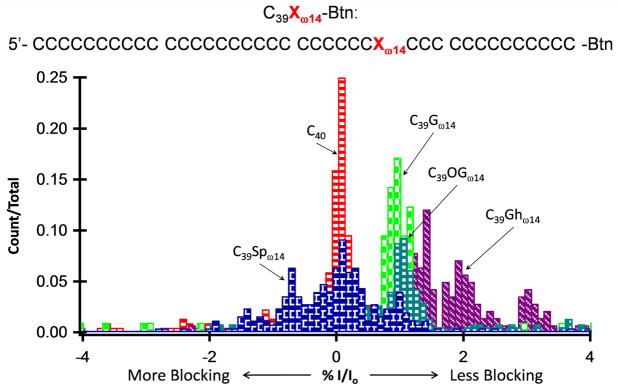
Our initial efforts focused on defining the electrical signature of a single oxidized guanine lesion suspended within the α-HL channel. Figure 3 shows histograms of the percent blockage current when G at position ω14, in a poly-dC background, was substituted by OG or by the further oxidation products Sp or Gh (guanidinohydantoin) (see the supporting information file for individual i-t traces and %I/Io histograms relative to C40 and native bases). C40 was used as a reference and current blockage for all molecules is reported referenced to the blockage level of C40, which is assigned a value of %I/Io = 0; all experiments were reproduced at least once, and peak positions relative to C40 are consistent in position (within 0.1%). In the poly-dC sequence context, G is about 1.2% less blocking than C, and OG yields an electrical signature nearly identical to G. The hydantoin products Sp and Gh are shifted from G and OG, and display broader histograms possibly due to the formation of diastereomeric pairs for each of these products as well as multiple conformations that might exist due to poor base stacking of hydantoins compared to normal bases.
Figure 3.
Current blockage histograms comparing guanine (G), 8-oxo-7,8-dihydroguanine (OG), spiroiminodihydantoin (Sp), and guanidinohydantoin (Gh) at position ω14 relative to C40. All oxidation products are within a poly-dC background, C39Xω14. C40 is used as a reference sample; the % I/Io for C40 was set equal to 0 and % I/Io for all other samples is relative to C40.
Although the oxidative lesions OG, Sp, and Gh give current blockage signatures significantly different from the native G base, we hypothesized that even larger spirocyclic adducts (Sp-X) created by the oxidation of OG in the presence of a primary amine (Figure 1) would modulate the ion channel current to an even larger extent. The amines selected to form adducts were chosen to modify the size and geometry of the DNA strand at position ω14.
Amine adducts introduced at position ω14 within the poly-dC background, C39Xω14, included: benzylamine (Bz), lysine (Lys), glucosamine (GlcN), spermidine (Spd), spermine (Spm), and Gly-Pro-Arg-Pro amide (GPRP). Figure 4 shows the structures for the spirocyclic adducts mentioned above and the %I/Io histograms relative to C40 (see the supporting information file for individual i-t traces and %I/Io histograms for spirocyclic adducts relative to C40 and native bases). The C39Spdω14 and C39Spmω14 adducts are long, linear, and flexible adducts, and both produce multiple, more blocking %I/Io peaks compared with C39Gω14, blocking up to 5% and 7% more current, respectively; Spm being the larger adduct compared with Spd, produced the deepest current blockades. C39Lysω14 produced multiple current blockage levels, with %I/Io peaks spread over a large range, roughly 1–5% more blocking than C39Gω14, with a prominent peak on top of C40. The C39Lysω14 peak that is approximately 1% more blocking than C39Gω14, has a similar %I/Io peak position and distribution to C39Spω14 (Figure 3), and is possibly a result of Lys having been hydrolyzed to Sp. The bulky spirocyclic adducts produced the most variable current blockage signals. For example, C39Bzω14, C39GlcNω14, and C39GPRPω14, all resulted in deeper current blockades compared with C39Gω14, and all produced signals with a broad distribution of current blockage levels and noise amplitudes (see the supporting information file). The various current blockage levels and noise amplitudes are thought to be associated with cyclic adducts adopting different conformations within the α-HL channel.
Figure 4.
