Abstract
We report the use of silicate nanofilms for on-plate desalting and subsequently direct laser desorption/ionization-mass spectrometric (LDI-MS) analysis of peptides. A hydrophobic octadecyltrichlorosilane (OTS) monolayer is formed on a calcinated nanofilm on a gold substrate to facilitate sample deposition and interaction with the surface that allows effective removal of MS-incompatible contaminants such as salts and surfactants by simple on-plate washing while the peptides are retained on the spot. By eliminating interferences from matrix-related ions and contaminants, sensitivity of MS analysis has been enhanced over ca. 20 times, leading to improved detection of peptides at the low-fmol level. A high recovery rate of the peptides is obtained by using relatively rough nanofilms, which are prepared through a modified layer-by-layer (LbL)-deposition/calcination process. The performance of the films has been investigated with peptide samples in the presence of high salts (NaCl and sodium acetate) and urea. Compared to MALDI analysis with CHCA-matrix, LDI with on-plate desalting offers marked improvement for analysis of peptides due to low background ions and reduction of sample complexity. Additionally, selective capture of the hydrophobic components of a protein can be achieved, providing a highly useful strategy for specific peptide enrichment. LDI with on-plate desalting approach has also been successfully applied to peptide analysis from protein digests.
INTRODUCTION
Matrix-assisted laser desorption/ionization-mass spectrometry (MALDI-MS) has become one of the most important tools for peptide and protein analysis in proteome research due to its high ionization efficiency, high sensitivity, rapidity, capability of high throughput and ease of automation1, 2. MALDI experiments, however, are known to depend on the co-crystallization condition for matrix-analyte mixtures on the sample plate. The existence of contaminants such as salts often impedes the crystallization process, resulting in significant decrease of sensitivity and reproducibility in MS detection. This has been a long and persistent problem in direct MALDI analysis of biological samples, where high salt matrixes and buffers must be used to stabilize the molecules and maintain their activities. Therefore, sample desalting has been a crucial step in MALDI-MS analysis of biomolecules including peptides and proteins.
HPLC, in particular reversed-phase HPLC, is frequently used for prefractionation of complex biological samples prior to MALDI-MS analysis3. However, consumption of large volumes of solvent for elution leads to sample dilution, and extra step of solvent evaporation can be time-consuming and often result in unexpected contamination and sample loss4, 5. In recent years, miniaturized sample preparation techniques, such as micro- or nano-scale solid phase extraction (SPE), have been developed to reduce the sample size and simplify the sample handling process4, 5. An effective approach is to carry out SPE in pipette tips packed with chromatographic materials such as ZipTip5 (Millipore, Bedfold, MA) and StageTips6 (Proxeon, Cambridge, MA). SPE with surface-functionalized magnetic nanoparticles is another interesting strategy7, 8. But these off-probe SPE approaches suffer from their inherent limitations including sample loss and potential contamination during multistep sample handling4, as well as a relatively high operational cost. Recently, several methods for on-plate sample concentration and desalting have been reported9, and a number of hydrophobic coatings such as Teflon10, self-assembled monolayers (SAMs)11, parafilm wax12, 13, poly(methyl methacrylate) materials14, 15, polysulfone-poly(ethylene oxide) block copolymer16, polyethylene and polypropylene17 have been used for MALDI analysis. Sensitivity of MS detection increases by reduction of sample spot size and removal of contaminants. However, many of these methods require complex preparation protocols and have sample carryover effects, leading to limited applications. In addition, problems arising from organic matrix, such as sample dilution, inhomogeneous crystallization and background noise in low mass range, can yield poor MALDI-MS results even after desalting process18.
To circumvent the matrix problem, there has been considerable interest in developing laser desorption ionization (LDI) techniques for MS analysis without the use of organic matrix19, while LDI is facilitated by the surface of substrate materials (SALDI)19, 20. Graphite21, carbon nanomaterials22, silicon nanoparticles23, silica24, metal oxide particles25, 26 and metal nanoparticles27–29 have been used as SALDI substrates where some of the nanomaterials have served as selective probes for analyte pre-concentration with their unique surface properties30, 31. But aggregation of nanomaterials on MALDI-plate often results in erratic LDI efficiency32 and there has been concern over co-desorption of nanomaterials in LDI that may contaminate the mass spectrometer29, 33. One of the most widely used substrates in SALDI is porous silicon (DIOS)34, 35 and functionalized DIOS substrates have enabled on-plate sample preparation for SALDI-MS detection of biological samples36, 37. Highly sensitive detection of peptides has been reported by using modified DIOS substrate with hydrophobic surface property37. More recently, hydrophobic initiators were utilized to facilitate desorption of analytes on DIOS substrate.38
We report here an on-plate desalting method on calcinated nanofilms for direct laser desorption/ionization mass spectrometric analysis of peptides containing complex salt background. The method is based on a novel silicate/Au substrate we recently developed that allows SALDI-MS analysis of biomolecules with remarkably high efficiency.39 The nanofilm was constructed through a layer-by-layer (LbL) deposition/calcination process and presented better stability towards air oxidation and aqueous hydrolysis39, 40. Moreover, it is inexpensive, easy to fabricate, and allows versatile functionalization by tailoring surface property with silylation chemistry. In this work, we expand the application of the silicate nanofilms and demonstrate on-plate desalting and subsequent SALDI analysis of peptides in high salt conditions on the same chip that substantially enhances the sensitivity and performance of the detection. To effectively carry out the desalting, the nanofilm surface is rendered exceedingly hydrophobic by use of a monolayer of octadecyltrichlorosilane (OTS), where varied degrees of interactions with biomolecules, in particular peptides, are manipulated to retain target analytes on the surface whereas major contaminants are selectively removed. The removal of contaminants that impede MALDI mass spectrometric analysis results in selective enrichment of the target molecules and suppression of the background signal. In addition, without the use of matrix, this approach offers clean MS spectrum in low mass range, leading to enhancement of sensitivity and signal-to-ratio ratio for analytes in MS detection. LDI with on-plate desalting on the hydrophobic calcinated surface has also been successfully applied to analysis of peptides in protein digests.
EXPERIMENTAL SECTION
Materials and instrument
3-Mercaptopropionic acid (3-MPA), trifluoroacetic acid (TFA), poly(allylamine hydrochloride) (PAH), DL-dithiothreitol (DTT), iodoacetamide (IAA), urea, [Sar1, Thr8]-angiotensin II (MW = 956.1), neurotensin (MW = 1672), bradykinin (MW = 1060.2), cytochrome c (bovine heart), β-casein, α-casein, TPCK-treated trypsin (bovine pancreas) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). Sodium silicate (SiOx), microscope glass slides, citric acid, octadecyltrichlorosilane (OTS) and acetonitrile were from Thermo-Fisher Scientific (Pittsburgh, PA). Stainless steel tape was purchased from LabelValue.com (Tampa, FL). Water was purified by a Milli-Q system. All other reagents were analytical grade and used without further purification.
Preparation of nanoscale calcinated films
Preparation of calcinated silicate layers on gold substrates was conducted by a modified procedure based on our previous works 40 39. In brief, a 46-nm thick gold layer was e-beam deposited onto cleaned stainless steel tapes and glass slides with pre-deposition of 2-nm Cr film as adhesion layer. Cleaned gold substrates were then immersed in a 5 mM 3-MPA ethanol solution overnight, followed by extensive rinsing with ethanol and DI water. PAH (1 mg/mL, pH 8.0) and sodium silicate solution (22 mg/mL, pH 9.5) were alternately deposited to the surface. This process was repeated until the designated number of layers was reached. The substrates were then washed by D.I. water and dried in air, calcinated in a furnace by heating to 450 °C at a rate of 17 °C per min and brought to room temperature after 4 h. To prepare a relatively flat silicate film, extensive rinsing of the substrate by DI water was performed between each deposition of polyelectrolytes. The reproducibility of silicate film preparation on gold was evaluated by using surface plasmon resonance (SPR) with “NanoSPR-3” (Morton Grove, IL) (support information).
To generate hydrophobic surface, OTS solution at a concentration of 10 mM in toluene was prepared and matured for 20 min before use. Calcinated chips were then immersed into the OTS solution for 2 min. After the dipping, the chips were rinsed by toluene, ethanol and DI water, and dried with compressed air.
