Abstract
Antagonistic host–parasite interactions can drive rapid adaptive evolution in genes of the immune system, and such arms races may be an important force shaping polymorphism in the genome. The RNA interference pathway gene Argonaute-2 (AGO2) is a key component of antiviral defense in Drosophila, and we have previously shown that genes in this pathway experience unusually high rates of adaptive substitution. Here we study patterns of genetic variation in a 100-kbp region around AGO2 in three different species of Drosophila. Our data suggest that recent independent selective sweeps in AGO2 have reduced genetic variation across a region of more than 50 kbp in Drosophila melanogaster, D. simulans, and D. yakuba, and we estimate that selection has fixed adaptive substitutions in this gene every 30–100 thousand years. The strongest signal of recent selection is evident in D. simulans, where we estimate that the most recent selective sweep involved an allele with a selective advantage of the order of 0.5–1% and occurred roughly 13–60 Kya. To evaluate the potential consequences of the recent substitutions on the structure and function of AGO2, we used fold-recognition and homology-based modeling to derive a structural model for the Drosophila protein, and this suggests that recent substitutions in D. simulans are overrepresented at the protein surface. In summary, our results show that selection by parasites can consistently target the same genes in multiple species, resulting in areas of the genome that have markedly reduced genetic diversity.
Keywords: RNAi, virus, arms race, selective sweep, AGO2, Drosophila
Background
When natural selection replaces one allele with another, the hitchhiking of nearby variants through the population leaves a characteristic footprint in genetic diversity (Maynard Smith and Haigh 1974; Braverman et al. 1995). Such ‘selective sweeps’ will significantly shape genomic variation, with the impact depending on the selective advantage of new mutations, and the frequency with which they arise (e.g., Maynard Smith and Haigh 1974; Durrett and Schweinsberg 2004; Pennings and Hermisson 2006; Hermisson and Pfaffelhuber 2008). Many such sweeps have been identified in nature and have contributed to our understanding of both the process and targets of recent selection (e.g., Nielsen et al. 2007). In particular, studies in Drosophila have provided a plethora of examples, including genes thought to be involved in adaptation to new or anthropogenic environments (Schlenke and Begun 2004; Pool et al. 2006) and genes likely to be engaged in intragenomic conflict, such as male–female conflict or meiotic drive (Nurminsky et al. 1998; Derome et al. 2004; Holloway and Begun 2004; Presgraves et al. 2009).
These findings reflect the broader observation that conflict within and between genomes maybe an important driver of adaptive molecular evolution (Begun et al. 2007; Haerty et al. 2007; Nielsen et al. 2007; Obbard, Welch, et al. 2009; Singh et al. 2009; Slotte et al. 2010). The conflict that occurs between host and parasite—requiring continual innovation on both sides as hosts evolve to resist their parasites and parasites evolve to evade host resistance—is thought to be of particular importance (e.g., Hurst and Smith 1999; Woolhouse et al. 2002; Schlenke and Begun 2003) and recent studies using Drosophila have confirmed this at the genome-wide scale, both by analyzing rates of nonsynonymous substitution across the Drosophila phylogeny and by inferring patterns of adaptive substitution for different components of the Drosophila immune system (Sackton et al. 2007; Obbard, Welch et al. 2009).
A key component of innate immunity in plants, fungi, and invertebrates is antiviral RNA interference (RNAi) (Ding and Voinnet 2007; Obbard, Gordon, et al. 2009). This defence mechanism is particularly well studied in Drosophila (Galiana-Arnoux et al. 2006; van Rij et al. 2006; Sabin et al. 2009; Saleh et al. 2009), where it evolves under unusually strong selective pressure: we have previously found that the antiviral RNAi genes Argonaute-2 (AGO2), Dicer-2, and R2D2 each show elevated rates of adaptive evolution over the long term in both Drosophila melanogaster and D. simulans (Obbard et al. 2006; Obbard, Welch, et al. 2009). Because many positive-sense RNA viruses express viral suppressors of RNAi (VSRs) that block antiviral RNAi, it has been hypothesized that this rapid adaptive evolution in RNAi genes is likely to be due to a molecular arms race with VSRs (Obbard et al. 2006; Marques and Carthew 2007; van Rij and Andino 2008; Obbard, Goldon, et al. 2009).
Here we examine the wider impact on the Drosophila genome of selective sweeps in one of these antiviral RNAi genes (AGO2) and discuss the evidence that this gene has experienced recurrent and recent selection in multiple species. To do this, we surveyed DNA sequence variation in natural populations of D. melanogaster, D. simulans, and D. yakuba across a 120-kbp region around AGO2 (fig. 1 ). Multiple lines of evidence suggest a recent selective fixation in, or near, AGO2 in all three species, and using these data, we investigate the timing of selective sweeps and the strength of selection associated with the most recently fixed alleles in D. simulans.
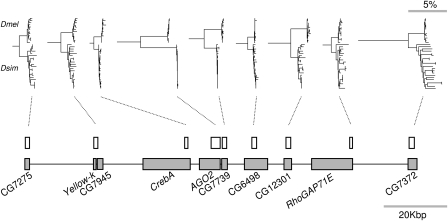
FIG. 1.
Genomic positions and gene trees for loci surrounding AGO2. In each tree, the upper clade is Drosophila melanogaster and the lower clade is D. simulans. The size and position of amplified regions are shown by white boxes, and gray boxes show the corresponding genes (note that the amplified fragment from Yellow-k partially overlaps locus CG7945). The total length of the surveyed region was approximately 123 kbp, and the total length of amplified sequence per individual was approximately 13.5 kbp. Gene trees were constructed using neighbor joining (MEGA v. 3.1, Kumar et al. 2004), based on coding sites only and were rooted using D. yakuba. All trees are drawn to the same scale. The shallow within-species genealogy associated with recent selective sweeps in AGO2 is clear in both species.
Materials and Methods
Choice of Loci
We analyzed polymorphism data at AGO2 and eight flanking loci, spread over a approximately 60-kbp region either side of AGO2 in three species of Drosophila (see supplementary table S1, Supplementary Material online). For clarity, we refer to loci by the name of the D. melanogaster ortholog throughout the article. These loci were chosen based on position, with more closely spaced markers being chosen near the putative site of selection where the gradient in diversity is expected to be steepest. In D. melanogaster, this region has an intermediate recombination rate (ca. 1.9cM Mbp−1). Known or hypothetical coding sequences were chosen so that synonymous and short-intron sites could be used for analysis as these are least likely to be under appreciable selection in Drosophila (e.g., Halligan and Keightley 2006). Following initial analysis of this region, 35 short haplotypes (1,325 bp) were additionally obtained from the center of AGO2 in D. simulans.
In D. melanogaster, the wider region spans the range 3L:15,492,244–15,615,246 (genome release r5.26), whereas in D. simulans, it spans 3L:14,833,076–14,954,658 (release r1.3). Relative spacing was taken from D. simulans genome release r1.3; however, incomplete assembly of the D. simulans genome (e.g., several hundred bases of AGO2 appear in unplaced fragment chrU_M_6024) means that absolute locus positions can only be treated as approximate. In D. yakuba, this region falls within synteny block 42 (as defined by Ranz et al. 2007) and loci span the range 3L:18,007,466–18,163,218. However, sequences nearly identical to some of these loci additionally appear in unplaced fragments, and absolute positions should again be treated as provisional.
Origin of Accessions
For the analysis of AGO2 and eight surrounding loci, flies were sourced as described in Obbard et al. (2006) and Jiggins and Kim (2007). Briefly, 21 East African D. simulans haplotypes (Nairobi, Kenya as described in Dean and Ballard 2004) were obtained either from lines that been inbred by sib mating for six to nine generations (a subset of these AGO2 sequences, but not other loci, are reported in Obbard et al. 2006) or by employing simulans-specific primers on artificial melanogaster–simulans interspecies hybrids. Twelve West African haplotypes were obtained from D. melanogaster (collected by B. Ballard and S. Charlat in Franceville, Gabon in 2002) using third chromosomes previously made isogenic by standard crosses to the balancer stock TM6/Sb. Seven to eleven D. yakuba (Gabon) sequences were obtained for each locus from isofemale lines that had been inbred by sib mating for six to nine generations. In addition, 35 shorter AGO2 haplotypes were also obtained from partially inbred North American and Madagascan D. simulans lines provided by P. Andolfatto and P. Haddrill. The Madagascan accessions are described in Dean and Ballard (2004) and North American accessions were collected in California (by A. Clark in 1999).
