INTRODUCTION
X-ray crystallographic studies reveal that CXCL12 is able to form multiple dimer types, a traditional CXC dimer and a ‘CC-like’ form. Phylogenetic analysis of all known human chemokines demonstrates CXCL12 is more closely related to the CC chemokine class than other CXC chemokines. These observations indicate that CXCL12 contains genomic and structural elements characteristic of both CXC and CC chemokines.
Chemokines are members of a superfamily of proteins involved in the migration of cells to the proper anatomical position during embryonic development or in response to infection or stress during an immune response1. There are two major (CC and CXC) and two minor (CX3C and XC) families based on the sequence around the first conserved cysteine. The topology of all structures is essentially identical with a flexible N-terminal region of 3-8 amino acids, a 10-20 residue N-terminal loop, a short 310-helix, three β-strands, and a α-helix. The major consequence of the subtle difference between the families occurs at the oligomeric level. Monomers of the CC, CXC, and CX3C families form dimers in a family-specific manner. The XCL1 chemokine is a monomer that can interconvert between two folded states2. All chemokines activate GPCRs according to family-specificity, however there are a few examples of chemokines crossing the family boundary to function as antagonists3,4. A two-stage mechanism for chemokine activation of GPCRs has been proposed5,6. The N-terminal region of the receptor interacts with the chemokine, followed by receptor activation by the chemokine N-terminal region. Monomeric chemokines have been demonstrated to be the active form for receptor function7. There are numerous examples of both chemokines and their receptors forming dimers8–11. While family-specific dimerization may be an attractive explanation for why specific chemokines only activate GPCRs within their own family, the role of dimers in the function of chemokines has not been resolved7,8,12–14.
Given that CXCL12 is in the CXC family, the CXC dimer is considered the physiologic dimer in all previous studies based on crystallographic evidence. NMR and mutational studies agree with the CXC dimer form in solution8,14,15. The CXC form of the dimer is seen in recent structures of CXCL12 bound to a heparin disaccharide16 and several CXCR41–38 peptides17. In one case, crystals of the CXC-type dimer were soaked in a heparin disaccharide solution to determine the interactions between this dimer and bound disaccharide. In another case, in order to overcome NMR chemical shift line broadening when CXCR41–38 peptides are added, a ‘locked’ dimer was constructed by introducing a cysteine mutant that linked subunits as a CXC dimer through an inter-subunit disulfide bond. The solution structures of the locked CXC dimer with CXCR41–38 peptides were determined. The locked CXC dimer retained Ca2+ mobilization yet lost chemotaxis activity, presumably because the monomer is the active form17. In addition to existing as a monomer18 and CXC dimer19, CXCL12 is now demonstrated to have the capacity to form CC type dimers in the presence of a CXCR41–27 peptide.
MATERIALS AND METHODS
X-RAY CRYSTALLOGRAPHY
CXCL12 was purified as previously described16. CXCR41–27 was synthesized at the W. M. Keck facility (Yale University). A mixture of CXCL12 (12 mg/ml) and CXCR41–27 (10 mg/ml) were screened with Hampton Crystal Screen 1 and 2 using a Mosquito liquid handling robot (TTP Labtech). Initial hits were found in two conditions. One condition containing 10% Jeffamine M-600, 0.01 M FeCl3, 0.1 M sodium citrate tribasic, pH 5.6 yielded crystals in the P212121 space group. Crystals were optimized by using an additive screening kit (Hampton Research) and by adjusting the ratio of protein-to-well solution. The crystals used for the data collection were grown with an additional 1% PEG 3350 at a 3:1 protein: well solution ratio. The second condition (#2) which yielded crystals in the P3221 space group consisted of 30% PEG 4000, 0.2 M lithium sulfate, and 0.1 M Tris HCl, pH 7.5. Optimization was not required.