Film characterization by SEM, AFM and contact angle measurements
Microscopic images were obtained from a Hitachi TM-1000 tabletop scanning electronic microscope (SEM) system (Tokyo, Japan). Atomic force microscope (AFM) images were collected by a Veeco Dimension 5000 atomic force microscope (Santa Barbara, CA) with manufacturer-provided software. All images were obtained in the tapping mode, and RMS surface roughness values were obtained by average multiple 5 µm2 area across the entire substrate at a scan rate of 1.5 Hz. Contact angle measurements were performed on a home built device with deionized water (1 µL). The images for water droplets on substrate were taken by a computer controlled 12-bit cooled QImaging RETIGA 2000R CCD camera (Surry, BC, Canada). All measurements were made in ambient atmosphere at room temperature.
Sample preparation for MS analysis
The stock solution for peptides was prepared by dissolving [Sar1, Thr8]-angiotensin II and neurotensin in 50% acetonitrile to a concentration of 200 µM, respectively. Standard peptide solution was prepared by diluting the stock solution. Cytocheme c, β-casein and α-casein solutions (40 µM) were prepared in 50 mM NH4HCO3 (pH 8.0) and denaturized at 50 °C for 1 h. BSA was firstly denaturized in 50 mM NH4HCO3 (pH 8.0) containing 8 M urea for 1 h and treated with DTT at 56 °C for 4 h and followed by treatment of IAA for 1 h in dark at room temperature. After reaction, 40 µM BSA solution was prepared by dilution of 50 mM NH4HCO3 (pH 8.0) for 10 times, giving urea concentration below 1 M. For protein digest experiments, proteins were digested by TPCK-treated trypsin at 37 °C for 24 h with a protein/enzyme ratio of 50:1. Formic acid was added to stop the reaction. The solutions of protein digests were stored at −20 °C before usage.
CHCA solution (10 mg/mL) was prepared in 60% ACN/water solution containing 0.1% TFA. When CHCA was used as the matrix, the sample solution was prepared in a 1:10 ratio of peptide solution to CHCA. For MALDI-MS analysis, 1.0 µL of sample solution was deposited onto the MALDI sample plate and dried at vacuum prior to MS detection.
Calcinated substrates were first washed by DI water and ethanol, and dried by compressed air. The cleaned substrates were attached onto an MALDI plate by adhesive polyimide film tape before samples deposition. An aliquot of 1 µL of 60% ACN aqueous solution containing 0.1% TFA and 10 mM citric acid was firstly deposited on the calcinated substrate to facilitate sample deposition on the hydrophobic surface. Then, sample solution was deposited on the top immediately. After dried in air or in vacuum, contaminants were re-dissolved in 2 µL of 0.1% TFA. Clean paper stick was used to removal the droplets. SALDI-MS analysis was performed direct on the calcinated surface.
Off-probe desalting for peptides was performed by using ZipTipC18 pipette tips (Millipore, Bedfold, MA) with the recommended standard protocol. In brief, one µL sample solution was aspirated in the pre-equilibrated tip, followed by washing with 5 µL 0.1% TFA in D.I. water for 3 times. Elution solution (60% acetonitrile in water with 0.1% TFA, 1 µL × 2) was used to directly elute sample to the substrate after aspirating and dispensing for several times.
LDI-TOF MS
Laser desorption and matrix assisted laser desorption/ionization mass spectra were obtained by using Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems, USA) operating in positive reflector mode. The mass spectrometer is equipped with a pulsed nitrogen laser operated at 337 nm with 3 ns-duration pulses. The accelerating voltage, grid voltage and extraction delay time were set as 20 kV, 65% and 190 ns, respectively. MS spectra were acquired as an average of 60 laser shots. The energy of each laser pulse was measured by using a FieldMax-P laser energy meter (Coherent, Santa Clara, CA)41. The effective range of laser intensity in our experiment is from 1600 to 2700 a.u. corresponding to laser fluence approximately from 20 to 130 mJ/cm2. In this range, the arbitrary laser intensity rises in a roughly linear relationship (R2 ~0.96) with the increase of laser fluence. FindPept tool (http://ca.expasy.org/tools/findpept.html) was used for peptide mapping analysis.
RESULTS AND DISCUSSION
Characterization of nanoscale silicate films
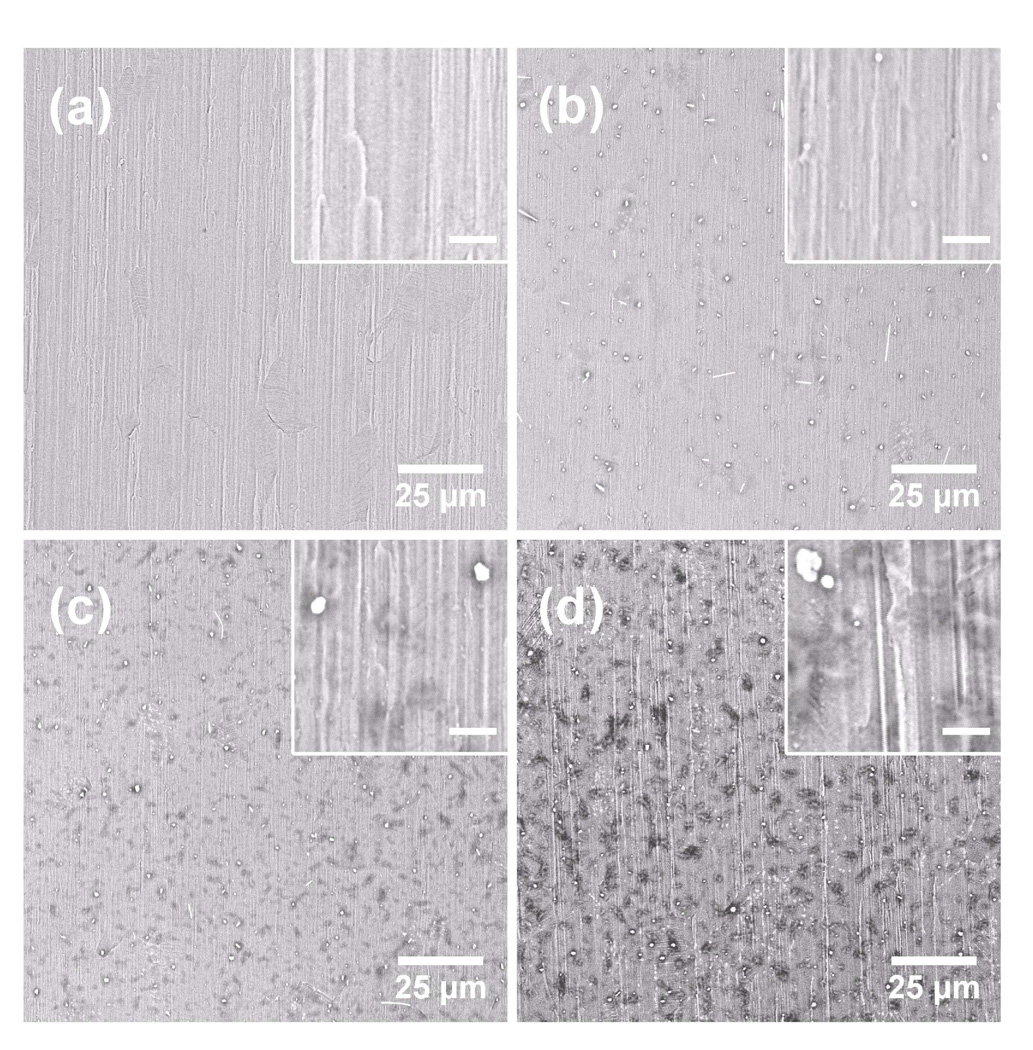
As surface roughness and wetting property are important parameters in LDI42, we first attempted to build a highly rough silicate nanofilm by modifying the LbL procedure. One approach is to use sodium silicate solution prepared at low pH as this condition is known to promote rapid growth of the polyelectrolyte layers39. However, aqueous sodium silicate tends to gel with time at low pH, resulting in poor reproducibility40. We found that by omitting the rinsing step and adopting a spraying-based LbL method43, ultrathin silicate films with varied roughness could be obtained within 10 circles of polyelectrolyte deposition. Figure 1 shows the SEM images of the calcinated films obtained under different conditions. Bare gold film on stainless steel tape shows a relative smooth surface except repeated ridges and wrinkles arising from blunt irregularities of the steel substrate (Figure 1a). Films with 8 layers of silicate fabricated with washing between depositions of polyelectrolytes prove that the washing process reduced polyelectrolyte aggregation, leading to a relative smooth surface with only some small features (average size ~280 nm) homogeneously scattered (Figure 1b). Figure 1c shows the film surface with 4 layers of silicate without washing steps. A relative rough surface with large features (average size ~700 nm) was produced with massive islands on the surface. When the number of silicate layer increased to 8, a highly rough silicate film was generated (Figure 1d). The average size of irregular feature (dots) increased to ~1 µm, while the number of silicate islands per square cm decreased from 5.5 × 107 in Figure 1c to 4.0 × 107 in Figure 1d. The island growth appeared to follow an Ostwald ripening process, where the exchange of species between the adsorbed islands could be involved44, 45. The high coverage of silicate islands on the surface, achieved by omitting the washing steps, led to the increase of the surface area needed for this work.