Drosophila erecta sequences (used to provide an outgroup for D. yakuba) were derived from published D. erecta genome sequence (r1.3). The D. sechellia AGO2 sequence (used to infer putative sites of recent selection in D. simulans AGO2) was derived from genome shotgun sequence chromatograms deposited in the NCBI Trace archive (Clark et al. 2007), complemented by new sequencing from a D. sechellia line provided by the Drosophila species stock center (University of California San Diego).
PCR and DNA Sequencing
Polymerase chain reaction (PCR) primers for AGO2 were designed from the published genome sequences of D. melanogaster, D. simulans, and D. yakuba. The 5' end of AGO2 was not sequenced as glutamine-rich repeat regions make alignment ambiguous and sequencing problematic. “‘Universal” PCR primers for the flanking loci were designed using consensus sequences from all three species. PCR failed for locus CG12031 in D. yakuba, and this region was not sequenced in this species. Where it was necessary to obtain D. simulans sequences from artificial simulans–melanogaster hybrids, a single simulans-specific PCR primer was paired with a universal primer for each locus. All primer sequences and reaction conditions are available from the authors on request. After PCR, unincorporated primers and dNTPs were removed using exonuclease I and shrimp alkaline phosphatase, and the products were then sequenced in both directions using BigDye v3.1 (Applied Biosystems) and using a ABI capillary sequencer (Gene Pool facility, University of Edinburgh). The sequence chromatograms were inspected by eye to confirm the validity of all variants within and between species and assembled using SeqManII (DNAstar Inc., Madison).
Tests for Selection
Unless otherwise specified, we used DNAsp (Rozas et al. 2003) to calculate summary statistics. Pairwise Hudson–Kreitman–Aguadé (HKA) tests (Hudson et al. 1987) between AGO2 and each neighboring locus were performed on synonymous sites only, using the program “HKA” (Hey 2004). Significance was assessed by coalescent simulation (as implemented in HKA), making the conservative assumptions that loci were unlinked and that no recombination occurred within loci. We ran 10,000 replicates for each test. In addition, we tested for a departure from neutrality in AGO2 as compared with all neighboring loci using the likelihood ratio test by Wright and Charlesworth (2004), based on the HKA approach. Starting parameters were taken from the standard HKA tests, and the Markov chain was run for 500,000 iterations. Each run was repeated three times to confirm convergence. Haplotype-based tests (K: number of haplotypes, M: frequency of the commonest haplotype, Hd: haplotype diversity, and the haplotype configuration) were performed using coalescent simulations implemented in “haploconfig” (Innan et al. 2005), conditional on estimated θ (Watterson 1975). To allow for some uncertainty in estimations of the local recombination rate, we followed Innan et al. (2005) in using a uniform distribution of recombination rates spanning the range ±50% either side of the map-based estimate for this region in place of a single recombination rate estimate. Where we wished to separate the effect of selection on the D. melanogaster and D. simulans lineages, we reconstructed hypothetical ancestral sequences using a maximum-likelihood codon-based approach (PAML; Yang 2007) and D. yakuba and D. erecta as outgroups. Tajima's D statistic (Tajima 1989) was calculated for synonymous and silent sites in the short central region of D. simulans AGO2 using DNAsp (Rozas et al. 2003).
Selective Sweep Models in D. melanogaster and D. simulans
We used three different approaches to model the selective sweep process. First, we applied the method of Kim and Stephan (2002) to AGO2 and the eight flanking loci, using the program CLSW (http://yuseobkim.net/Programs/KimCLA0906/). This approach calculates the composite likelihood ratio (CLR) for a single sweep model versus a standard neutral model, based on the full site frequency spectrum and assuming that sites are independent. We inferred the ancestral or derived status of variants by maximum likelihood (PAML; Yang 2007) and applied test LR1, which uses the unfolded frequency spectrum. Statistical significance was inferred by comparing the CLR to an empirical null distribution derived from 1,000 standard neutral coalescent simulations, implemented in “ms” (Hudson 2002). To reduce the time needed to run the neutral simulations, the number of potentially recombining sites was taken to be one-tenth of the sequence length, rather than the number of bases in the sequence.
To mitigate the potential impact of segregating deleterious variants that will skew allele frequencies, we limited our analysis to short introns and four-fold degenerate positions, both which should more closely approximate neutrality (e.g., Halligan and Keightley 2006). This was done by simulating the whole 120-kbp region as if all sites were neutral and later retaining only those variants that fall in positions which correspond to our four-fold and short-intron sites. For D. melanogaster, we ran simulations assuming that θ = 0.008 (the average for the loci analyzed here), the recombination rate r = 1.92 cM Mbp−1 (updated for locations in genome release 5 from Singh, Arndt, and Petrov 2005), and the mutation rate μ = 3.5 × 10−9 bp−1 generation−1 (Keightley et al. 2009). For D. simulans, we used θ = 0.0237 (the average for the loci analyzed here), r = 4.2 cM Mbp−1 (third chromosome rate, taken from Wall et al. (2002)), and assumed that the mutation rate was identical to that of D. melanogaster. Because some demographic processes can lead to a high rate of false positives (Jensen et al. 2005), we also compared CLR statistics to null distributions generated under various demographic scenarios simulated using “ms” (Hudson 2002). Rather than simultaneously inferring the demographic parameters from our data, or using a demographic model inferred from a different data set (e.g., Li and Stephan 2006), we chose to simulate six simple population growth models which cover the range of likely scenarios for African populations (Haddrill et al. 2005; Li and Stephan 2006): 10-fold and 2.5-fold step-change increases in population size, each occurring 10, 50, and 100 kya (assuming 10 generations per year). We did not include a bottleneck scenario, for which there is little evidence in African populations (Haddrill et al. 2005; Li and Stephan 2006).
Second, we applied the maximum likelihood method of Li and Stephan (test L1 in Li and Stephan 2005) to both synonymous and nonsynonymous sites in AGO2 and eight flanking loci using the program MOSY (http://www.zi.biologie.uni-muenchen.de/∼li/mosy/). This method differs from the approach of Kim and Stephan (2002) in that it conditions on the expected branch lengths of the genealogy and uses data from the compact site frequency spectrum that considers only the frequencies 1, 2, and ≥3, in place of the full-site frequency spectrum. Significance was inferred by comparison of the likelihood ratio with an empirical null distribution derived from constant-size neutral coalescent simulations performed with MOSY. For the neutral simulations, θ was taken to be the average of the loci analyzed here (θ = 0.0038 and 0.0135 for D. melanogaster and D. simulans, respectively) and Ne was estimated from previously published synonymous site diversity in these populations (Obbard, Welch, et al. 2009) and the neutral mutation rate in D. melanogaster (Keightley et al. 2009) (estimated Ne = 1.9 × 106 for D. simulans and 1.3 × 106 for D. melanogaster). The time since the sweep occurred (τ), the strength of selection (s), and the location of the site under selection were set as free parameters to be estimated. To reduce the time taken to generate null distributions, only 100 (D. simulans) and 200 (D. melanogaster) simulation replicates were performed.
Third, to improve our estimate of the timing and geographic scope of the sweep in D. simulans, we applied the Bayesian rejection sampling algorithm of Przeworski (2003) to estimate s, the selective coefficient, and T, the time since fixation of the selected allele (in units of 4Ne generations). This approach aims to sample the posterior distribution of T and s conditional on three summary statistics (the number of segregating sites, the number of haplotypes, and Tajima's D) by simulating a sweep model under random draws from the parameter priors and retaining those draws in which the simulated summary statistics deviate from the observed statistics by less than a specified threshold. Based on the analyses above, the selected site was taken to be immediately adjacent to the sequenced region (parameter K = 1), and priors were gamma distributed with mean mutation rate μ = 3.5 × 10−9, recombination rate r = 4.2 × 10−8, and Ne = 1.9 × 106 (as used in MOSY, above). T was sampled from a uniform prior over the interval [0,1] and s from a uniform prior over the interval [50/Ne,0.05]. The acceptance threshold was set to 0.1, and 500 samples from the posterior were used to estimate parameters T and s; all other options were set to the defaults. This analysis was performed on 56 short (1,325 bp, including 288 bp of intronic sequence) D. simulans haplotypes in addition to the sequences used in the analyses above. The additional sequences comprised 11 accessions from California, which is thought to have been colonized relatively recently by D. simulans (Capy and Gibert 2004), and 24 accessions from Madagascar, which is argued to be its ancestral range (Dean and Ballard 2004).