DATA COLLECTION
Data were collected at NSLS at BNL on beamline X29A using the Quantum ADSC Q315 CCD detector. Crystal condition # 1 was soaked in cryoprotectant solution containing the heavy atom derivative (25% Jeffamine M-600, 0.01 M FeCl3, 0.1 M Sodium citrate Tribasic pH 5.6, 1% PEG-3350, 10 mM Potassium tetrachloroaurate) for 18 hours at 18 °C then flash frozen in liquid nitrogen and transferred to the −180 °C cold stream. A wavelength florescence scan was done around the theoretical peak wavelength for Au3+ absorption. 360 frames were collected at 1 degree per frame with a 1 second exposure time at 1.04Å (the measured peak wavelength for the gold soaked crystal). Crystal condition #2 was flash frozen without an additional cryoprotectant in liquid nitrogen then transferred to a −180 °C cold stream. 180 frames were collected at 1 degree per frame with a 1 second exposure time at 1.0809Å.
DATA PROCESSING, PHASING AND REFINEMENT
Data frames from both crystal conditions were processed using HKL200020. The gold soaked crystal condition #1 was processed for anomalous diffraction measurements.
Initial molecular replacement using CXCL12 as a search model for crystal form 1 failed due to translational symmetry with a NCS axis parallel to one edge of the unit cell. This NCS was identified by constructing a native Patterson map. There was a strong peak in the native Patterson map at (0, 0.5, 0.36). Crystal form #1 was phased using SAD with the programs Sharp/Autosharp21 and Solve22 in parallel to locate the tetrachloroaurate ions. Phased MR using Molrep23 located the position of five chains of CXCL12, 5-fold NCS averaging of the maps allowed phased MR to identify three other chains. Eight-fold NCS averaging then allowed the final two chains to be located. Refinement was done using phenix.refine24. Manual fitting and addition of water molecules and gold compounds using composite omit maps and model-bias removed maps (http://tuna.tamu.edu) as guides was continued in combination with TLS and restrained refinement including 10-fold NCS with loose restraints in refmac25 until there was no further reduction in Rfree.
Phasing on crystal form #2 was done by molecular replacement in Phaser26 using chain A from PDB ID code 2NWG as a search model. Refinement was done using phenix.refine24, followed by several rounds of manual adjustment and model building using Coot27 and automatic restrained refinement in refmac from Coot.
Final refinement of both structures was done using TLS and restrained refinement in refmac25. Validation of both structures was done using SFcheck28, Coot and the RCSB validation server29. Molprobity validation30 was done and clash scores were calculated. Crystallography statistics are included in Table 1.
Table 1.
Summary of crystallography statistics
| Data Collection | ||
| PDB ID code | 3HP3 | 3GV3 |
| Space group | P212121 | P3221 |
| Cell dimensions | ||
| a, b, c (Å) | 41.83, 117.46, 134.50 | 55.51, 55.51, 45.96 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Wavelength (Å) | 1.04 | 1.0809 |
| Resolution rangea (Å) | 29.2 – 2.20 (2.28 – 2.20) | 30.0 – 1.60 (1.66 – 1.60) |
| <I>/<σ I> | 47.8 (4.1) | 26.8 (4.3) |
| Completeness | 99.9% (99.9%) | 98.8% (100%) |
| Rmerge | 0.063 (0.667) | 0.074 (0.491) |
| Redundancy | 14.0 (14.1) | 10.4 (10.5) |
| Refinement | ||
| Number of refelections | 1418341 | 113767 |
| Unique reflections | 39406 (3382) | 10951 (1057) |
| Rfactor | 0.22 | 0.20 |
| Rfree | 0.27 | 0.25 |
| Protein chains | 10 | 1 |
| Number of atoms | 5484 | 562 |
| Protein | 5241 | 514 |
| Water | 231 | 58 |
| Gold | 12 | NA |
| B-factor average | 29.7 | 23.6 |
| R.m.s. deviations | ||
| Bong lengths (Å) | 0.008 | 0.031 |
| Bond angles (°) | 1.229 | 2.405 |
| Molprobity clash score | 12.6 | 16.2 |
| Ramachandran plot | ||
| Most favored (%) | 94.33 | 98.36 |
| Allowed (%) | 4.21 | 1.64 |
| Generously Allowed (%) | 1.46 | 0 |
Highest resolution shell is shown in parenthesis.