Figure 1.
SEM photograms of bare gold-covered substrate (a) and calcinated substrates fabricated with different methods: 8 layers of silicate fabricated with water washing between deposition of each polyelectrolytes (b); 4 layers of silicate fabricated without water washing (c); 8 layers of silicate fabricated without water washing (d). Amplification is ×1000. The insert pictures show substrates with 10,000-fold amplification. The scale bar represents 2 µm.
To acquire detailed morphological properties of the silicate films, AFM characterization was utilized and the results are summarized in Supporting Information. The mean roughness value (RMS) for bare gold on steel tapes is 16.0 ± 1.3 nm, which is much higher than gold deposited on glass slides (2.01 ± 0.29 nm)43 and is attributed to the blunt irregularities of the substrates. For silicate films prepared with a washing step, low density of scattered dots in nanometer scales produced a relative smooth surface. The average roughness for this surface is 20.2 ± 4.2 nm. For the films prepared without a washing step, the size of dots grew rapidly, resulting in a fast increase in thickness and surface roughness: 27.7 ± 5.4 nm for 4-layer silicate and 48.9 ± 8.1 nm for the 8-layer film. Additionally, large particles/islands that arise from aggregation of silicate were observed, which we believe are the primary source of deviation in the roughness measurement. The increase in deviation and roughness indicates the extent of aggregation aggravates as the thickness of silicate film grows larger.
SALDI-MS on OTS-modified calcinated films
The hydrophobic calcinated films were then investigated for effective SALDI analysis of peptides. Optimization of OTS-modification was carried out by varying OTS concentration and reaction time (supporting information). Concentrated OTS (>100 mM) yielded hydrophobic surface quickly (<30 s), but the SALDI-activity of substrate was reduced, suggesting that a highly compact OTS layer may obstruct the heat transfer from substrate to analyte in LDI process39. Modification with 10 mM OTS solution for 2 to 5 min appeared to give satisfactory LDI performance for peptide detection as well as good surface hydrophobicity. Surface wetting is crucial to the success of LDI analysis. Since hydrophobic surface is not friendly to aqueous solution, small amount of organic solvents, such as ACN, methanol and ethanol, are typically utilized to facilitate deposition of aqueous sample solution. In our experiment, 1 µL of 60% ACN aqueous solution containing 0.1% TFA and 10 mM citric acid was deposited on the calcinated substrate to facilitate sample deposition on the hydrophobic surface.
We compared the SALDI performance for a 5-layer silicate nanofilm before and after OTS coating to show the benefit of modification by the hydrophobic layer (supporting information). The laser threshold for LDI was about 170 a.u. (~9 mJ/cm2) lower on an OTS-modified surface than non-modified one, which can be explained by the reduction of thermo-conductivity of silicate nanofilm with the coatings28. The MS signal for peptide was enhanced by about 2–5 times after OTS modification, which arises from the reduction of sample spot size on the hydrophobic surface, thus leading to an increase in ion signal as well as S/N ratio. The improvement of SALDI detection was also obtained with gold surface after C18 coating using 1-octadecanethiol, resulting in more than 4 folders of enhancement of ion signal for peptides. However, the MS signal on gold is relatively low due to the poor SALDI activity of gold as compared to the calcinated films (supporting information). OTS is not a SALDI-active material. No visible ion signal was produced from the OTS-modified glass substrate.
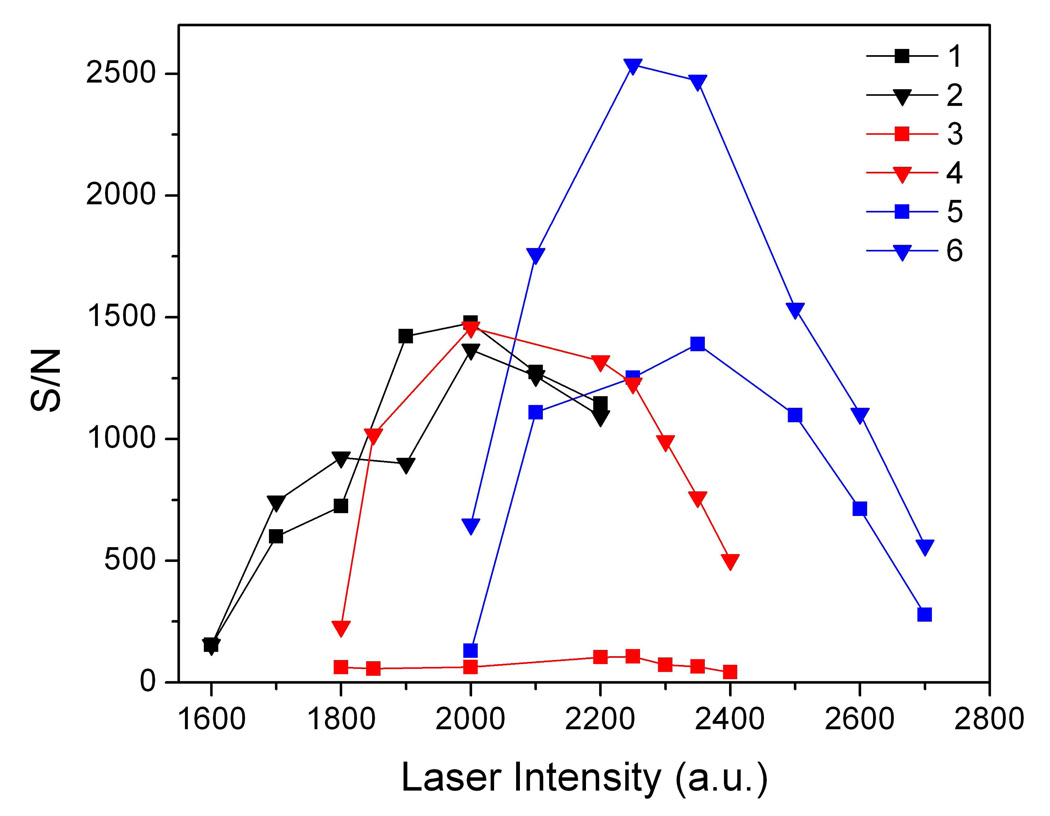
To study the surface roughness effect, two substrates coated with 8-layer silicate nanofilms with and without washing steps in the preparation were used and referred as smooth film (S-film, with washing) and rough film (R-film, without washing), respectively, based on their difference in surface roughness. Figure 2 shows the signal-to-noise (S/N) ratios of peptide protonated ions as a function of laser intensity for a mixture of two peptides, [Sar1, Thr8]-angiotensin II and neurotensin, and the MALDI results with CHCA for comparison. The laser threshold was low in MALDI-MS with CHCA matrix, in which laser intensity of 1600 a.u. (~20 mJ/cm2) was strong enough to yield sizable signal (curve 1 and 2). For SALDI-MS on the calcinated surface, the laser thresholds for OTS-modified S-film and R-film increased to about 1800 (~27 mJ/cm2, curve 3 and 4) and 2000 a.u. (~39 mJ/cm2, curve 5 and 6), respectively. It suggested a relatively inefficient energy transferring on calcinated surfaces. This result is in fact consistent with the studies using DIOS and SALDI-MS on metals46. The increased laser threshold on the R-film substrate could possibly arise from the increase in thickness of the silicate film, which may weaken the absorption of UV-energy and subsequent heat transfer from the Au-layer to analytes. On the other hand, the increase in thickness enhances the stability of the silicate films towards laser. The highest laser intensity that this film can endure increases up to 50 a.u. (~7 mJ/cm2) compared to the S-film.
Figure 2.
Dependency of signal-to-noise ratio for peptide protonated ions on laser intensity in MS detection with CHCA matrix (black, curve 1 and 2), with 8 layers of silicate fabricated with water washing between deposition of each polyelectrolytes (red, curve 3 and 4), and with 8 layers of silicate fabricated without water washing (blue, curve 5 and 6). Silicate films were coated with OTS after calcination. The sample was a mixture of [Sar1, Thr8]-angiotensin II (squares) and neurotensin (triangles), 1 pmol each. Error bars are omitted for simplicity.