Timing the Most Recent Selective Sweep in D. simulans
In addition to estimating the time since fixation under a sweep model, we also used BEAST (Drummond and Rambaut 2007) to estimate the age of common ancestry for the 56 D. simulans AGO2 haplotypes under a simple gene-tree model. This was done for all sites, and also for a subset of the data comprising only short-intron and four-fold degenerate positions, as these should more closely conform to the neutral substitution rate. We assumed a strict molecular clock with the D. melanogaster mutation rate of 3.5 × 10−8 bp−1 yr−1 (i.e., 10 generations per year) and that no recombination had occurred within this region since the common ancestor of the sequences. This corresponds to a model in which all the mutations seen have arisen since the sweep started. If some of the segregating variants actually predate the sweep and are present because of recombination into the selected background during the sweep process, or if recombination has occurred between variants since the sweep completed, then this approach will tend to overestimate the time that has elapsed. We used an HKY substitution model with no rate heterogeneity between sites, and default priors were used for all parameters except tree shape, which followed a Yule process. We ran the Markov chain for 107 steps, recording a total of 1,000 states, of which we discarded the first 10% as burn-in. Traces suggested the chain mixed well and had reached stationarity. The effective sample size for each parameter estimate was >500.
McDonald–Kreitman Tests
The proportion (or number) of nonsynonymous substitutions attributable to positive selection rather than genetic drift can be estimated from counts of polymorphisms and substitutions at synonymous and nonsynonymous sites (reviewed in Eyre-Walker 2006). This forms the basis of the McDonald–Kreitman (MK) test (McDonald and Kreitman 1991), which can be extended to estimate the rate of nonsynonymous adaptive substitution in more sophisticated model-based frameworks (Bierne and Eyre-Walker 2004; Welch 2006). These methods assume that synonymous sites are neutral and that all nonsynonymous mutations are neutral, advantageous, or strongly deleterious. We used the approach of (Welch 2006; see also Obbard, Welch, et al. 2009) to estimate the number of adaptive substitutions per nonsynonymous site that have occurred in each of the sequenced loci, for each of the three lineages independently. We compared three models: in the first, all loci in the analyzed region were constrained to have the same number of adaptive substitutions per site; in the second, each of the loci had a different adaptive rate; and in subsequent models each locus in turn was allowed to differ from the others (which shared a rate). Each locus was assigned an independent parameter for constraint (f in Welch 2006) and all loci shared the same population-scaled diversity (4Neμ) and divergence (μt). We evaluated model fit using Akaike weights derived from the Akaike Information Criterion corrected for small sample size (Burnham and Anderson 2002).
Structural Modeling of AGO2
To better understand the nature of recent selection on AGO2, we used fold-recognition and homology modeling to build a three-dimensional structural model of the AGO2 protein and pinpoint recent amino acid substitutions within the structure of this protein. We excluded the extreme 5' end of the gene as the length-variable glutamine-rich repeats cannot be aligned between species. Residue positions are given relative to the start of our alignment (residue 431 of FlyBase CG7439-PB). A fold-recognition search (Bennett-Lovsey et al. 2008) identified the closest structural homolog to full-length D. simulans AGO2 as the Argonaute protein from Pyrococcus furiosus (PHYRE E value = 3.15 × 10−15, estimated precision = 100%). Its highest resolution structure (PDB ID: 1U04, Song et al. 2004) was used as a template, along with part of a related structure (PDB ID: 1Z26) and the experimentally determined PAZ domain from D. melanogaster AGO2 (PDB ID: 1R6Z, Song et al. 2003). Target-template alignment was based on a multiple sequence alignment of individual domains of the Drosophila proteins (N-terminal/stalk, PAZ, anchor, mid, and PIWI), using the program PROMALS-3D (Pei et al. 2008) to improve indel positioning. After generating initial models of individual domains using MODELLER 9.7 (Sali and Blundell 1993), alignments were subjected to further manual editing based on predicted (PSIPRED 2.4 McGuffin et al. 2000) and known (STRIDE Frishman and Argos 1995) secondary structure. Some strongly predicted secondary structure elements absent in the template were restrained during model building. Four 14–28 residue segments lacking template-guided restraints were modeled on other protein segments (from PDB IDs: 2GJU, 2WZI, 3I3L, and 3KZ1) with primary and secondary structures similar to those predicted for Drosophila AGO2. The extreme C-terminus was not modeled due to lack of an appropriate template. Twenty models of D. simulans AGO2 were generated. From the five with the lowest objective function score (Sali and Blundell 1993), the best was selected based on feasible domain–domain interaction and orientation within the intact overall structure, valid stereochemistry using a Ramachandran plot (selected representative model dihedral angle statistics: favored 86.6%; allowed 8.8%; outliers 4.6%; Morris et al. 1992), coarse packing quality (WHATIF average quality control score: −1.508, Vriend and Sander 1993), and the model validation program, ProQ (LGscore: 2.659, MaxSub: 0.209, Wallner and Elofsson 2003). Although actual position and orientation of side chains might be tentative given the low target–template sequence identity, the derived model should be suitable for inferring residues that are surface exposed or buried, as determined using GETAREA (Fraczkiewicz and Braun 1998). We also confirmed that the mid-domain of our model was similar to one derived from the recently published human AGO2 Mid-domain (Frank et al. 2010) (supplementary fig. S1, Supplementary Material online).
Results
Diversity Is Reduced around AGO2
In all three species (D. melanogaster, D. simulans, and D. yakuba), we found a substantial reduction in genetic diversity surrounding the antiviral gene AGO2 (figs 1 and 2 and supplementary table S2, Supplementary Material online). This effect is reflected in the shallow within-species gene trees seen for AGO2 and the closest neighboring loci in both D. melanogaster and D. simulans (fig. 1 ). In each case, there is a ∼100-kbp valley of reduced genetic diversity centered on AGO2, and in all three species, AGO2 has a lower genetic diversity than any of the other loci analyzed (fig. 2). Across the nine loci analyzed in D. melanogaster, Watterson's (1975) θ at synonymous sites ranged from 0.004 at AGO2 to 0.025 at the edge of the sampled region; in D. simulans from 0.003 at AGO2 up to 0.069; and in D. yakuba from 0.006 up to 0.054 (fig. 2 and supplementary table S2 in supporting information, Supplementary Material online). The genetic variation observed at AGO2 is substantially less than is typical for other genes in the genome. In previous studies, the mean θsyn was 0.018 and 0.038 in the same populations of D. melanogaster and D. simulans, respectively (chromosome 3L, data from Obbard, Welch, et al. 2009) and 0.030 in D. yakuba (autosomal loci with intermediate recombination, Llopart et al. 2005)
FIG. 2.
Genetic diversity around AGO2. Genetic diversity at all sites (upper row) and synonymous sites (middle row) is considerably reduced around AGO2 (positioned at zero on the x axis) in all three species. This is also reflected in the diversity/divergence ratio at synonymous sites (lower row: synonymous site diversity within species, θs, divided by divergence between species, KS). Loci that are significantly different from AGO2 in individual pairwise HKA tests (Hudson et al. 1987) are marked on the diversity/divergence graphs with asterisks: *P < 0.05, **P < 0.01, ***P < 0.001.