PHYLOGENETIC TREE
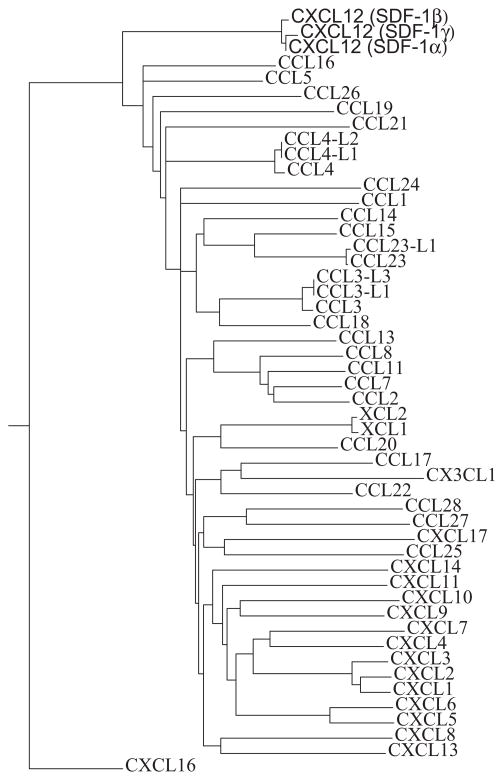
The protein sequences of all human chemokines were downloaded from NCBI web site (http://www.ncbi.nlm.nih.gov/). A multiple sequence alignment was done using T-Coffee at the EBI server (http://www.ebi.ac.uk/Tools/t-coffee/index.html) with all the default parameters. The tree in Figure was calculated also using T-Coffee at the EBI server. The algorithm used PID as the distance measure (PID = number of equivalent aligned non-gap symbols * 100/smallest number of non-gap positions in either of both sequences), which is essentially the “number of identical residues per 100 residues”. The neighbor-joining method was used to calculate the tree. The NJ method is a greedy algorithm to find the tree with the shortest branch lengths.
RESULTS AND DISCUSSION
It was suggested in the original publication of the X-ray structure of CXCL12 that if the CXC portion of the sequence was ignored, the sequence and structure of CXCL12 was the most divergent from all known CXC chemokines known at that time19. In the present study we observe CXCL12 forming as a CC-chemokine dimer. Using the full repertoire of human chemokines known from the human genome sequence, phylogenetic analysis is able to further establish a closer relationship between human CXCL12 and CC chemokines than to CXC chemokines [Figure 1].
Figure 1.
Phylogenetic tree calculated using the neighbor-joining (NJ) method representing all human chemokines and their isoforms. The tree is based on the multiple sequence alignment. The three isoforms of CXCL12 (SDF-1α, β, and γ) are indicated at the uppermost portion of the figure.
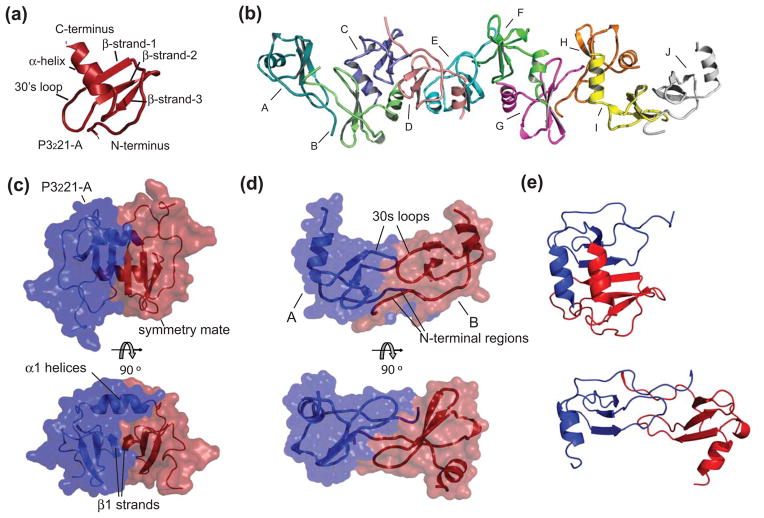
Previous crystallographic studies resulted in CXC dimers of CXCL1216,19,31 but in solution monomers predominate2,15,18 and are dimerized by basic pH, a heparin disaccharide, the introduction of an additional cysteine that forms an inter-subunit disulfide, and peptides from the CXCR4 N-terminal region8,14,17. In this study crystals in two different space groups are presented [Table 1]. One crystal form (P3221) contains a monomer in the asymmetric unit [Figure 2(a)], and forms a typical CXC dimer when symmetry is applied [Figure 2(b)]. Inter-subunit interactions are identical to that observed previously consisting of three β-strands from each subunit forming a six stranded β-sheet and contacts between the two α-helices. The second crystal form (P212121) has a decamer of CXCL12 in the asymmetric unit [Figure 2(c)]. The decamer is formed from five dimers of CXCL12. These dimers are elongated and cylindrical with symmetric subunit-subunit hydrogen bonding and hydrophobic interactions primarily involving the N-terminal regions (residues 4-15) and the 30’s loops. This interaction strongly resembles a typical CC chemokine dimer [Figure 2(d)]. A comparison of both dimers is presented with Chain A from both dimers held in the same orientation [Figure 2(e)]. These crystals formed in an attempt to co-crystallize CXCL12 in the presence of the CXCR41–27 peptide. Electron density for the CXCR4 peptide was not observed in three-dimensional structure of either crystal form. The peptide acts as a precipitant or induces dimerization and/or nucleation, yet is displaced during the crystallization.