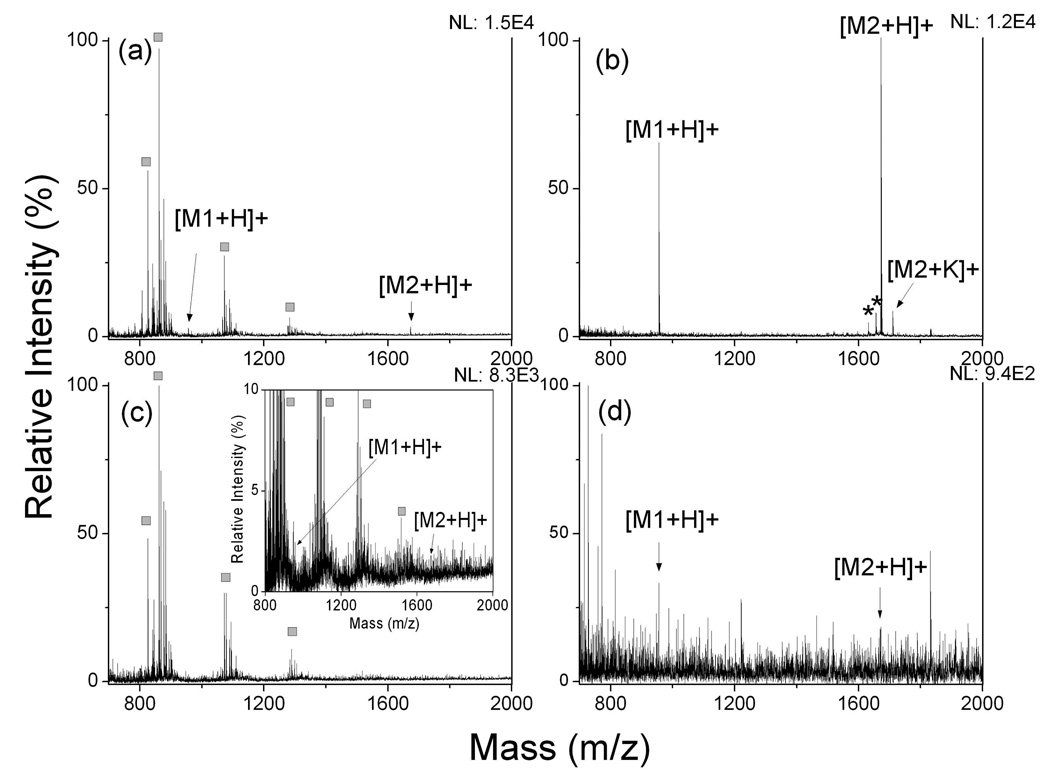
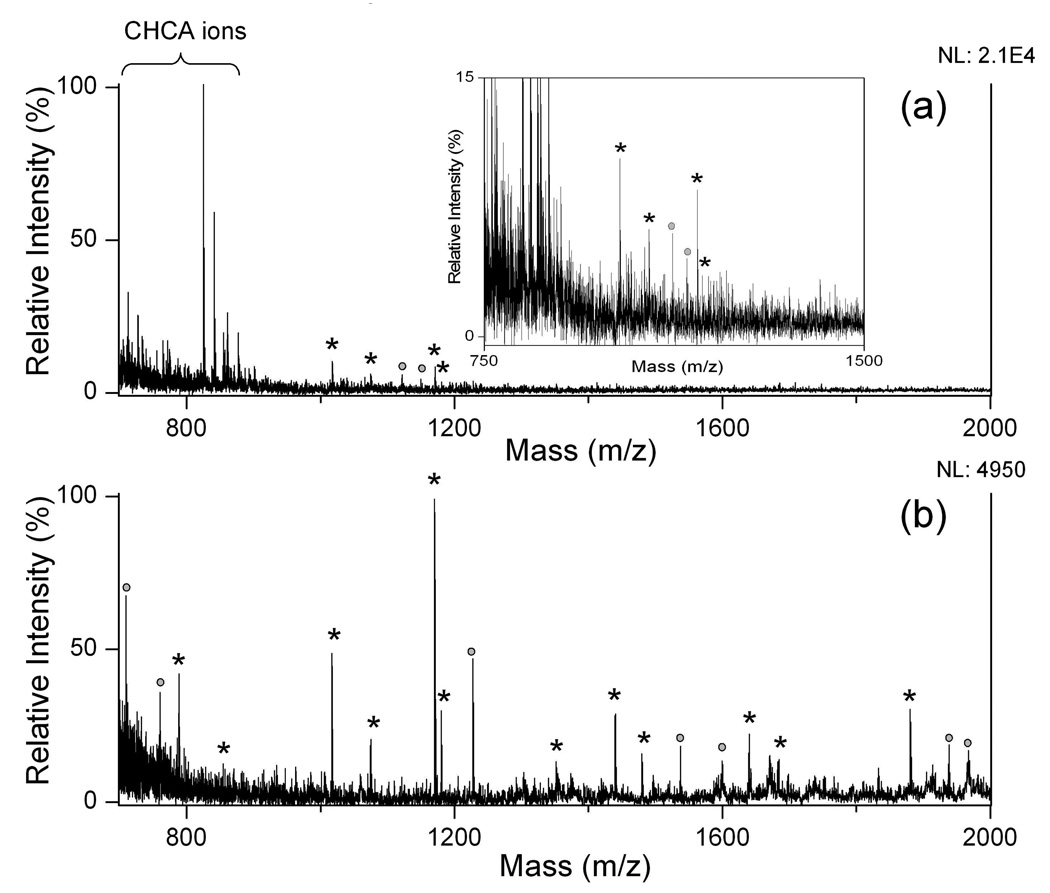
We then assessed the performance of OTS-modified R-films for analysis of trace amount peptides in a mixture in the femtomole range. Figure 3 showed MALDI- and SALDI-MS spectra for 25 fmol and 5 fmol of peptides, respectively. MALDI-MS yielded complicated spectra and matrix-related ions due to excessive amount of CHCA (~50 nmol) dominating in the spectra, leading to suppression of LDI of peptide ions. SALDI-MS on this surface, however, provided much better performance than MALDI-MS. Over twenty-folder increase of sensitivity in MS detection can be achieved when 25 fmol of peptides was used as sample (Figure 3b). When peptide amount was lowered to 5 fmol, the SALDI peaks for peptides were still highly distinguishable as compared to the background noise (Figure 3d) while ion signal of analytes was totally submerged by CHCA-related ions in MALDI detection. The results clearly indicated a low femtomole LOD for peptides on the OTS-modified substrate. Similar analysis was carried out with the S-films. The sensitivity was nearly two orders of magnitude lower as compared to that on the R-film (data not shown).
Figure 3.
MALDI and SALDI mass spectra of peptide mixture. (a) 25 fmol peptides with CHCA matrix; (b) 25 fmol peptides on OTS-modified calcinated surface; (c) 5 fmol peptides with CHCA matrix; (d) 5 fmol peptides on OTS-modified calcinated surface; The analyte was the mixture of [Sar1, Thr8]-angiotensin II (M1) and neurotensin (M2), equal amount in sample. Asterisks (*) represent peaks assigned to fragment ions of peptides; squares (□) represent peaks from CHCA related ions. NL: normalized level.
It is interesting to note that although ion intensity of peptide ions increases with the increase of laser intensity, the corresponding S/N ratios of peptide peaks tend to decrease after the laser intensity passes a threshold value in both MALDI- and SALDI-MS detection. The decrease in MALDI is caused by suppression of matrix-related ions, which become dominant at high laser intensity. The decrease in SALDI at high laser intensity, however, is due to surface damage. Additionally, the protonated ion of neurotensin yielded a comparable S/N value to MALDI-MS analysis at the laser intensity close to 2000 a.u., while the ratio for angiotensin II ions remained low. This difference may stem from varied retention efficiency by the two surfaces towards analytes, as will be discussed in details below.
The maximum laser intensity the chip can endure was also slightly higher for the OTS modified surface. For example, at laser intensity of 2500 a.u. (~96 mJ/cm2), the MS signal for peptide almost completely disappeared on the unmodified calcinated surface, while on OTS-modified surface the MS signal reduced about 50% in magnitude at its maximum intensity, suggesting better stability of the calcinated films after OTS coating. The up mass limit we tested for the OTS substrate is ca. 3500 Da, which provides an enough mass window for common peptide analysis.