This reduction in genetic variation is suggestive of recent selective sweeps of new advantageous AGO2 alleles but might also be due to differences in mutation rate. To test whether the reduction in diversity is significant, we used the HKA test, which identifies differences in diversity between loci given the divergence between species for those loci, thereby correcting for mutation rate (Hudson et al. 1987). In D. melanogaster, HKA tests found a significant reduction in synonymous site diversity at AGO2 relative to each of the three loci in the 5' flank of AGO2 and one on the 3' flank (fig. 2 and supplementary table S3, Supplementary Material online). In D. simulans, all the flanking loci had significantly higher diversity than AGO2. In D. yakuba, the diversity of the two loci immediately neighboring AGO2 was not significantly higher than that of AGO2, but the remaining six loci were significantly higher. An HKA-based maximum likelihood approach to identify differences in diversity (Wright and Charlesworth 2004) also found that AGO2 had significantly reduced diversity in all three species (table 1). These tests are highly conservative as they use neighboring loci as the neutral standard, despite the fact that they too appear to have been affected by the sweep (fig. 2).
Table 1.
HKA Likelihood Ratio Tests.
| k (AGO2) | lnL | 2ΔLnL | P | |
| Drosophila melanogaster (vs. ancestral sequence) | ||||
| No selection | 1 | −49.2 | ||
| Selection on AGO2 | 0.22 | −46.5 | 5.4 | 0.0200 |
| D. simulans (vs. ancestral sequence) | ||||
| No selection | 1 | −62.6 | ||
| Selection on AGO2 | 0.08 | −56.7 | 11.8 | 0.0006 |
| D. yakuba (vs. D. erecta) | ||||
| No selection | 1.00 | −53.3 | ||
| Selection on AGO2 | 0.22 | −50.6 | 5.4 | 0.0206 |
Note.—k is the estimated reduction in diversity due to selection on AGO2 (Wright and Charlesworth 2004). lnL is the log-likelihood of the model, and 2ΔLnL is the log-likelihood test statistic.
AGO2 and Neighboring Loci Display Unusual Haplotype Structure
Following a selective sweep, linked alleles from neighboring sites spread together, increasing the extent to which polymorphisms from different loci co-occur within individuals (i.e., linkage disequilibrium) and reducing the number of haplotypes (e.g., Kim and Nielsen 2004; Innan et al. 2005). We tested D. simulans and D. melanogaster for deviations in haplotype structure from the standard neutral model but did not include D. yakuba in this analysis because of our small sample size (n = 7) and a lack of information on the local recombination rate. In D. simulans, four different tests detected unusual haplotype structure at AGO2 and a neighboring locus (table 2). Both loci had significantly fewer haplotypes than expected under a neutral model (Innan et al. 2005) (K, table 2) and correspondingly lower haplotype diversity Hd. In addition, the frequency of the most common haplotype (M) was significantly higher than expected and the haplotype configuration contained too many high-frequency haplotypes to be compatible with a standard neutral model (Innan et al. 2005). Within the D. melanogaster data set, AGO2 deviated from a neutral model only under the haplotype configuration test (Innan et al. 2005), displaying too many intermediate-frequency haplotypes (table 2). Three out of four tests additionally identified a departure for a third locus in D. simulans (table 2).
Table 2.
Haplotype Configuration Tests.
| K (95%) | M (95%) | Hd (Haplotype configuration) | P | |
| Drosophila melanogaster | ||||
| CG7275 | 12 (7, 12) | 1 (1, 5) | 0.917 (12, 0, 0, 0, ,0, 0, 0, 0, 0, , 0) | ns |
| yellow-k | 11 (7, 12) | 2 (1, 5) | 0.903 (10, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0) | ns |
| CrebA | 11 (6, 12) | 2 (1, 5) | 0.903 (10, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0) | ns |
| AGO2 | 7 (7, 12) | 3 (1, 4) | 0.820 (4, 1, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0) | 0.025 |
| CG7739 | 9 (5, 10) | 2 (2, 6) | 0.875 (6, 3, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0) | ns |
| CG6498 | 7 (6, 11) | 5 (2, 5) | 0.764 (5, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0) | ns |
| CG12301 | 9 (6, 11) | 2 (2, 5) | 0.875 (6, 3, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0) | ns |
| RhoGAP71E | 8 (4, 9) | 3 (2, 7) | 0.847 (5, 2, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0) | ns |
| CG7372 | 9 (6, 12) | 3 (1, 5) | 0.861 (7, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0) | ns |
| D. simulans | ||||
| CG7275 | 14 (14, 21) | 4 (1, 4) | 0.903 (10, 2, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…) | ns |
| yellow-k | 16 (14, 20) | 3 (2, 4) | 0.921 (13, 1 , 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…) | ns |
| CrebA | 20 (13, 20) | 2 (2, 5) | 0.948 (19, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…) | ns |
| AGO2 | 8* (10, 18) | 13** (2, 8) | 0.594** (6, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0,…) | <0.005 |
| CG7739 | 6** (9, 17) | 14** (1, 8) | 0.530** (3, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1,…) | <0.005 |
| CG6498 | 18 (12, 20) | 3 (2, 5) | 0.934 (16, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…) | ns |
| CG12301 | 14 (14, 21) | 6** (1, 4) | 0.875* (11, 2, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…) | 0.011 |
| RhoGAP71E | 17 (12, 19) | 4 (2, 6) | 0.921 (15, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…) | ns |
| CG7372 | 18 (16, 21) | 2 (1, 4) | 0.939 (15, 3, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…) | ns |
Note.—K (95%) is the number of haplotypes (95% bounds under neutrality from simulation), M (95%) the frequency of the most common haplotype (95% bounds under neutrality from simulation), and Hd the haplotype diversity (as in Depaulis and Veuille 1998). Asterisks denote significance. The haplotype configuration is a vector that records the frequency of haplotypes occurring n times in the sample where n = (1,2,3, …, x) and x is the sample size (Innan et al. 2005). Note that the fragment from Yellow-k partly overlaps locus CG7945).
Model Fitting Identifies a Recent Selective Sweep in D. simulans
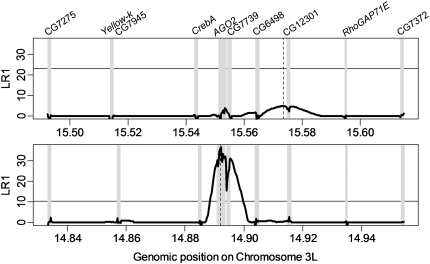
To obtain estimates of the strength of selection, the timing of the sweeps, and the location of the site under selection in D. simulans and D. melanogaster, we applied the CLR method of Kim and Stephan (2002), which uses information from genetic diversity and the site-frequency spectrum. For D. melanogaster, this method found no evidence for a recent selective sweep (fig. 3). This was true under both the constant-size population model (CLR = 5.0 vs., nominal 5% significance threshold of CLR = 21) and all the population-growth models (observed CLR = 5.0 vs. significance thresholds of CLR = 21–25 for the six population-growth models). For D. simulans, there was evidence for a recent selective sweep (fig. 3) under both the constant-size model (CLR = 36.7 vs. 5% significance threshold of CLR = 10) and the population-growth models (CLR = 36.7 vs. significance thresholds of CLR = 10–12 across the six growth models) (see Materials and Methods for details). This approach identified position 59041 of the analyzed region, which is within the coding sequence of AGO2 (approximately genomic position 3L:14,892,118) as the most likely focal site of the sweep and the strength of selection (2Nes) as 21482, implying a selective advantage of roughly 0.8%. The detection of a sweep in D. simulans but not in D. melanogaster could in principle result from the lower power in D. melanogaster (n = 12) compared with D. simulans (n = 21). However, an identical analysis of this region in the Drosophila Population Genomics Project data set (n = 37, derived from North American samples, http://www.dpgp.org/) gives an extremely similar CLR profile and similarly fails to detect a significant sweep in D. melanogaster, suggesting that our result is not merely due to reduced power in D. melanogaster (data not shown).
Fig. 3.
Composite likelihood profile. The CLR between a standard neutral model and selective sweep model, considering each site in turn (Li and Stephan 2005). The region surrounding AGO2 is shown for Drosophila melanogaster (upper panel) and D. simulans (lower panel). Gray regions are those for which sequence data are available, and the thin horizontal line shows the most stringent 5% significance threshold for this statistic derived a range of plausible population-growth scenarios (see Materials and Methods). The maximum likelihood estimate of the focal site for the sweep is given by a vertical dashed line; note that under this model there is no significant evidence of a recent sweep in D. melanogaster but that D. simulans shows strong evidence of a sweep in AGO2.