Figure 2.
Crystal structures of CXCL12. (a) Cartoon representation of the single protein chain of the asymmetric unit in the P3221 space group CXCL12 crystals. Chemokine structural elements are labeled. (b) The protein composition of the asymmetric unit in the P212121 space group CXCL12 crystals. Ten monomers are arranged as an elongated decamer and form five CC-like dimers. (c) The semi-transparent surface representation of the asymmetric unit of the P3221 space group of CXCL12 and a symmetry mate forms a CXC dimer that interact through their α-helices and β-strand-1. (d) Semi-transparent surface of two representative chains in the decamer of CXCL12 which form a CC chemokine-like dimer and interact with their amino termini and 30’s loops. (e) CXC dimer of CXCL12 (upper panel) compared to CC dimer of CXCL12 (lower panel). Both dimers were aligned using only Chain A of each in the alignment algorithm.
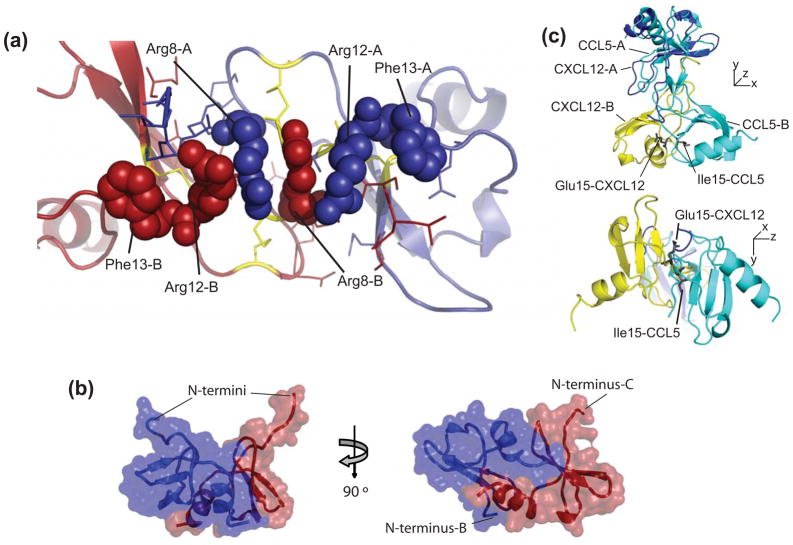
Within the decamer, an alternative dimer interface is observed in addition to the CC-type dimer. The additional dimer form is composed of alternating CXCL12 monomers from adjacent CC dimers [Figure 3(a)]. The formation of this potential dimer under physiological conditions is currently excluded based on the absence of this quaternary structure in the extensive structural biology of chemokines32. To more fully characterize the significance of these two dimers within the decamer, the Protein Interfaces, Surfaces and Assemblies server was used (Table 2)33. Both types of dimers had negative ΔiG’s upon formation of an interface, but the contributions of hydrogen bonds and salt bridges are not included in this calculation33. It is important to note that the CC dimers have an average of 19 hydrogen bonds versus 8 for the alternate dimer. The significance scores (CCS) are 1.0 all CC-like chemokines and vary from 0 to 1.0 for the alternate dimer. These results are consistent with the possibility that the CC-type dimer can form in environments other than this crystal form and the alternative dimer cannot.
