Abstract
Programmed cell death (PCD) during neuronal development and disease has been shown to require de novo RNA synthesis. However, the time course and regulation of target genes is poorly understood. By using a brain-biased array of over 7,500 cDNAs, we profiled this gene expression component of PCD in cerebellar granule neurons challenged separately by potassium withdrawal, combined potassium and serum withdrawal, and kainic acid administration. We found that hundreds of genes were significantly regulated in discreet waves including known genes whose protein products are involved in PCD. A restricted set of genes was regulated by all models, providing evidence that signals inducing PCD can regulate large assemblages of genes (of which a restricted subset may be shared in multiple pathways).
Programmed cell death (PCD) is an essential component of neuronal development and has been associated with many forms of neurodegeneration (1, 2). During development and disease, death signals are transduced through multiple pathways in a highly regulated process that involves transcription-dependent and -independent mechanisms (3–5). These pathways converge on caspase-mediated proteolytic cascades and may be integrated with regulation of energy, redox-state, ion homeostasis in the mitochondria, and the control of cell-cycle.
In the cerebellum, granule cell development occurs postnatally. The final number of cerebellar granule neurons (CGNs) represents the combined effects of migration, cell division, and target-related PCD that removes neuronal precursors failing to establish appropriate synaptic connections. This PCD can be modeled in vitro where depolarizing amounts of extracellular potassium (K+) in the culture medium promote the survival of CGNs and low concentrations induce apoptosis (6, 7). The resulting PCD can be blocked by inhibitors of protein or RNA synthesis (8, 9). Possible target genes comprising this gene expression component include c-jun, cyclin D1, c-fos, and caspase 3, whose mRNAs are regulated during CGN PCD (10).
Because transcription can be an obligate component of neuronal PCD, we took advantage of cDNA array technology to broadly characterize mRNA expression during time courses of transcription-dependent and -independent models of CGN PCD. As has been elegantly demonstrated for basic cellular processes such as cell-cycle regulation (11), our goal was to identify temporally coupled genes that may delineate functional pathways coordinately mobilized by neuronal PCD. Our directed approach used bioinformatics to specifically design an array representing ≈7,600 rat genes appropriate for the study of neuronal apoptosis. Expression was monitored after combined K+ and serum-withdrawal (KS-W), K+-withdrawal alone (K-W), and kainic acid (KA) treatment. The former two induce PCD that is actinomycin D (actD)-sensitive, whereas the latter induces toxicity that is actD-insensitive. These models involve distinct and overlapping signaling mechanisms, including those mediated by neurotrophic factors, by death factors, by cAMP, and by calcium (9, 12–14). Regulated genes were characterized by time course expression pattern and by overlap between the various models. Our results indicated that known pro- and anti-apoptotic proteins were coordinately regulated, including transcription factors, receptors, Bcl-2 family members, and caspases.
Materials and Methods
blast Sequence Comparison Analysis.
Expressed sequence tags (ESTs) determined for the 5′ end of cDNA clones picked from two libraries [rat frontal cortex (8,304 clones) and nerve growth factor (NGF)-deprived differentiated PC12 cells (5,680)] ranged from 100–1,000 nt and averaged 500 nt. To minimize redundancy, Basic Local Alignment Search Tool (blast; ref. 15) was used to identify sequence clusters consisting of contiguous matches. Of the 1,620 clusters and 5,779 singletons found, representative clones were chosen and 7,295 prepared for construction of Smart Chip. By comparison to the public databases, ≈70% of Smart Chip appeared to be unannotated ESTs. Of the controls, 289 included genes with known function in the central nervous system, housekeeping genes, and negative hybridization controls such as vector, poly(A), or Cot sequences.
cDNA Array Construction.
Bacterial cultures of the 7,295 individual EST clones from the two libraries were consolidated by using a Genesis RSP 150 robotic sample processor (Tecan AG, Hombrechtikon, Switzerland). A subset of 202 chosen randomly for sequence validation all matched their seed sequence implicating 100% fidelity in tracking. To prepare templates for array elements, oligonucleotide primers flanking the cloning site were used to amplify by PCR the cDNA insert using KlenTaq1 DNA polymerase (Ab Peptides, St. Louis). Following ethanol precipitation, concentration (to 1–10 mg/ml), and resuspension in 3 × SSC (1 × SSC: 150 mM sodium chloride, 15 mM sodium citrate, pH 7.0), 20 nl from each template was arrayed onto nylon filters (Biodyne B, Life Technologies, Rockville, MD) at a density of ≈64/cm2 by using a 96-well format pin robot (CRS Robotics, Burlington, ON, Canada). After the filters were dry, the arrayed DNA was denatured in 0.4 M sodium hydroxide, neutralized in 0.1 M Tris⋅HCl, pH 7.5, rinsed in 2 × SSC, and dried to completion.