As a second approach to fit a selective sweep model, we applied the method of Li and Stephan (test L1 in Li and Stephan 2005). This method also found no evidence for a recent selective event in D. melanogaster as the difference in log-likelihood between the neutral and selective models was ΔlogL = 16.9, as compared with a nominal 5% significance threshold from constant-population neutral simulation of ΔlogL = 36.5. However, there was again evidence for a recent selective sweep in D. simulans, where ΔlogL = 151.3 compared with a nominal 5% significance threshold from constant-population neutral simulation of ΔlogL = 93.6. The inferred site of selection was position 60787 bp of the analyzed region, which falls very close to the 3' end of AGO2, and the estimated strength of selection was s = 0.3%
Both these approaches assume a single recent sweep has occurred at an unknown location within the sequenced region, and both use information from the site frequency spectrum to draw inferences. However, recurrent sweeps and soft sweeps (from standing variation) alter the expected site frequency spectrum (e.g., Kim 2006; Pennings and Hermisson 2006), and it is unclear how well single-sweep analyses perform if recurrent selection means that patterns of genetic diversity had not reached equilibrium prior to the most recent sweep. The unknown outcome of erroneously applying single-sweep models means that these results should be treated with some caution. However, if this effect does result in reduced power, this may explain why they failed to detect selection on AGO2 in D. melanogaster, despite other analyses being consistent with recent selection.
Date and Scale of the Selective Sweep in D. simulans
The maximum likelihood method of Li and Stephan (2005) dated the sweep in D. simulans to 0.05 × 4Ne generations ago. This corresponds to approximately 38 kya, assuming 10 generations per year and Ne ∼ 1.9 × 106, and suggests that the sweep occurred much more recently than D. simulans’ common ancestry with D. sechellia or D. mauritiana (∼250 kya, see McDermott and Kliman 2008) and possibly before D. simulans’ expansion out of Africa into Europe, which is itself thought to be more recent than the spread of D. melanogaster (Capy and Gibert 2004) that occurred ∼10–16 kya (Stephan and Li 2007).
To improve estimates of the timing and geographic scope of the sweep, we sequenced a short central region (1.3 kbp) adjacent to the putative site of selection from 11 Californian and 24 Madagascan accessions, in addition to those sampled from Kenya. Across the fifty-six 1325 bp haplotypes, we found only 16 silent- and synonymous-site polymorphisms (πs = 0.0035) and 6 nonsynonymous polymorphisms (πa = 0.0006), and Tajima's D statistic (silent and synonymous sites only) was D = −1.96 (P < 0.008, assessed by coalescent simulation assuming no recombination). All three populations had individually low diversity (πsilent = 0.0018, 0.0019, 0.0054, respectively, for Kenya, California, and Madagascar), and although there was some genetic divergence between populations, it was extremely low and there were no fixed differences: Hudson's (2000) Snn = 0.49 and Kst = 0.07, P < 0.01 for both. These observations suggest that the sweep is likely to have affected the whole extant D. simulans population.
Using these 56 D. simulans partial AGO2 sequences, we also inferred the approximate date of the sweep using the rejection-sampling algorithm of Przeworski (2003) and a tree-based approach implemented in BEAST (Drummond and Rambaut 2007). The selective sweep model (Przeworski 2003) estimated the time since fixation to be 13.5 kya (95% Highest Posterior Density (HPD) interval 2.1–45) and s = 0.01 (95% HPD interval: 0.0006–0.046), and the BEAST analysis estimated it to be 45 kya (95% HPD interval: 25–70) when using all sequenced sites or 57 kya (95% HPD interval: 16 to 105) when limited to putatively neutral sites. Note that the BEAST values are expected to be overestimates (see Materials and Methods).
Sites of Recent Substitution in D. simulans AGO2
Both model-fitting approaches (Kim and Stephan 2002; Li and Stephan 2005) suggest the most likely target of selection during the most recent sweep falls close to, or within, the coding sequence of AGO2; however, given these data, neither approach can infer the position with sufficient resolution to determine which site within AGO2 was substituted. Nonetheless, all analyses indicate the sweep occurred much more recently than the split between D. simulans and D. sechellia (estimated to be 250 kya), suggesting that the selected site will appear as a substitution on the D. simulans lineage alone. We used a codon-based phylogenetic model (PAML) to infer the ancestral D. simulans–D. sechellia sequence by maximum likelihood, and thereby identify 8 candidate nonsynonymous substitutions that have been fixed in D. simulans since its common ancestor with D. sechellia. However, there appear to be no clear patterns in the structural locations of these recent substitutions (fig. 4).
FIG. 4.
Recent amino acid substitutions in D. simulans AGO2. The surface structure of Drosophila AGO2 derived from published archean and Drosophila Argonaute structures by fold-recognition and homology modeling (see Materials and Methods). Moving down the figure, the four panels are successive 90° rotations about the vertical axis. The PAZ domain is indicated in green, the PIWI domain is indicated in blue, and the amino acid substitutions that occurred in D. simulans since the split from D. sechellia (ca. 250 kya) are shown in red. The two remaining substitutions at L106 and S404 are buried within the structure (see also supplementary fig. S2B, Supplementary Material online).
Argonaute proteins are defined by the presence of PAZ and PIWI domains, which are thought to interact with the 3' end of single-stranded RNA and to catalyze Argonaute endonuclease activity, respectively. Two of the eight candidates for recent substitution in the D. simulans lineage appear in the region 5' of the PAZ domain, two fall within the PAZ domain, three between the PAZ and PIWI domains, and one near the center of the PIWI domain. The majority of the substitutions (M106L, A185G, D194E, I238V, P404S, S428P) are conservative and do not alter residue charge or size (though P404S is located in an alpha-helix, which may be altered by the substitution). The most 5' site (K15T) and the site near the center of the PIWI domain (K636T) are more radical substitutions in terms of residue size and charge, and the loss of charge on the surface may be of functional relevance (fig. 4). When mapped on the structural model of Drosophila AGO2, three of the amino acid substitutions are exposed on the surface of the protein, two partially exposed, and three buried.
Compared with the total number of modeled residues that fall in each category (183 surface, 187 partially exposed, and 383 buried), this indicates a slight but nonsignificant excess of surface substitutions (63% exposed or partially exposed vs. 49%). When all the substitutions between D. melanogaster and D. simulans are identified (Supplementary fig. S2, Supplementary Material online), the slight bias toward surface substitutions is still present and becomes significant (64% of 94 substitutions are exposed or partially exposed, compared with 49% of 753 residues in the structural model, P = 0.008 using Fisher's exact test). This is in line with previous studies that have identified significantly higher rates of evolution and lower levels of constraint in surface residues (Bustamante et al. 2000; Conant and Stadler 2009) and does not suggest that AGO2 is unusual in displaying an excess of surface substitutions. Indeed, in Saccharomyces, the average ratio of exterior:interior constraint may be as high as 0.1 (Conant and Stadler 2009), suggesting that surface substitution in AGO2 could even be underrepresented compared with other genes.
As with the eight most recent substitutions in D. simulans, the complete set of substitutions that separate D. melanogaster and D. simulans exhibit no clear structural patterns. Three substitutions lie in proximity to the conserved residues involved in binding the 5′ end of RNA (Frank et al. 2010), two of which are conservative (V486I, V507I), whereas the other (S464F) introduces an additional hydrophobic residue in close proximity to Y468 and may well affect, and potentially even be involved in, binding to the 5' end of RNA. Three substitutions are in close proximity to the aromatic residues of the PAZ domain involved in binding the 3' end of RNA (Song et al. 2004), one (I238V) is conservative, whereas the others are nonconservative in terms of polarity (S235N) or charge (Q215K). Their sizes are similar, however, and based on their predicted location and orientation they appear unlikely to interfere with RNA binding, although a minor effect for these changes cannot be ruled out. Similarly, there are substitutions in close proximity to the key polar residues of the catalytic site in the PIWI domain (Parker 2010), two conservative (T574A, T583S) and one that varies in terms of size (Y582T) but is not expected to alter protein function.