Figure 3.
Intersubunit contacts of CXCL12 dimers. (a) Arginine zipper motif forms the interactions in the amino terminal regions of the CC-like CXCL12 dimer. Cysteines are colored yellow, arginines from chains A and B are colored purple and red, respectively. (b) Crystallographic packing between CC-dimers. Cartoon representation with semi-transparent surface of two representative chains in the decamer of CXCL12 in between each CC chemokine-like dimer. (c) Overlay of the CC-like dimer of CXCL12 and the CC dimer of CCL5. Chain A from each dimer was aligned. Glutamic acid 15 from CXCL12 and isoleucine 15 from CCL5 are shown as sticks to indicate the 180° rotation of the carbon backbone.
Table 2.
Summary of subunit-subunit interfaces1
| Chains | Number of residues in interface2 | Interface Area (Å2) | ΔiG kcal/mol3 | Hydrogen Bonds | Salt Bridges | CSS4 | |
|---|---|---|---|---|---|---|---|
| CXCL12 CXC dimer | A, sym | 48 | 1103.2 | −4.1 | 22 | 0 | 1.00 |
| CXCL12 CC dimer 1 | A, B | 45 | 889.2 | −9.3 | 12 | 0 | 1.00 |
| CXCL12 CC dimer 2 | C, D | 46 | 1015.9 | −5.7 | 21 | 0 | 1.00 |
| CXCL12 CC dimer 3 | E, F | 48 | 1101.0 | −4.1 | 22 | 0 | 1.00 |
| CXCL12 CC dimer 4 | G, H | 45 | 977.4 | −6.8 | 19 | 0 | 1.00 |
| CXCL12 CC dimer 5 | I, J | 52 | 1299.8 | −8.9 | 20 | 1 | 1.00 |
| average of CC dimers | 47 | 1056.7 | −7.0 | 19 | <1 | 1.00 | |
| between CC dimers 1 | B,C | 49 | 842.0 | −9.1 | 4 | 0 | 0.27 |
| between CC dimers 2 | D,E | 33 | 531.0 | −8.5 | 3 | 0 | 0.00 |
| between CC dimers 3 | F,G | 42 | 780.2 | −5.2 | 6 | 0 | 1.00 |
| between CC dimers 4 | H,I | 42 | 854.0 | −10.7 | 8 | 0 | 1.00 |
| between CC dimers 5 | A, J sym | 38 | 624.5 | −8.8 | 3 | 0 | 0.15 |
| average between dimers | 41 | 726.3 | −8.5 | 5 | 0 | 0.48 | |
Calculated using the Protein interfaces, surfaces and assemblies service PISA at European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), authored by E. Krissinel and K. Henrick
Includes residues from both subunits
Corresponds to solvation free energy from hydrophobic forces. This does not include satisfied hydrogen bonds and salt bridges
Complexation Significance Score, which indicates how significant for assembly formation the interface is.
One interesting aspect of the subunit-subunit interactions of the CC-like dimer is a zipper of four arginine side chains, Arg8 and Arg12 from each subunit, which is terminated on each side with Phe13 [Figure 3(b)]. A comparison with CCL5 reveals that the CXCL12 CC-like dimer has one distinct difference. Alignment of one subunit (chain A) for each protein leads to a different orientation for subunit B [Figure 3(c)]. The difference is initiated by the way the amino terminal regions come together for each protein. The zipper of arginine residues forms hydrogen bonds with N-terminal backbone atoms. This precludes the N-terminal region of each subunit to form complementary H-bonds resulting in a two-stranded β-sheet typical of dimeric CC chemokines. Consequently, due the presence of the N-terminal H-bonds to the Arg zipper (instead of the complementary N-terminal H-bonds that form the β-sheet) there is nearly rotation of 180° in the N-terminal backbone atoms of chain B in CXC12 as compared to CCL5. This results in a displacement of chain B of CXCL12 relative to CCL5 allowing the 30s loops to come together and interact with each other. This displacement is not unique in the structural biology of chemokines. For example, CCL2 also forms a typical CC dimer, yet there is a significant variation in the position of the ‘B’ chains when the ‘A’ chains are aligned from independent structural determinations34–36, including PDB ID code 3IFD (unpublished).