On-plate desalting and subsequent SALDI-MS detection of peptides
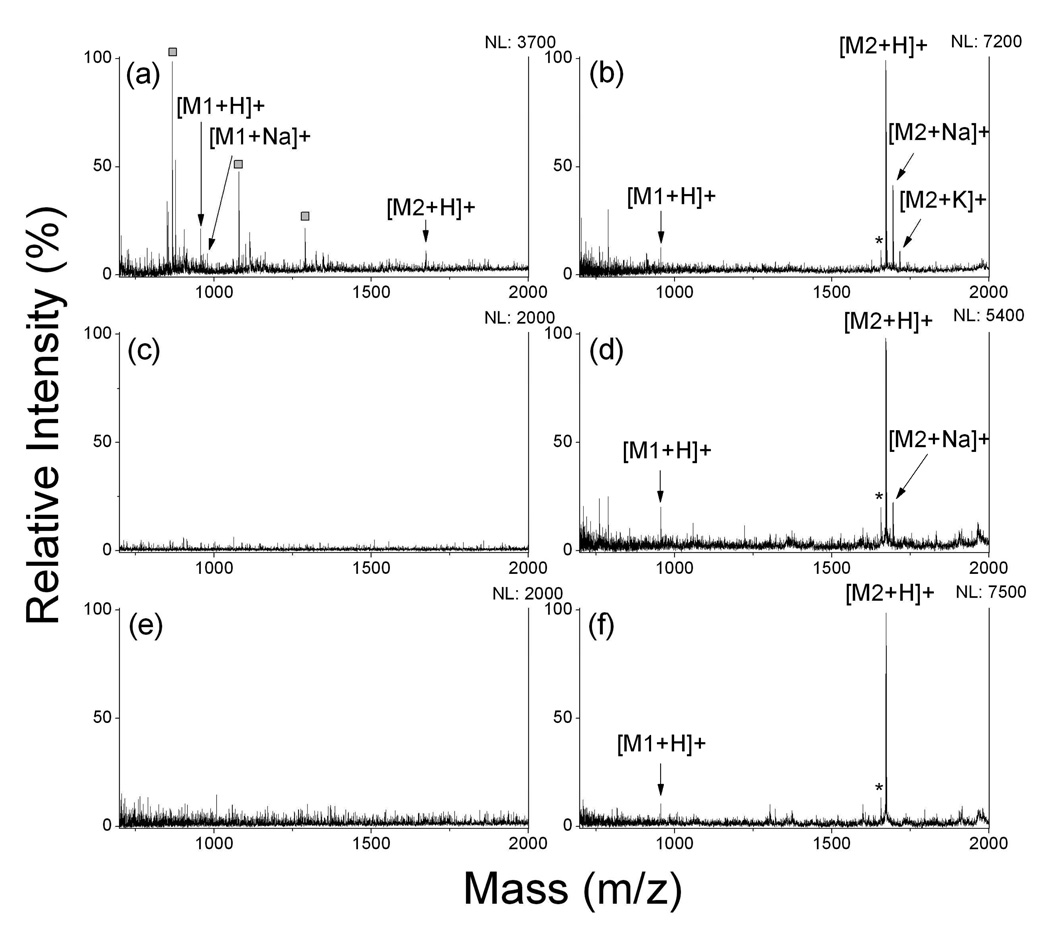
Hydrophobic surfaces have been used for sample concentration and on-target desalting before MALDI-MS detection was applied9. Here, we studied the on-plate desalting and SALDI-MS analysis with the OTS-modified, rough calcinated films with high hydrophobicity. The contact angle of the modified surface was very high, larger than 160 degree after OTS coating (Supporting Information). On-plate desalting for peptides were tested using different salt solutions including 1 M NaCl, 200 mM NaAc and 8 M urea. Figure 4 shows MS spectra without and with on-plate desalting for 100 fmol of the peptides. For MALDI-MS analysis, as co-crystallization of matrix molecules with analytes is crucial, high salts often hamper the co-crystallization process as many of these salts are incompatible with organic matrix solutions. Although MS signal was still detectable by the “sweet-spots” searching (Figure 4a), the high background noises from alkaline adducted CHCA-related ions were dominant in the spectrum, yielding poor detection sensitivity. Direct LDI-MS detection of NaCl-containing peptide samples was also performed without a desalting process. However, no signal was obtained and a layer of salt precipitates was generated after sample drying. This layer is thought to seriously impede the UV-absorption and the energy transfer on the calcinated film. After removal of salts by washing, enhanced peptide signal was obtained without “sweet-spot” searching, which indicated the homogeneous distribution of analyte on the film that leads to a better reproducibility than MALDI analysis. Figure 4b shows the MS spectrum after desalting on the OTS-modified calcinated film and S/N ratio increased about 1.8 and 4.1 times for [Sar1, Thr8]-angiotensin II (M1) and neurotensin (M2), respectively. The presence of organic solvent-compatible salts such as NaAc and urea allows co-crystallization of salts with matrix and analytes and makes it rather difficult to obtain quality mass spectra. No peaks were observed in the presence of NaAc and urea in MALDI-MS detection (Figure 4c and 4e). But with on-plate desalting, peaks for two peptides were clearly measurable and the improvement in sensitivity was evidently demonstrated (Figure 4d and 4f). In addition, OTS-modified calcinated substrate has also been successfully applied to removal of surfactants, including 0.1% CHAPs and 0.1–5% SDS, from peptide mixtures before LDI-MS (support information).
Figure 4.
MALDI and SALDI mass spectra of peptide mixture with addition of different salts. (a) 1 M NaCl, CHCA matrix; (b) 1 M NaCl, desalting&SALDI on OTS-modified calcinated surface; (c) 200 mM NaAc, CHCA matrix; (d) 200 mM NaAc, desalting&SALDI OTS-modified calcinated surface; (e) 8 M urea, CHCA matrix; (f) 8 M urea, desalting&SALDI OTS-modified calcinated surface. The analyte was the mixture of [Sar1, Thr8]-angiotensin II (M1) and neurotensin (M2), 100 fmol each. Asterisks (*) represent peaks assigned to fragment ions of peptides; squares (□) represent peaks from CHCA related ions. NL: normalized level.
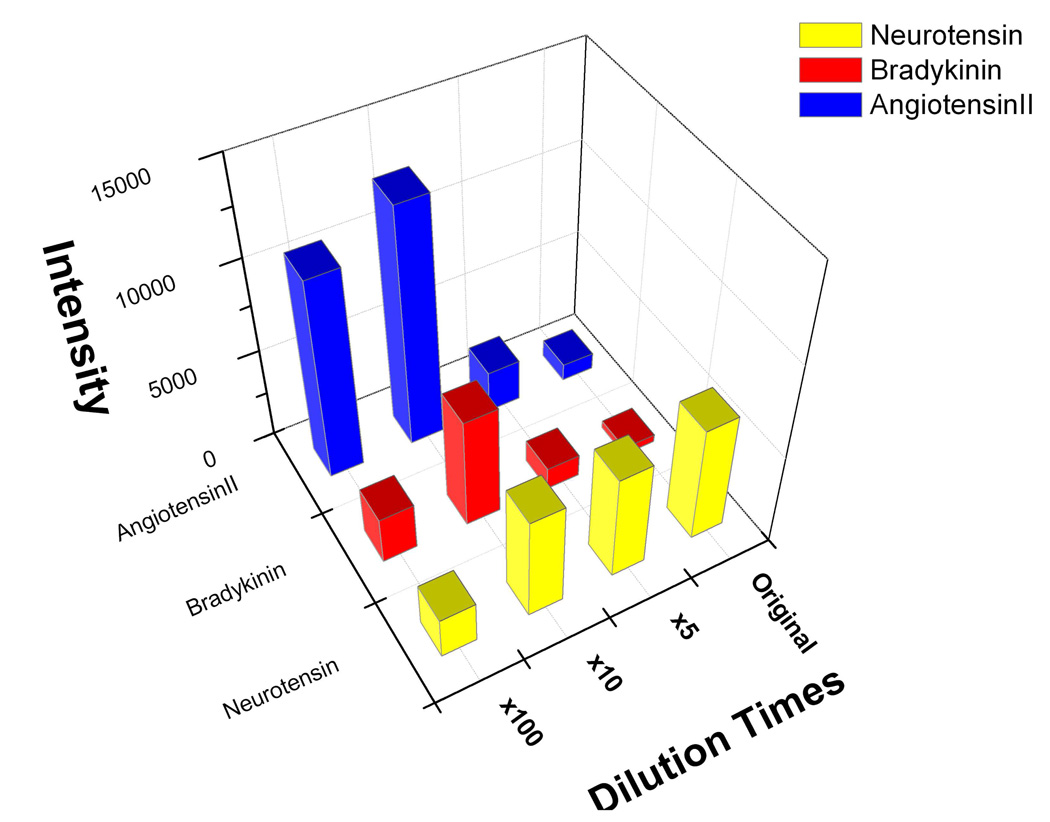
We noticed that the recovery rate of on-plate desalting for peptides is relatively low by comparing LDI-MS analysis of analytes before and after desalting. The sample loss may arise from co-elution of analytes during intensive washing. Compared to the S-films, neurotensin analysis was less affected by the rinsing step on the OTS-modified R-films. This may be explained by the enlarged surface area for analyte capture for the R-films. Higher S/N ratio for protonated ions of neurotensin was also observed on this substrate, which could be explained by the reduced size of sample spots after OTS modification (support information). To further evaluate the performance of on-plate approach, off-probe desalting of peptides followed by SALDI-MS was performed by using C18-ZipTips. It turns out that the two desalting methods produced comparable recovery rate for peptides (support information), suggesting the on-plate approach offers similar desalting efficiency as the ZipTip method. However, operation of ZipTip is laborious due to multistep sample handling, while on-plate approach is much simpler and amenable to high throughput analysis. In addition, better recovery for relatively hydrophobic peptide (i.e., neurotensin) on the calcinated films suggests the selectivity of the surface towards hydrophobic species. To further verify this selectivity, a mixture of three