Long-Term Adaptive Evolution in AGO2
To explore the possibility that strong directional selection on protein-coding loci other than AGO2 may contribute to the low diversity seen in this region, we studied the action of long-term selection using an approach based on the MK test (McDonald and Kreitman 1991). Following Welch (2006, see also Obbard, Welch, et al. 2009), we estimated the number of adaptive substitutions per nonsynonymous site that have occurred in each of the sequenced loci, for each of the three lineages. For each species, the best-supported model was one in which AGO2 had a higher rate of adaptive substitution than the other genes (Akaike weights of 1.00 for D. melanogaster, 0.84 for D. simulans and 0.98 for D. yakuba; see table 3 for MK data and see supplementary table S3, Supplementary Material online, for details of model selection). The only other model to receive appreciable support was for D. simulans, in which all genes had different rates (Akaike weight 0.16). These results corroborate previous analyses, suggesting that AGO2 experiences an unusually high rate of adaptive evolution (Obbard et al. 2006; Obbard, Welch, et al. 2009) and suggest that, in addition to having the lowest diversity of all the genes in this region (above), AGO2 has also experienced the strongest selection for change over the long term.
Table 3.
Interspecific Divergence and McDonald–Kreitman Analysis.
| n | Ln | Ls | KA | KS | KA/KS | Ds | Ps | Dn | Pn | a | a/bp | |
| Drosophila melanogaster | ||||||||||||
| CG7275 | 12 | 941 | 280 | 0.002 | 0.054 | 0.042 | 9 | 21 | 2 | 1 | 0 | 0.000 |
| yellow-k | 12 | 882 | 252 | 0.005 | 0.059 | 0.082 | 9 | 16 | 3 | 6 | −6 | −0.007 |
| CrebA | 12 | 761 | 247 | 0.003 | 0.04 | 0.079 | 8 | 8 | 2 | 3 | −3 | −0.004 |
| AGO2 | 12 | 1884 | 567 | 0.032 | 0.068 | 0.476 | 34 | 5 | 54 | 4 | 48 | 0.025 |
| CG7739 | 12 | 1055 | 322 | 0.009 | 0.082 | 0.11 | 23 | 7 | 9 | 3 | 4 | 0.004 |
| CG6498 | 12 | 1074 | 336 | 0.004 | 0.038 | 0.103 | 11 | 6 | 4 | 2 | 1 | 0.001 |
| CG12301 | 12 | 1064 | 275 | 0.022 | 0.027 | 0.819 | 7 | 5 | 21 | 8 | 8 | 0.008 |
| RhoGAP71E | 12 | 537 | 171 | 0.001 | 0.062 | 0.017 | 8 | 6 | 0 | 2 | −3 | −0.006 |
| CG7372 | 12 | 974 | 265 | 0.017 | 0.065 | 0.263 | 15 | 5 | 14 | 11 | −3 | −0.003 |
| D. simulans | ||||||||||||
| CG7275 | 21 | 941 | 280 | 0.005 | 0.081 | 0.061 | 8 | 56 | 4 | 7 | 2 | 0.002 |
| yellow-k | 21 | 883 | 251 | 0.008 | 0.071 | 0.11 | 7 | 43 | 3 | 17 | −3 | −0.003 |
| CrebA | 21 | 761 | 247 | 0.001 | 0.031 | 0.018 | 6 | 8 | 0 | 2 | −1 | −0.001 |
| AGO2 | 21 | 1889 | 565 | 0.041 | 0.067 | 0.632 | 37 | 6 | 75 | 1 | 75 | 0.040 |
| CG7739 | 21 | 1058 | 328 | 0.011 | 0.04 | 0.27 | 11 | 10 | 11 | 3 | 10 | 0.009 |
| CG6498 | 21 | 1072 | 338 | 0.002 | 0.033 | 0.064 | 6 | 26 | 2 | 6 | 0 | 0.000 |
| CG12301 | 21 | 1121 | 295 | 0.009 | 0.051 | 0.179 | 10 | 36 | 4 | 33 | −7 | −0.006 |
| RhoGAP71E | 21 | 531 | 171 | 0.007 | 0.046 | 0.162 | 2 | 28 | 1 | 15 | −4 | −0.008 |
| CG7372 | 21 | 948 | 258 | 0.028 | 0.077 | 0.359 | 7 | 64 | 6 | 94 | −26 | −0.027 |
| D. yakuba | ||||||||||||
| CG7275 | 7 | 939 | 282 | 0.014 | 0.149 | 0.091 | 29 | 37 | 11 | 3 | 4 | 0.005 |
| yellow-k | 8 | 880 | 254 | 0.019 | 0.166 | 0.113 | 34 | 13 | 14 | 9 | −5 | −0.006 |
| CrebA | 7 | 759 | 249 | 0.017 | 0.091 | 0.183 | 21 | 4 | 12 | 2 | 7 | 0.010 |
| AGO2 | 11 | 1882 | 566 | 0.04 | 0.215 | 0.186 | 11 | 8 | 70 | 4 | 63 | 0.033 |
| CG7739 | 8 | 1056 | 327 | 0.015 | 0.19 | 0.079 | 52 | 12 | 15 | 1 | 13 | 0.012 |
| CG6498 | 8 | 1009 | 323 | 0.01 | 0.15 | 0.068 | 37 | 19 | 10 | 1 | 8 | 0.008 |
| RhoGAP71E | 7 | 557 | 178 | 0.015 | 0.184 | 0.079 | 26 | 12 | 8 | 0 | 8 | 0.014 |
| CG7372 | 7 | 1034 | 283 | 0.112 | 0.245 | 0.458 | 59 | 22 | 96 | 17 | 58 | 0.056 |
Note.—n is the number of alleles sampled; Ln and Ls the number of nonsynonymous and synonymous sites, respectively; KA and KS are the nonsynonymous and synonymous divergence; Ds, Ps, Dn, and Pn are counts of fixed differences (D) and polymorphisms (P) at synonymous and nonsynonymous sites; and “a” is the maximum-likelihood estimate of the number of nonsynonymous adaptive substitutions per gene under the model described in Obbard, Welch, et al. (2009) and Materials and Methods (above). Divergence is measured from their common ancestor in the case of D. melanogaster and D. simulans and from D. erecta in the case of D. yakuba.
Maximum likelihood estimates for the total number of adaptive substitutions at nonsynonymous sites in the sequenced region of AGO2 are 48, 75, and 63 for D. melanogaster, D. simulans, and D. yakuba, respectively (measuring from the common ancestor of D. melanogaster and D. simulans and from D. erecta). If we assume the most recent common ancestor of D. melanogaster and D. simulans lived 2.3 Ma (Russo et al. 1995), and a total of 7.2 My of evolution separate D. yakuba and D. erecta, then these numbers correspond to rates of 0.011, 0.017, and 0.005 adaptive substitutions per nonsynonymous site per million years in AGO2 or sweeps occurring roughly every 50, 30, and 110 thousand years.
Discussion
Evidence for Recurrent Selection on AGO2
We found that genetic diversity was greatly reduced around AGO2 in three species of Drosophila. There is compelling evidence that the reduced variation is caused by selection on AGO2 itself, as in all the species, the valley of low diversity was centered on AGO2, and in D. simulans, a model-fitting approach identified AGO2 as the target of selection. It is hard to envisage any process other than recurrent natural selection that could account for our results. For example, local variation in mutation rate is accounted for by reference to outgroup divergence, and gene conversion and changes in population size are unlikely to affect the same candidate gene (chosen a priori) across different species. Furthermore, sequences were derived from populations that do not appear to have experienced a strong bottleneck, which may lead to a spurious inference of selection (Haddrill et al. 2005). Finally, there is also evidence for long-term selection on AGO2 since these species shared a common ancestor as, in lineages leading to all three species, there is a significant excess of amino acid substitutions in AGO2 relative to neighboring genes, suggesting that the protein-coding sequence of AGO2 has experienced an elevated rate of adaptive substitution (table 3).