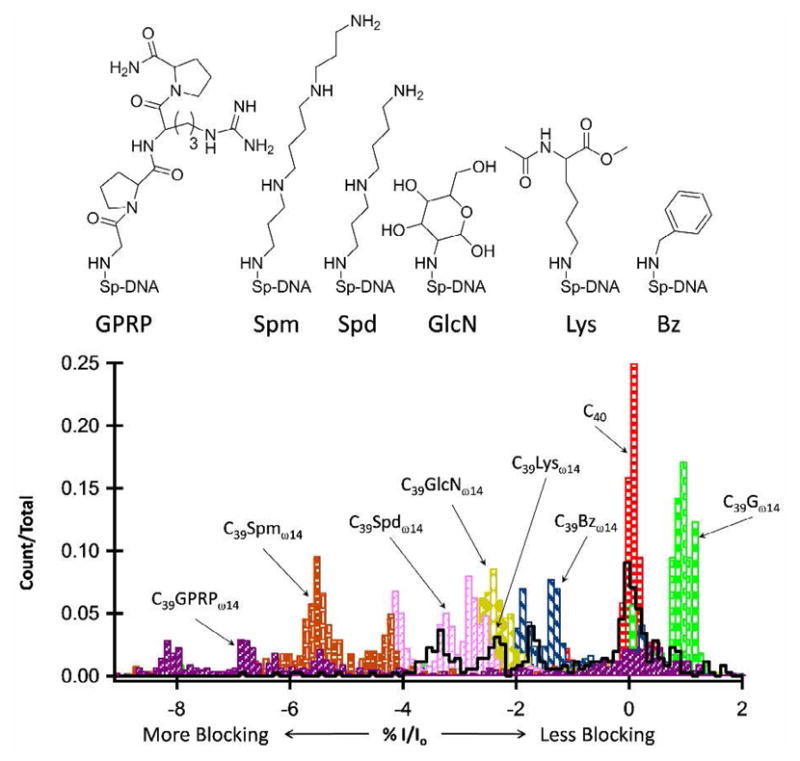
%I/Io histograms comparing benzylamine (Bz), lysine (Lys), glucosamine (GlcN), spermidine (Spd), spermine (Spm), and Gly-Pro-Arg-Pro amide (GPRP) adducts at position ω14 relative to C40. All adducts are within a poly-dC background, C39Xω14. C40 was used as a reference sample; the %I/Io for C40 was set equal to 0, and %I/Io for all other samples is relative to C40.
C39Bzω14 and C39GlcNω14 produced an approximately 3 and 4% negative shift in %I/Io, respectively, relative to C39Gω14. The C39GPRPω14 adduct produced events with the deepest current blockades, up to 10% compared with C39Gω14, but the distribution was very broad and contained a prominent population of events with current blockage levels similar to that of C40. This is interpreted to suggest the molecule is not readily entering the sensing regions of the channel, resulting in a current blockage signal arising from the surrounding poly-dC sequence. To further test the ability to detect oxidative damage within a natural DNA sequence, base modifications were examined within an immobilized heterosequence.
The oxidatively damaged species OG, Sp, and Gh, as well as the spermine adduct to Sp (Spm) were selected for immobilization studies within a heterosequence background to determine the extent to which a DNA damage site can be detected in a sequence of biological relevance. These lesions permit the study of primary DNA damage products as well as a simple adduct to OG (Spm), all of which produced strong %I/Io peaks rather than a dispersed population of blockage currents. The heterosequence selected for study was a portion of the K-ras gene near codon 12 embedded in a poly-dT background while maintaining the 40-mer length (Figure 5). A poly-dT background is used to avoid secondary structures which occur as the poly-dC interacts with the G-rich K-ras sequence. Point mutations within codon 12 of the K-ras proto-oncogene have been shown to cause uncontrolled cell growth and loss of cell differentiation, leading to various human adenocarcinomas.34
Figure 5.