peptides and their diluted solutions were used for direct SALDI-MS analysis. The peptide concentration in the original mixture is 10, 5 and 2.5 µM for [Sar1, Thr8]-angiotensin II, bradykinin and neurotensin, respectively. The retention of three peptides in C18 reversed-phase HPLC is: neurotensin>bradykinin>[Sar1, Thr8]-angiotensin II. As shown in Figure 5, with ×100 dilution, [Sar1, Thr8]-angiotensin II showed relatively high ion signal and was dominant in mass spectrum due to its relatively high content in the mixture. The relative ratio of ion intensity is 4.8:1.1:1.0 for [Sar1, Thr8]-angiotensin II, bradykinin and neurotensin. When the concentration increased by 10 times, all three peptide showed enhanced ion signal. In this case, [Sar1, Thr8]-angiotensin II remained dominant in the mass spectrum, but its relative ion intensity was reduced, and the relative ratio of ion intensity has changed to about 2.3:1.1:1.0. Further increase of the concentration resulted in neurotensin becoming dominant in the spectrum. The relative ratio of ion intensity changed to about 1.9:1.0:4.7 and 2.2:1.0:15.5 for ×5 dilution and the original sample, respectively. This change may arise from the competitive binding of peptides to the limited adsorption sites on the surface. Neurotensin, relatively hydrophobic among the three peptides, has better accessibility to the hydrophobic surface, and thus inclines to occupy the available binding sites preferentially. As a consequence, the ion signal of neurotensin increases with the rise of peptide concentration in solution. In contrast, [Sar1, Thr8]-angiotensin II and bradykinin are relatively hydrophilic peptides, which have relatively poor retention on a hydrophobic surface. Therefore, these hydrophilic peptides are easily displaced by neurotensin from the hydrophobic surface when abundant sample is applied, resulting in the decrease of ion intensity with increase of peptide concentration. This result clearly demonstrates the good selectivity of OTS-modified substrate towards hydrophobic species, which would be very useful for selectively detection of relatively hydrophobic components from a complex sample by this on-plate competition and isolation approach. Compared to OTS-modified substrate, C18-ZipTip tends to yield high recovery for hydrophilic peptide after desalting, which can be attributed to the high amount of SPE absorbent used in the C18-ZipTip. Excess absorbent, however, could result in sample loss owing to the irreversible binding, especially when small-size samples were tested. Therefore, on-plate desalting can serve as a complementary method to C18-ZipTip for efficient purification of small-size samples before MS detection.
Figure 5.
A 3D graphic presentation of the analysis for the peptide mixture solutions with different dilutions. Original peptide mixture consists of 10 µM [Sar1, Thr8]-angiotensin II, 5 µM bradykinin and 2.5 µM neurotensin. The deposition volume is 1.0 µL.
Application for the analysis of protein digests
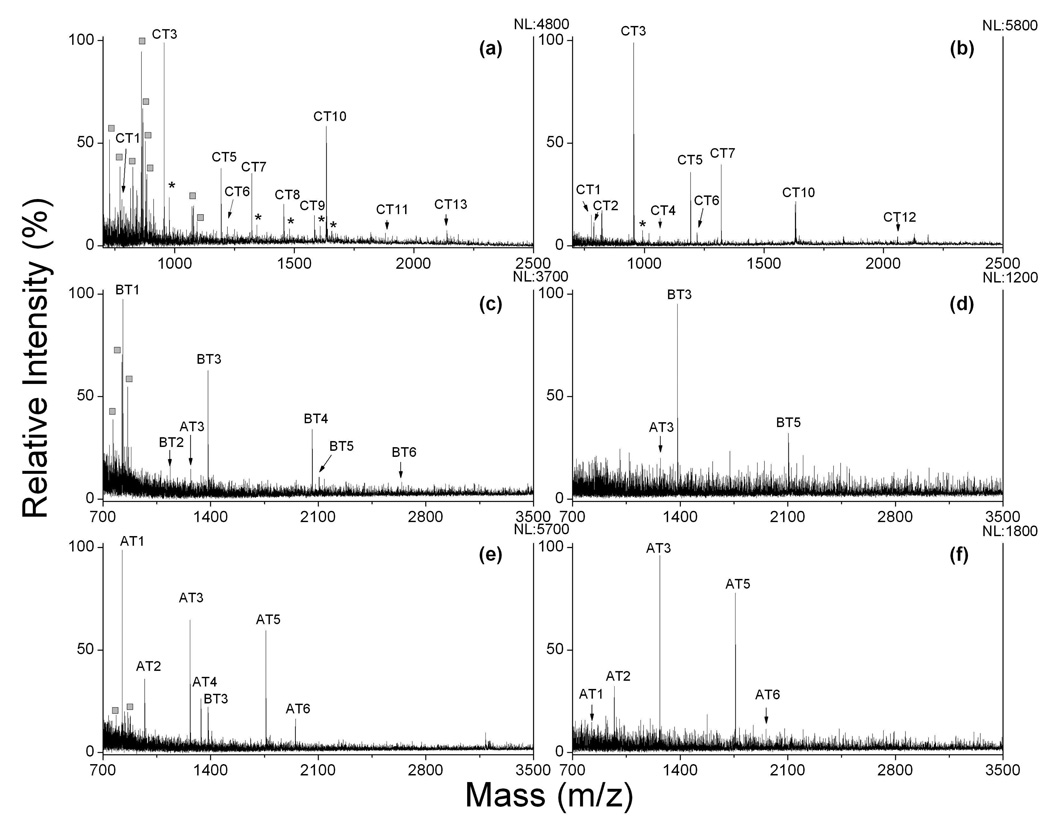
On-plate desalting and SALDI-MS was further applied to the analysis of protein digests. Figure 6a and b show MS spectra of 40 fmol of tryptic digest from cytochrome c in the presence of 50 mM NH4HCO3. The identified peptides were listed in Table S1 in the support information. MALDI-MS analysis yielded abundant matrix-related ions, and the S/N values for peptides were low because of the suppression of LDI by the production of matrix ions. In the SALDI analysis with on-plate washing, a clean spectrum was achieved with much improved S/N ratio for peptides. For instance, peak CT3, CT5 and CT7 at m/z 955, 1194 and 1322, respectively, showed relative higher abundance in both MALDI- and SALDI-MS detection. S/N enhancement factors for these peaks by SALDI were determined to be 2.4, 1.7 and 2.0, respectively, as compared to MALDI analysis. On-plate desalting/LDI analysis for β-casein and α-casein digests showed similar enhanced results for the peptides. As shown in Figure 6c–f, β-casein peptide BT3 and BT5 at m/z 1384 and 2107 gave 1.7 and 3.8 times of increase in S/N ratio with SALDI analysis, respectively, as compared to the MALDI analysis, while S/N ratio for α-casein fragment AT2, AT3 and AT5 was enhanced by 1.2, 2.0 and 1.8 times, respectively. Our results clearly show that on-plate desalting/LDI approach can enhance MS detection for peptides in a complex sample, such as protein digest. The enhancement may be attributed to elimination of matrix-related ions and removal of contaminants.
Figure 6.