Two further independent observations from other studies also support recent selection on AGO2. First, while screening for transposable element (TE) insertions in derived D. melanogaster populations, Gonzalez et al. (2008) identified a potential selective sweep associated with an S-element insertion in the first intron of AGO2. Because the insertion allele is associated with altered expression levels of AGO2, it was argued that this may represent an adaptive regulatory change associated with the TE insertion. However, given the high rate of adaptive amino acid substitution in AGO2 (above, and Obbard et al. 2006; Obbard, Welch, et al. 2009), it is hard to exclude the possibility that the TE-insertion hitchhiked to its presently high (but not fixed) frequency on a selected amino acid variant. Second, in a genome-wide survey of changes in expression level, Graze et al. (2009) found AGO2 expression to significantly differ between D. simulans and D. melanogaster (see also McManus et al. 2010), and changes in expression tend to be correlated with rapid evolution (Nuzhdin et al. 2004).
These results add to a growing literature showing that parasite-mediated selection is an important cause of molecular evolution. However, the extent to which this process determines variation in extant phenotype remains unclear as selection can fix alleles with selective advantages much smaller than can be measured in the laboratory. Our analyses suggest that the most recently fixed D. simulans AGO2 allele had a selective advantage of somewhat less than 1% and may therefore have had experimentally measurable phenotypic effects. However, although we estimate 90–100% of nonsynonymous substitutions in AGO2 have been driven by selection (compared with a genome average of ca. 45%, Bierne and Eyre-Walker 2004; Welch 2006; Obbard, Welch, et al. 2009), this only corresponds to a selective sweep in this gene once every 30–100 thousand years, with the most recent sweep in D. simulans occurring 13–57 thousand years ago. Thus, although this gene is one of the most strongly selected in the Drosophila immune system (Obbard, Welch, et al. 2009), such arms races are unlikely to be driving allelic replacement with a frequency likely to be observed on an ecological timescale.
Likely Selective Agents
RNA interference mediated by the Dicer2-R2D2-AGO2 pathway is a major antiviral defence mechanism in Drosophila (e.g., Galiana-Arnoux et al. 2006; van Rij et al. 2006) and other insects (e.g., Campbell et al. 2008), and several insect viruses—including Drosophila C virus—carry genes that actively suppress RNAi (Chao et al. 2005; van Rij et al. 2006; Wang et al. 2006; Nayak et al. 2010). VSRs act in several ways: by sequestering short-interfering RNAs (siRNAs), by competing with siRNAs for the active sites of RNAi pathway genes, by blocking cell-to-cell movement of siRNAs, or by destabilizing or degrading key proteins in the pathway (Diaz-Pendon and Ding 2008). The last class includes VSRs that interact directly or indirectly with Argonaute proteins (e.g., Csorba et al. 2010) including one that interacts with D. melanogaster AGO2 (Nayak et al. 2010). Our structural model of Drosophila AGO2 suggests that recent substitutions in D. simulans have primarily occurred at the protein surface. Although this is true of many proteins, should these substitutions indeed be the result of recent selective sweeps, then these data may be indicative of AGO2 evolving to alter its ability to interact with other molecules, potentially VSRs.
Nevertheless, the Dicer2 and AGO2 not only mediate antiviral defence but also target transcripts from TEs (see Obbard and Finnegan 2008 for references), particularly during colonization by a new TE (Rozhkov et al. 2010). Like viruses, TEs are costly to their hosts, and although no TE-encoded suppressors of RNAi have been characterized, they may exist (Blumenstiel and Hartl 2005), and evolutionary conflict with TEs has the potential to drive a host–parasite molecular arms race (Lee and Langley 2010; Lu and Clark 2010). It is therefore striking that the Piwi-interacting RNAi pathway, which modulates TE transcript levels in germline tissues and is thought to target heterochromatin formation to TE insertions (Klattenhoff and Theurkauf 2008), also contains several genes which show a high rate of adaptive substitution (reviewed in Obbard, Gordon, et al. 2009). These include the heterochromatin protein Rhino, the putative exonucleases Maelstrom and Krimper, the helicases Spindle-E and Armitage, and Piwi-family Argonaute proteins Aubergine and Piwi (Vermaak et al. 2005; Heger and Ponting 2007; Obbard, Gordon, et al. 2009; Obbard, Welch, et al. 2009). Therefore, it remains possible that the recent selection on AGO2 is associated with its role in TE suppression (potentially during the invasion of new TEs, Rozhkov et al. 2010) rather than its antiviral function. Nevertheless, because we expect host–TE conflict to be limited to reproductive tissues (Charlesworth and Langley 1986), the opportunity for TEs to be the driving force in recurrent selection at AGO2 may depend on the relative importance of the Dicer2- AGO2 pathway in suppressing germline versus somatic TE expression.
Finally, the distinction between viruses and retrotransposons such as gypsy (an “endogenous retroviruses”) is a subtle one (Huszart and Imler 2008; Llorens et al. 2008), and there may be considerable mechanistic overlap in their control. For example, Drosophila lacking functional piwi-interacting (pi) RNA pathway genes Armitage, Piwi, or Aubergine appear to be compromised in their ability to resist the double-stranded RNA birnavirus DXV (Zambon et al. 2006), and piRNAs derived from viruses including DAV, DXV, DCV, and Nora have been reported from Drosophila cell culture that expresses Piwi (Wu et al. 2010). If it is confirmed that components of the piRNA pathway can play a role in antiviral defence, then viruses may be the selective force in both the piRNA and the Dicer2-R2D2-AGO2 pathways.
Conclusions
The evolution of many of some the most rapidly evolving genes in animal genomes appears to be driven by evolutionary arms races, where there is a battle of adaptation and counter-adaptation with parasites (e.g., Hurst and Smith 1999; Schlenke and Begun 2003; Sackton et al. 2007; Obbard, Welch, et al. 2009), or during sexual reproduction (Begun et al. 2007; Haerty et al. 2007; Slotte et al. 2010). In line with this, we have previously shown that the antiviral RNAi pathway of Drosophila, which is known to be targeted by viral suppressor molecules, contains three genes under exceptionally strong selection (Dcr-2, AGO2, and R2D2) (Obbard et al. 2006). Here we have shown that selection on AGO2 has left its mark across a large genomic region in three different species of Drosophila (figs 1 and 2). As far as we are aware, comparable examples—such as artificial selection and drug or pesticide resistance (e.g., Schlenke and Begun 2004)—tend to stem from recent human action. Thus, the finding of recent selective sweeps in AGO2 represents a particularly interesting example of how recurrent parasite-mediated selection may have a significant impact on the genome.
Supplementary Material
Supplementary figures S1 and S2 and tables S1, S2, and S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Bill Ballard, Sylvain Charlat, Penny Haddrill, and Peter Andolfatto for the original fly collections and/or permission to use their fly lines; Kang-Wook Kim for performing DNA extractions; Jill Lovell and the Edinburgh Gene Pool for DNA sequencing; Dinesh Soares for help and advice with structural modeling; Haipeng Li, Pavlos Pavlidis, and Molly Przeworski for advice on the use of their software and/or help with computational approaches to selective-sweep analysis. We thank John Welch, Andrea Bettancourt, Pavlos Pavlidis, John McDonald, and an anonymous reviewer for comments which improved the manuscript considerably. We also thank Nick Barton for comments on the first version of this article and Toby Johnson for performing a selective-sweep analysis presented in that version. Author contributions: T.J.L. and F.M.J. initiated this study of dipteran immunity genes. D.J.O. and F.M.J. designed this analysis of AGO2 and D.J.O. performed all PCR, DNA sequencing, and sequence analyses. F.M.J. provided fly stocks, screened D. melanogaster for inversions, and performed crosses. N.J.B. performed the structural modeling and its interpretation. D.J.O., F.M.J., and T.J.L. wrote the article, with contributions from N.J.B. This work was funded by Wellcome Trust (grant 073210/Z/03/Z to T.J.L.). D.J.O. is currently funded by Wellcome Trust RCD Fellowship (085064/Z/08/Z) and F.M.J. was supported by a Royal Society research fellowship and Wellcome Trust (grant WT081279MA). N.J.B. was funded by Wellcome Trust (grant 088179/A/09/Z).