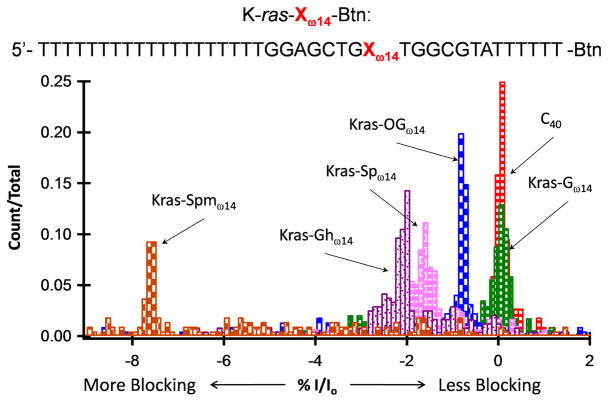
%I/Io histograms for G, OG, Sp, Gh, and Spm base modifications at position ω14 relative to C40. All adducts are within a poly-dT background, Btn-K-ras-Xω14. C40 was used as a reference sample; the %I/Io for C40 was set equal to 0, and %I/Io for all other samples was relative to C40. The %I/Io peak positions relative to C40 and amongst modified DNA molecules have shifted blocking order compared with modifications within a homopolymer background, indicating that the current blockage level is not only influenced by the modification itself, but also the surrounding sequence.
Specifically, in lung cancer, the K-ras gene predominantly undergoes a G → T transversion mutation (GGT → GTT) that could result from the failure to repair guanine oxidative damage before replication.34,35 Thus, we positioned the K-ras sequence within the α–HL channel in such a way that the central G of codon 12, or its oxidized derivatives, was located at the ω14 position. Results for detecting the native gene sequence (K-ras-Gω14), oxidation products OG (K-ras-OGω14), Sp (K-ras-Spω14), and Gh (K-ras-Ghω14), as well as the Spm adduct (K-ras-Spmω14), are displayed in Figure 5.
When comparing the %I/Io peaks for modifications within the poly-dC background against the K-ras sequence, it is apparent that the surrounding sequence has a profound influence on the current signature and event population distribution. Specifically, K-ras-Gω14 and K-ras-OGω14 produced %I/Io peaks separated by approximately 1% from each other, with the K-ras-Gω14 producing the lesser blockage. Whereas G and OG produced overlapping histograms in the C40 background, their signatures are quite distinct and show narrower distributions in the K-ras sequence. K-ras-Spω14 resulted in a single, strong %I/Io peak, approximately 1.5% more blocking than K-ras-Gω14 and again distinct from the OG signal; in the poly-dC background the Sp oxidation product produced a dispersed population of current blockage levels yielding multiple %I/Io peaks with positions similar to C40, C39Gω14, and C39OGω14. The K-ras-Ghω14 sample produced the greatest change from its C39Ghω14 analog; K-ras-Ghω14 has a single %I/Io peak approximately 2% more blocking compared with K-ras-Gω14. In the poly-dC background the Gh oxidation product produced multiple %I/Io peaks that are 0.5–2% less blocking than C39Gω14.
Although %I/Io peak position, dispersion, and relative amplitude are sequence dependent, if the structural modification is large enough, the current blockade from the modification will dominate the influence from the surrounding sequence. The spermine adduct now proves to be such a modification as it consistently produced a more blocking %I/Io peak shift of around 8% relative the unmodified (Gω14) sequence in both the K-ras and poly-dC backgrounds. Overall, %I/Io peaks appear sharper and less dispersed for oxidation products and adducts within the K-ras sequence than when present in the poly-dC background.
The immobilization studies presented above are preliminary experiments toward translocation studies; it will be necessary to examine the most blocking adducts for the ability to readily pass through the constriction of α–HL and translocate the pore. It was previously demonstrated by the Howorka laboratory that peptide tags can be used to change the current blockage amplitude and event duration of translocating DNA strands,36,37 and by the same principal, manipulating the size, geometry, and electrical characteristics of the DNA damage site should lead to detection via nanopore translocation. Future works will focus on characterizing those adducts that most readily distinguish ssDNA strands with oxidative damage from undamaged strands. The translocation for adducts will be evaluated for unique current blockage levels, current signatures, event durations, and translocation velocities as a function of applied voltage, and reported at a later date.
These initial studies have shown the power of ion channel recordings for the detection of DNA damage and highlighted the ability to detect a single lesion within both homo- and hetero-polymer DNA sequences. We have taken a first step toward damage detection through ion channel recording translocation experiments. In addition to being able to detect DNA damage, this work has introduced many options for being able to identify the damage as each adduct produced a unique current blockage level and/or population distribution and noise signature, with a general trend of larger adducts producing the most blocking current blockage levels.