MALDI and SALDI mass spectra of protein digests. (a) 40 fmol cytochrome c digest with CHCA matrix; (b) 40 fmol cytochrome c digest with on-plate desalting/SALDI on an OTS-modified calcinated surface. (c) 100 fmol β-casein digest with CHCA matrix; (d) 100 fmol β-casein digest with on-plate desalting/SALDI on an OTS-modified calcinated surface. (e) 100 fmol α-casein digest with CHCA matrix; (f) 100 fmol α-casein digest with on-plate desalting/SALDI on an OTS-modified calcinated surface. CT, BT and AT represent identified peptides from tryptic digests from cytochrome c, β-casein and α-casein, respectively, and sequence information is given in Supporting Information. Asterisks (*) represent peaks assigned to alkaline adducted ions of peptides; squares (□) represent peaks from CHCA related ions. NL: normalized level.
However, it was found that the number of identified peptide from protein digests with SALDI-MS detection was less than that with MALDI analysis (supporting information). This leads to a relatively low amino acid sequence coverage score for protein identification. To understand the reason, we compared the results from MALDI and SALDI. With cytochrome c digest as an example, peptide CT2 and CT4 were only detectable with SALDI after desalting. These peptides tend to be swamped by background from CHCA-related ions in MALDI analysis. But SALDI failed to detect a few other cytochrome c peptides, including CT8, CT9, CT11 and CT13. The amino acid composition for these peptides were taken into account, and the calculated scores of grand average of hydropathicity (GRAVY) index were in the range from −0.961 to −1.187 (support information), which indicates a relative strong hydrophilicity for these peptides. As a result, desorption of these peptides with salts occurs during the desalting process, resulting in loss of MS signal in the detection. Additionally, the missing β-casein and α-casein fragments in SADLI-MS analysis also showed strong hydrophilicity (supporting information). For instance, the GRAVY scores for β-casein peptide BT1, BT2, BT4 and BT6 were in the range from −0.929 to −2.331, while α-casein peptide AT4 has a GRAVY score of −1.755. It suggests that hydrophobic surface is unfavorable to the binding of hydrophilic species, leading to loss of hydrophilic components during on-plate washing process. The result further suggests that the OTS-modified glassy substrate has selectivity towards analytes with hydrophobic properties.
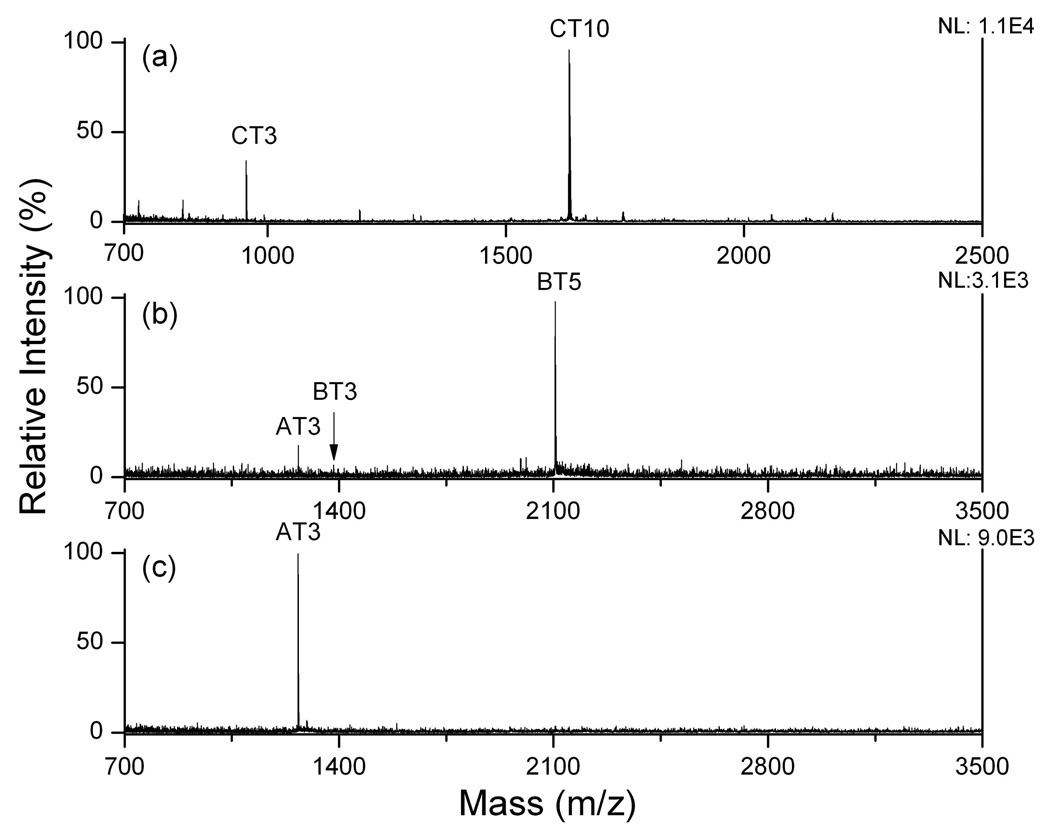
We further studied the SALDI analysis of higher concentration of protein digests after on-plate desalting. As shown in Figure 7a to c, enhanced signal was observed for cytochrome c peptide CT3 and CT10, for β-casein peptide BT3 and BT5, and for α-casein peptide AT3, and the GRAVY scores for these peptides were 0.067, 0.021, 0.283, 0.832 and 0.070, respectively. The positive GRAVY scores indicate the relatively high hydrophobicity of these peptides, which further suggests the selectivity of OTS-substrate towards hydrophobic peptides. The concentration and isolation of relative hydrophobic peptides can be explained again by the competition model, in which peptides tend to compete for the limited binding sites on the surface when excess protein digests are applied. Hydrophobic peptides preferably occupy the available binding sites on the C18 surface, leading to their concentration while other less competitive materials are washed away. The ability to analyze hydrophobic peptides is advantageous and practically useful as separation and concentration process can be carried out directly on the substrate. Additionally, the simplicity in sample preparation of SALDI analysis can reduce the sample dilution and background ions arising from the organic matrix.
Figure 7.
SALDI mass spectra of protein digests with high amount. (a) 400 fmol cytochrome c digest; (b) 4 pmol β-casein digest; (c) 4 pmol α-casein digest. NL: normalized level. Sequence information is given in the supporting information.
On-plate desalting and SALDI-MS have also been performed for the analysis of 40 pmol BSA digests with presence of 800 mM urea. Figure 8a shows the MALDI results for comparison. LDI for analytes was highly suppressed by the existence of salts with CHCA matrix. Only 4 peaks were assigned to BSA peptides. The sequence coverage was only 4.6%. SALDI, on the other hand, provides a much improved MS background and sensitivity with the OTS-modified substrate by a simply rinsing to remove contaminants (Figure 8b). As a result, 13 peptides were detected, and the sequence coverage was increased to 16.8%. This result indicated the potential of combination of SALDI and calcinated films in the application of high throughput proteome analysis. It should be noted that the matched BSA peptides had relative high GRAVY scores (support information) except LSHKDDSPDLPK and QEPERNECFLSHK, which showed relative low ion intensity in spectrum.
Figure 8.
MALDI and SALDI mass spectra of 40 pmol BSA tryptic digests. Asterisks (*) represent peaks assigned to protonated peptides of protein digests; circles (○) represent unidentified ions. NL: normalized level.
CONCLUSIONS
We have demonstrated on-plate desalting/direct LDI-MS analysis of peptides containing high salt background by using OTS-coated calcinated nanofilms on Au. The enhanced LDI of peptides in a digestive mixture has also been established. Sample complexity has been vastly reduced by removal of MS-incompatible contaminants, such as salts, surfactants and denaturing agent (urea), leading to improvement of LDI-MS analysis of peptides. The improvement of LDI-MS on the new surface was attributed to the following factors: (1) the “buffer” effect of the OTS layer to reduce the thermo-conductivity of nanofilm, (2) improved stability of nanofilm under laser shot, and (3) reduction of sample size by hydrophobic property of the surface. The surface roughness and thickness of the silicate layer are precisely tailored by modifying the LbL procedure and solution composition. High surface roughness improves on sample recovery after on-chip clean-up due to higher surface area and sample capacity. Optimal nanofilm thickness is important as thicker films are found to impair heat transfer from gold to the analyte, leading to elevation of LDI threshold. OTS coating also improves on sample deposition and facilitates on-plate pre-preparation prior to LDI-MS analysis. The on-plate clean-up procedure is simple, rapid and easy to perform without the need of eluting the analytes off the surface. The selective capture of hydrophobic components from analytes makes it possible to analyze hydrophobic peptides in complex proteomic samples including protein digest.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the National Science Foundation (CHE-0719224) and the National Institute of Health (1R21EB009551-01A2). We thank Dr. Richard Kondrat, Lei Xiong and Yongsheng Xiao for the help in mass spectrometric analysis.