References
- Begun DJ, Holloway AK, Stevens K, et al. (13 co-authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJE, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multi-model inference: a practical information-theoretic approach. New York: Springer; 2002. [Google Scholar]
- Bustamante CD, Townsend JP, Hartl DL. Solvent accessibility and purifying selection within proteins of Escherichia coli and Salmonella enterica. Mol Biol Evol. 2000;17:301–308. doi: 10.1093/oxfordjournals.molbev.a026310. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegyptiuses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Gibert P. Drosophila melanogaster, Drosophila simulans: so similar yet so different. Genetica. 2004;120:5–16. doi: 10.1023/b:gene.0000017626.41548.97. [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by flock house virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. The evolution of self-regulated transposition of transposable elements. Genetics. 1986;112:359–383. doi: 10.1093/genetics/112.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, et al. (417 co-authors) Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Conant GC, Stadler PF. Solvent exposure imparts similar selective pressures across a range of yeast proteins. Mol Biol Evol. 2009;26:1155–1161. doi: 10.1093/molbev/msp031. [DOI] [PubMed] [Google Scholar]
- Csorba T, Lózsa R, Hutvágner G, Burgyán J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010;62:463–472. doi: 10.1111/j.1365-313X.2010.04163.x. [DOI] [PubMed] [Google Scholar]
- Dean MD, Ballard JWO. Linking phylogenetics with population genetics to reconstruct the geographic origin of a species. Mol Phylogenet Evol. 2004;32:998–1009. doi: 10.1016/j.ympev.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Depaulis F, Veuille M. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol Biol Evol. 1998;15:1788–1790. doi: 10.1093/oxfordjournals.molbev.a025905. [DOI] [PubMed] [Google Scholar]
- Derome N, Metayer K, Montchamp-Moreau C, Veuille M. Signature of selective sweep associated with the evolution of sex-ratio drive in Drosophila simulans. Genetics. 2004;166:1357–1366. doi: 10.1534/genetics.166.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Ding SW. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu Rev Phytopathol. 2008;46:303–326. doi: 10.1146/annurev.phyto.46.081407.104746. [DOI] [PubMed] [Google Scholar]
- Ding S-W, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett R, Schweinsberg J. Approximating selective sweeps. Theor Popul Biol. 2004;66:129–138. doi: 10.1016/j.tpb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol Evol. 2006;21:569. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19:319–333. [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5[prime]-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Gonzalez J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 2008;6:2109–2129. doi: 10.1371/journal.pbio.0060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics. 2009;183:547–561. doi: 10.1534/genetics.109.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Jagadeeshan S, Kulathinal RJ, et al. (11 co-authors) Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Ponting CP. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophilagenomes. Genome Res. 2007;17:1837–1849. doi: 10.1101/gr.6249707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pfaffelhuber P. The pattern of genetic hitchhiking under recurrent mutation. Electron J Probab. 2008;13:2069–2106. [Google Scholar]
- Hey J. HKA—a computer program for tests of natural selection [Internet] 2004. [cited 2010 November]. Available from: http://genfaculty.rutgers.edu/hey/software#HKA. [Google Scholar]
- Holloway AK, Begun DJ. Molecular evolution and population genetics of duplicated accessory gland protein genes in Drosophila. Mol Biol Evol. 2004;21:1625–1628. doi: 10.1093/molbev/msh195. [DOI] [PubMed] [Google Scholar]
- Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–11709. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Smith NGC. Do essential genes evolve slowly? Curr Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- Huszart T, Imler JL. Drosophila viruses and the study of antiviral host-defense. Adv Virus Res. 2008;72:227–265. doi: 10.1016/S0065-3527(08)00406-5. [DOI] [PubMed] [Google Scholar]
- Innan H, Zhang K, Marjoram P, Tavare S, Rosenberg NA. Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites. Genetics. 2005;169:1763–1777. doi: 10.1534/genetics.104.032219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD, Kim Y, DuMont VB, Aquadro CF, Bustamante CD. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics. 2005;170:1401–1410. doi: 10.1534/genetics.104.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Kim KW. A screen for immunity genes evolving under positive selection in Drosophila. J Evol Biol. 2007;20:965–970. doi: 10.1111/j.1420-9101.2007.01305.x. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter ML. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. Allele frequency distribution under recurrent selective sweeps. Genetics. 2006;172:1967–1978. doi: 10.1534/genetics.105.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee YCG, Langley CH. Transposable elements in natural populations of Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 2010;365:1219–1228. doi: 10.1098/rstb.2009.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. Maximum-likelihood methods for detecting recent positive selection and localizing the selected site in the genome. Genetics. 2005;171:377–384. doi: 10.1534/genetics.105.041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2006;2:e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A, Lachaise D, Coyne JA. Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics. 2005;171:197–210. doi: 10.1534/genetics.104.033597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens JV, Clark JB, Martinez-Garay I, Soriano S, de Frutos R, Martinez-Sebastian MJ. Gypsy endogenous retrovirus maintains potential infectivity in several species of Drosophilids. BMC Evol Biol. 2008;8:11. doi: 10.1186/1471-2148-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark AG. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010;20:212–227. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Carthew RW. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet. 2007;23:359. doi: 10.1016/j.tig.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitchhiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McDermott SR, Kliman RM. Estimation of isolation times of the island species in the Drosophila simulanscomplex from multilocus DNA sequence data. PLoS One. 2008;3:e2442. doi: 10.1371/journal.pone.0002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, O'Duff M, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophilarevealed by mRNA-seq. Genome Res. 2010 doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AL, Macarthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- Nayak A, Berry B, Tassetto M, et al. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Finnegan DJ. RNA interference: endogenous siRNAs derived from transposable elements. Curr Biol. 2008;18:R561–R563. doi: 10.1016/j.cub.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim K-W, Jiggins FM. Quantifying adaptive evolution in the Drosophilaimmune system. PLoS Genet. 2009;5:e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS. How to slice: snapshots of Argonaute in action. Silence. 2010;1 doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei JM, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2006;2:1998–2012. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, DuMont VB, Mueller JL, Aquadro CF. A scan of molecular variation leads to the narrow localization of a selective sweep affecting both afrotropical and cosmopolitan populations of Drosophila melanogaster. Genetics. 2006;172:1093–1105. doi: 10.1534/genetics.105.049973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Gerard PR, Cherukuri A, Lyttle TW. Large-scale selective sweep among segregation distorter chromosomes in African populations of Drosophila melanogaster. PLoS Genet. 2009;5(5):e1000463. doi: 10.1371/journal.pgen.1000463. doi:10.1371/journal.pgen.1000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. Estimating the time since the fixation of a beneficial allele. Genetics. 2003;164:1667–1676. doi: 10.1093/genetics/164.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, Ashburner M, Bergman CM. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:1366–1381. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R, McCombie WR, Hannon GJ, Evgen'ev MB. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA. 2010;16:1634–1645. doi: 10.1261/rna.2217810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M. Molecular phylogeny and divergence times of Drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Saleh M-C, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Strong selective sweep associated with a transposon insertion in Drosophila simulans. PNAS. 2004;101:1626–1631. doi: 10.1073/pnas.0303793101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Arndt PF, Petrov DA. Genomic heterogeneity of background substitutional patterns in Drosophila melanogaster. Genetics. 2005;169:709–722. doi: 10.1534/genetics.104.032250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Larracuente AM, Sackton TB, Clark AG. Comparative genomics on the Drosophila phylogenetic tree. Annu Rev Ecol Evol Syst. 2009;40:459–480. [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27(8):1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu JD, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song J-J, Smith SK, Hannon GJ, Joshua-Tor L. Crystal Structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Stephan W, Li H. The recent demographic and adaptive history of Drosophila melanogaster. Heredity. 2007;98:65–68. doi: 10.1038/sj.hdy.6800901. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Andino R. The complex interactions of viruses and the RNAi machinery: a driving force in viral evolution. In: Domingo E, Parrish CR, Holland JJ, editors. Origin and evolution of viruses. London: Academic press; 2008. [Google Scholar]
- van Rij RP, Saleh M-C, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G, Sander C. Quality-control of protein models of —directional atomic contact analysis. J Appl Crystallogr. 1993;26:47–60. [Google Scholar]
- Wall JD, Andolfatto P, Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Welch JJ. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- Wright SI, Charlesworth B. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li W-X, Ding S-W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci U S A. 2010;107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.