Supplementary Material
Acknowledgments
This work was supported by a seed grant from the University of Utah as well as funds from the NIH (HG005095). Instruments and software donated by Electronic Bio Sciences, San Diego, are gratefully acknowledged as well as helpful discussions with Drs. Geoffrey Barrall, Eric Ervin and John Watkins of EBS. The authors thank Dr. James Muller (Univ. of Utah) for assistance with mass spectrometry.
Footnotes
Supporting Information Available: Experimental methods for synthesis, purification and characterization of all oligomer adducts, immobilization i-t traces and %I/Io histograms, native base immobilization %I/Io histograms, and complete Ref. 10. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cooke MS, Olinski R, Evans MD. Clinica Chimica Acta. 2006;365:30–49. doi: 10.1016/j.cca.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Douki T, Ravanat JL. Free Radic Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Delaney JC, Essigmann JM. Chem Res Toxicol. 2008;21:232–252. doi: 10.1021/tx700292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gedik CM, Collins A. FASEB Journal. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 5.Mangal D, Vudathala D, Park JH, Lee SH, Penning TM, Blair IA. Chem Res Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azqueta A, Shaposhnikov S, Collins AR. Mutat Res. 2009;674:101–108. doi: 10.1016/j.mrgentox.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Cadet J, Poulsen H. Free Radic Biol Med. 2010;48:1457–1459. doi: 10.1016/j.freeradbiomed.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Muller JG, Duarte V, Hickerson RP, Burrows CJ. Nucleic Acids Res. 1998;26:2247–2249. doi: 10.1093/nar/26.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hada M, Sutherland BM. Radiat Res. 2006;165:223–230. doi: 10.1667/rr3498.1. [DOI] [PubMed] [Google Scholar]
- 10.Branton D, et al. Nat Biotech. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer D. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deamer DW, Branton D. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 15.Fologea D, Gershow M, Ledden B, McNabb SD, Golovchenko JA, Li J. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astier Y, Braha O, Bayley H. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 17.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Nature Nanotech. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 18.Meller A, Nivon L, Branton D. Phys Rev Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 19.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Phys Rev Lett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 20.Nakane J, Wiggin M, Marziali A. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purnell RF, Mehta KK, Schmidt JJ. Nano Lett. 2008;8:3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 22.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Proc Natl Acad Sci. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purnell RF, Schmidt JJ. ACS Nano. 2009;9:2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 24.Stoddart D, Maglia G, Mikhailova E, Heron AJ, Bayley H. Angew Chem Int Ed. 2009;48:1–5. doi: 10.1002/anie.200905483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace EVB, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Chem Commun. 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steenken S, Jovanovic SV, Bietti M, Bernhard K. J Am Chem Soc. 2000;122:2372–2374. [Google Scholar]
- 27.Luo W, Muller JG, Rachlin EM, Burrows CJ. Org Lett. 2000;2:613–617. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 28.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 29.Hosford ME, Muller JG, Burrows CJJ. Am Chem Soc. 2004;126:9540–9541. doi: 10.1021/ja047981q. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Muller JG, Ye Y, Burrows CJ. J Am Chem Soc. 2008;130:703–709. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 31.Zhang G, Zhang Y, White HS. Anal Chem. 2004;76:6229–6238. doi: 10.1021/ac049288r. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Galusha J, Shiozawa PG, Wang G, Bergren AJ, Jones RM, White RJ, Ervin EN, Cauley CC, White HS. Anal Chem. 2007;79:4778–4787. doi: 10.1021/ac070609j. [DOI] [PubMed] [Google Scholar]
- 33.White RJ, Ervin EN, Yang T, Chen X, Daniel S, Cremer PS, White HS. J Am Chem Soc. 2007;129:11766–00775. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer GP, Besaratinia Hum Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David SS, O’Shea VL, Kundu S. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell N, Howorka S. Angew Chem Int Ed. 2008;47:5565–5568. doi: 10.1002/anie.200800183. [DOI] [PubMed] [Google Scholar]
- 37.Borsenberger V, Mitchell N, Howorka S. J Am Chem Soc. 2009;131:7530–7531. doi: 10.1021/ja902004s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.