Reference
- 1.Aebersold R, Goodlett DR. Chem. Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 2.Karas M, Bahr U, Giessmann U. Mass Spectrom. Rev. 1991;10:335–357. [Google Scholar]
- 3.Murray KK. Mass Spectrom. Rev. 1997;16:283–299. [Google Scholar]
- 4.Pan CS, Xu SY, Zhou HJ, Fu Y, Ye ML, Zou HF. Anal. Bioanal. Chem. 2007;387:193–204. doi: 10.1007/s00216-006-0905-4. [DOI] [PubMed] [Google Scholar]
- 5.Pluskal MG. Nat. Biotechnol. 2000;18:104–105. doi: 10.1038/71778. [DOI] [PubMed] [Google Scholar]
- 6.Rappsilber J, Ishihama Y, Mann M. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 7.Horak D, Babic M, Mackova H, Benes MJ. J. Sep. Sci. 2007;30:1751–1772. doi: 10.1002/jssc.200700088. [DOI] [PubMed] [Google Scholar]
- 8.Mornet S, Vasseur S, Grasset F, Duguet E. J. Mater. Chem. 2004;14:2161–2175. [Google Scholar]
- 9.Xu YD, Bruening ML, Watson JT. Mass Spectrom. Rev. 2003;22:429–440. doi: 10.1002/mas.10064. [DOI] [PubMed] [Google Scholar]
- 10.Yuan XL, Desiderio DM. J. Mass Spectrom. 2002;37:512–524. doi: 10.1002/jms.307. [DOI] [PubMed] [Google Scholar]
- 11.Brockman AH, Dodd BS, Orlando N. Anal. Chem. 1997;69:4716–4720. doi: 10.1021/ac970650u. [DOI] [PubMed] [Google Scholar]
- 12.Hung KC, Rashidzadeh H, Wang Y, Guo BC. Anal. Chem. 1998;70:3088–3093. doi: 10.1021/ac980090e. [DOI] [PubMed] [Google Scholar]
- 13.Wang JH, Chen RB, Ma MM, Li LJ. Anal. Chem. 2008;80:491–500. doi: 10.1021/ac701614f. [DOI] [PubMed] [Google Scholar]
- 14.Jia WT, Wu HX, Lui HJ, Li N, Zhang Y, Cai RF, Yang PY. Proteomics. 2007;7:2497–2506. doi: 10.1002/pmic.200601029. [DOI] [PubMed] [Google Scholar]
- 15.Muck A, Nesnerova P, Pichova I, Svatos A. Electrophoresis. 2005;26:2835–2842. doi: 10.1002/elps.200400014. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Fang J, Kuang Y, Guo X, Lu H, Yang P. Chem. Commun. 2007:4468–4470. doi: 10.1039/b710741b. [DOI] [PubMed] [Google Scholar]
- 17.Worrall TA, Cotter RJ, Woods AS. Anal. Chem. 1998;70:750–756. doi: 10.1021/ac970969e. [DOI] [PubMed] [Google Scholar]
- 18.Cohen LH, Gusev AI. Anal. Bioanal. Chem. 2002;373:571–586. doi: 10.1007/s00216-002-1321-z. [DOI] [PubMed] [Google Scholar]
- 19.Peterson DS. Mass Spectrom. Rev. 2007;26:19–34. doi: 10.1002/mas.20104. [DOI] [PubMed] [Google Scholar]
- 20.Guo Z, Ganawi AAA, Liu Q, He L. Anal. Bioanal. Chem. 2006;384:584–592. doi: 10.1007/s00216-005-0125-3. [DOI] [PubMed] [Google Scholar]
- 21.Sunner J, Dratz E, Chen YC. Anal. Chem. 1995;67:4335–4342. doi: 10.1021/ac00119a021. [DOI] [PubMed] [Google Scholar]
- 22.Pan CS, Xu SY, Zou HF, Guo Z, Zhang Y, Guo BC. J. Am. Soc. Mass Spectrom. 2005;16:263–270. doi: 10.1016/j.jasms.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Wen XJ, Dagan S, Wysocki VH. Anal. Chem. 2007;79:434–444. doi: 10.1021/ac061154l. [DOI] [PubMed] [Google Scholar]
- 24.Hoang TT, Chen YF, May SW, Browner RF. Anal. Chem. 2004;76:2062–2070. doi: 10.1021/ac020707z. [DOI] [PubMed] [Google Scholar]
- 25.Lo CY, Lin JY, Chen WY, Chen CT, Chen YC. J. Am. Soc. Mass Spectrom. 2008;19:1014–1020. doi: 10.1016/j.jasms.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Bi HY, Qiao L, Busnel JM, Devaud V, Liu BH, Girault HH. Anal. Chem. 2009;81:1177–1183. doi: 10.1021/ac8024448. [DOI] [PubMed] [Google Scholar]
- 27.Castellana ET, Russell DH. Nano Lett. 2007;7:3023–3025. doi: 10.1021/nl071469w. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki H, Sugitani T, Watanabe T, Yonezawa T, Moriwaki H, Arakawa R. Anal. Chem. 2008;80:7524–7533. doi: 10.1021/ac800789t. [DOI] [PubMed] [Google Scholar]
- 29.McLean JA, Stumpo KA, Russell DH. J. Am. Chem. Soc. 2005;127:5304–5305. doi: 10.1021/ja043907w. [DOI] [PubMed] [Google Scholar]
- 30.Huang YF, Chang HT. Anal. Chem. 2007;79:4852–4859. doi: 10.1021/ac070023x. [DOI] [PubMed] [Google Scholar]
- 31.Huang YF, Chang HT. Anal. Chem. 2006;78:1485–1493. doi: 10.1021/ac0517646. [DOI] [PubMed] [Google Scholar]
- 32.Duan JC, Linman MJ, Chen CY, Cheng QJ. J. Am. Soc. Mass Spectrom. 2009;20:1530–1539. doi: 10.1016/j.jasms.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Ren SF, Zhang L, Cheng ZH, Guo YL. J. Am. Soc. Mass Spectrom. 2005;16:333–339. doi: 10.1016/j.jasms.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Wei J, Buriak JM, Siuzdak G. Nature. 1999;399:243–246. doi: 10.1038/20400. [DOI] [PubMed] [Google Scholar]
- 35.Shen ZX, Thomas JJ, Averbuj C, Broo KM, Engelhard M, Crowell JE, Finn MG, Siuzdak G. Anal. Chem. 2001;73:612–619. doi: 10.1021/ac000746f. [DOI] [PubMed] [Google Scholar]
- 36.Go EP, Uritboonthai W, Apon JV, Trauger SA, Nordstrom A, O'Maille G, Brittain SM, Peters EC, Siuzdak G. J. Proteome Res. 2007;6:1492–1499. doi: 10.1021/pr060608s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trauger SA, Go EP, Shen ZX, Apon JV, Compton BJ, Bouvier ESP, Finn MG, Siuzdak G. Anal. Chem. 2004;76:4484–4489. doi: 10.1021/ac049657j. [DOI] [PubMed] [Google Scholar]
- 38.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Nature. 2007;449 doi: 10.1038/nature06195. 1033-U1033. [DOI] [PubMed] [Google Scholar]
- 39.Duan JC, Linman MJ, Cheng Q. Anal. Chem. 2010;82:5088–5094. doi: 10.1021/ac100132x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips KS, Han JH, Martinez M, Wang ZZ, Carter D, Cheng Q. Anal. Chem. 2006;78:596–603. doi: 10.1021/ac051644y. [DOI] [PubMed] [Google Scholar]
- 41.Xiao YS, Retterer ST, Thomas DK, Tao JY, He L. J. Phys. Chem. C. 2009;113:3076–3083. [Google Scholar]
- 42.Alimpiev S, Nikiforov S, Karavanskii V, Minton T, Sunner J. J. Chem. Phys. 2001;115:1891–1901. [Google Scholar]
- 43.Linman MJ, Culver SP, Cheng Q. Langmuir. 2009;25:3075–3082. doi: 10.1021/la803835a. [DOI] [PubMed] [Google Scholar]
- 44.Thiel PA, Shen M, Liu DJ, Evans JW. J. Phys. Chem. C. 2009;113:5047–5067. [Google Scholar]
- 45.Krost A, Christen J, Oleynik N, Dadgar A, Deiter S, Blasing J, Krtschil A, Forster D, Bertram F, Diez A. Appl. Phys. Lett. 2004;85:1496–1498. [Google Scholar]
- 46.Okuno S, Arakawa R, Okamoto K, Matsui Y, Seki S, Kozawa T, Tagawa S, Wada Y. Anal. Chem. 2005;77:5364–5369. doi: 10.1021/ac050504l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.