Many studies of chemokines indicate that protein dynamics37, subtle conformational changes within a dimer38, or major conformational changes such as conversion from dimer to monomer2,12 are important for biological function. The results in this study reveal that only in the presence of CXCR41–27, CXCL12 crystallizes in two forms, each containing a different dimeric state. Whether formation of the new quaternary structure or a conversion between CXC and CC-forms has any significance in CXCL12 function are improbable but remains to be determined. The present study, however, demonstrates formation of a CC-like dimer is possible for CXCL12 and, more importantly, brings attention to an unappreciated phylogenetic and structural relationship between CXCL12 and CC chemokines.
Acknowledgments
This study was supported by the US National Institutes of Health (AI082295 to E.J.L.). We thank X29 beamline personnel at the National Synchrotron Light Source at Brookhaven National Synchrotron Light Source for help with X-ray data collection. Support for beamline X29 comes from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. We also thank G. Crichlow for helpful crystallographic advice and M. Hodsdon for helpful comments.
Footnotes
Accession codes. Coordinates and structure factors of CC-CXCL12 and CXC-CXCL12 have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB entries 3HP3 and 3GV3).
References
- 1.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 2.Veldkamp CT, Ziarek JJ, Su J, Basnet H, Lennertz R, Weiner JJ, Peterson FC, Baker JE, Volkman BF. Monomeric structure of the cardioprotective chemokine SDF-1/CXCL12. Protein Science. 2009;18(7):1359–1369. doi: 10.1002/pro.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viola A, Luster AD. Chemokines and Their Receptors: Drug Targets in Immunity and Inflammation. Annual Review of Pharmacology and Toxicology. 2008;48(1):171–197. doi: 10.1146/annurev.pharmtox.48.121806.154841. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez EJ, Wilken J, Thompson DA, Peiper SC, Lolis E. Comparison of the structure of vMIP-II with eotaxin-1, RANTES, and MCP-3 suggests a unique mechanism for CCR3 activation. Biochemistry. 2000;39(42):12837–12844. doi: 10.1021/bi001166f. [DOI] [PubMed] [Google Scholar]
- 5.Siciliano SJ, Rollins TE, DeMartino J, Konteatis Z, Malkowitz L, Van Riper G, Bondy S, Rosen H, Springer MS. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proc Natl Acad Sci U S A. 1994;91(4):1214–1218. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells TN, Power CA, Lusti-Narasimhan M, Hoogewerf AJ, Cooke RM, Chung CW, Peitsch MC, Proudfoot AE. Selectivity and antagonism of chemokine receptors. J Leukoc Biol. 1996;59(1):53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 7.Rajarathnam K, Sykes BD, Kay CM, Dewald B, Geiser T, Baggiolini M, Clark-Lewis I. Neutrophil activation by monomeric interleukin-8. Science. 1994;264(5155):90–92. doi: 10.1126/science.8140420. [DOI] [PubMed] [Google Scholar]
- 8.Veldkamp CT, Peterson FC, Pelzek AJ, Volkman BF. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 2005;14(4):1071–1081. doi: 10.1110/ps.041219505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vila-Coro AJ, Rodriguez-Frade JM, Martin De Ana A, Moreno-Ortiz MC, Martinez AC, Mellado M. The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. Faseb J. 1999;13(13):1699–1710. [PubMed] [Google Scholar]
- 10.Babcock GJ, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. Journal of Biological Chemistry. 2003;278(5):3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, He L, Combs CA, Roderiquez G, Norcross MA. Dimerization of CXCR4 in living malignant cells: control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Mol Cancer Ther. 2006;5(10):2474–2483. doi: 10.1158/1535-7163.MCT-05-0261. [DOI] [PubMed] [Google Scholar]
- 12.Fernando H, Chin C, Rosgen J, Rajarathnam K. Dimer dissociation is essential for interleukin-8 (IL-8) binding to CXCR1 receptor. J Biol Chem. 2004;279(35):36175–36178. doi: 10.1074/jbc.C400283200. [DOI] [PubMed] [Google Scholar]
- 13.Williams G, Borkakoti N, Bottomley GA, Cowan I, Fallowfield AG, Jones PS, Kirtland SJ, Price GJ, Price L. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J Biol Chem. 1996;271(16):9579–9586. doi: 10.1074/jbc.271.16.9579. [DOI] [PubMed] [Google Scholar]
- 14.Veldkamp CT, Seibert C, Peterson FC, Sakmar TP, Volkman BF. Recognition of a CXCR4 Sulfotyrosine by the Chemokine Stromal Cell-derived Factor-1[alpha] (SDF-1[alpha]/CXCL12) Journal of Molecular Biology. 2006;359(5):1400–1409. doi: 10.1016/j.jmb.2006.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gozansky EK, Louis JM, Caffrey M, Clore GM. Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha. J Mol Biol. 2005;345(4):651–658. doi: 10.1016/j.jmb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Murphy JW, Cho Y, Sachpatzidis A, Fan C, Hodsdon ME, Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1alpha) binding to heparin. J Biol Chem. 2007;282(13):10018–10027. doi: 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veldkamp CT, Seibert C, Peterson FC, De la Cruz NB, Haugner JC, 3rd, Basnet H, Sakmar TP, Volkman BF. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1(37):ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell- derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. Embo J. 1997;16(23):6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dealwis C, Fernandez EJ, Thompson DA, Simon RJ, Siani MA, Lolis E. Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 “fusin” coreceptor. Proc Natl Acad Sci U S A. 1998;95(12):6941–6946. doi: 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods In Enzymology. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 24.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods in molecular biology (Clifton, NJ) 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 29.Lin D, Manning NO, Jiang J, Abola EE, Stampf D, Prilusky J, Sussman JL. AutoDep: a web-based system for deposition and validation of macromolecular structural-information. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 7):828–841. doi: 10.1107/s0907444900005655. [DOI] [PubMed] [Google Scholar]
- 30.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi Y, Senda T, Nandhagopal N, Sugimoto K, Shioda T, Nagal Y, Mitsui Y. Crystal structure of recombinant native SDF-1alpha with additional mutagenesis studies: an attempt at a more comprehensive interpretation of accumulated structure-activity relationship data. J Interferon Cytokine Res. 2000;20(8):691–700. doi: 10.1089/10799900050116390. [DOI] [PubMed] [Google Scholar]
- 32.Lolis E, Murphy JW. The Structural Biology of Chemokines. In: Harrison JK, Lukacs NW, editors. The Chemokine Receptors. Totowa, N.J.: Humana Press; 2007. [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. Journal of Molecular Biology. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Blaszczyk J, Coillie EV, Proost P, Damme JV, Opdenakker G, Bujacz GD, Wang JM, Ji X. Complete crystal structure of monocyte chemotactic protein-2, a CC chemokine that interacts with multiple receptors. Biochemistry. 2000;39(46):14075–14081. doi: 10.1021/bi0009340. [DOI] [PubMed] [Google Scholar]
- 35.Handel TM, Domaille PJ. Heteronuclear (1H, 13C, 15N) NMR assignments and solution structure of the monocyte chemoattractant protein-1 (MCP-1) dimer. Biochemistry. 1996;35(21):6569–6584. doi: 10.1021/bi9602270. [DOI] [PubMed] [Google Scholar]
- 36.Lubkowski J, Bujacz G, Boque L, Domaille PJ, Handel TM, Wlodawer A. The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions. Nat Struct Biol. 1997;4(1):64–69. doi: 10.1038/nsb0197-64. [DOI] [PubMed] [Google Scholar]
- 37.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proceedings of the National Academy of Sciences. 2008;105(13):5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baryshnikova OK, Sykes BD. Backbone dynamics of SDF-1{alpha} determined by NMR: Interpretation in the presence of monomer-dimer equilibrium. Protein Sci. 2006;15(11):2568–2578. doi: 10.1110/ps.062255806. [DOI] [PMC free article] [PubMed] [Google Scholar]