Array Hybridization.
Rat poly(A)+ RNA was purchased from CLONTECH or was isolated as total RNA from cultured CGNs by using RNA STAT-60 (Tel-Test, Friendswood, TX) and prepared by using Oligotex (Qiagen, Chatsworth, CA). mRNA (1 μg) and oligo(dT)30 (1 μg) were incubated with SuperScript II (Life Technologies) at 50°C for 30 m in the presence of 0.5 mM each deoxynucleotide dATP, dGTP, and dTTP, and 100 μCi (1 Ci = 37 GBq) [α-33P]dCTP (2,000–4,000 Ci/mmol; NEN). After purification over CHROMA SPIN+TE-30 columns (CLONTECH), the labeled cDNA was annealed at 65°C for 1 h with 10 μg poly(dA)>200 (Amersham Pharmacia) and 10 μg rat Cot 10 DNA (16). At 2 × 106 cpm/ml, the annealed cDNA mixture was added to array filters in preannealing solution containing 100 mg/ml sheared salmon sperm DNA in 7% SDS, 0.25 M sodium phosphate, 1 mM EDTA, and 10% formamide. Following overnight hybridization at 65°C, the filters were washed twice for 15 m at 65°C in 2 × SSC, 1% SDS, twice for 30 m at 65°C in 0.2 × SSC, 0.5% SDS, and twice for 15 m at 22°C in 2 × SSC. Dried filters were exposed to phosphoimage screens for 60 h. Radioactive hybridization signals captured by a Fuji BAS 2500 phosphoimager (Fuji Medical Systems, Stamford, CT) were quantified by using array vision software (Imaging Research, St. Catherine's, ON, Canada). Array hybridizations performed in triplicate were highly reproducible (see Fig. 3, which is published as supplemental data on the PNAS web site, www.pnas.org) as reflected by the coefficient of variation (CV; standard deviation/mean) averaging less than 0.2 for genes whose intensities were above a detection threshold (of less than 1 in 100,000). The dynamic range of detection spanned three orders of magnitude. We validated apparent mRNA regulation by semiquantitative reverse transcription–PCR; of 51 Smart Chip genes tested, 41 were regulated as predicted (data not shown).
CGN Cell Culture.
CGNs prepared from 7-day-old rat pups were plated into basal medium Eagle (BME; Life Technologies) supplemented with 25 mM KCl, 10% dialyzed FBS (Summit Biotechnology, Ft. Collins, CO), 100 units/ml penicillin, and 100 μg/ml streptomycin (17). Aphidicolin (Sigma), added to the cultures 24 h after initial plating, reduced GFAP positive cells to <2% of the total. After 7 days in culture, medium was switched to 5 mM KCl, no serum, resulting in 60% cell death by 24 h postwithdrawal that was completely prevented by actD at 2 μg/ml. A gene expression response by serum-add-back was observed when the medium was replaced with nonconditioned BME, 25 mM KCl, and 10% serum (supplemental data, Fig. 4). For K-W and KA treatment, the cells were switched on day 2 to neurobasal medium (Life Technologies) supplemented with 25 mM KCl, 0.5% dialyzed FBS, B27 supplement (Life Technologies), 0.5 mM l-glutamine (Life Technologies), 0.1 mg/ml AlbuMAX I (Life Technologies), penicillin, streptomycin, and aphidicolin, as above. On day 7, K-W was initiated by switching to 5 mM KCl. KA treatment involved 30 m of 5 mM KCl, 100 μM kainic acid (Sigma) in sodium-free Locke's buffer (18).
Transcription Profiling Data Analysis.
For replicate hybridizations, the distribution of intensities across rat genes was normalized to a median of 100 and then averaged. The detection threshold for each experiment was found by graphing the average coefficient of variation (CV) versus mean gene expression intensity and was defined as the intensity at which lower intensities exhibited an average CV greater than 0.3. For most experiments, this ranged from 10 to 40, and the number of Smart Chip genes detected above threshold was 70% to 95%.
Expression Data Clustering Algorithms.
Stringent standards were used for choosing significantly regulated genes to minimize poor cluster recovery due to noise in the data. After averaging and normalization, the maximum (max) and minimum (min) intensity detected for each gene through the time course were used to define the following criteria: (i) detection, max intensity > threshold; (ii) noise filter, the difference between max and min intensity > threshold; and (iii) regulation, at least 3-fold induction between max and min intensity. By the above criteria, >90% of the replicate measurements varied by <30%. Therefore, a ratio of 3-fold was predicted to actually be within an interval of 2- to 5-fold with >99% confidence, even for gene expression intensities at or near threshold. Hierarchical clustering was performed by using Euclidean distance and the average linkage algorithm (19). matlab software (Mathworks, Natick, MA) was used to perform clustering and generate dendrograms and tiled gene expression figures. The functions of the regulated ESTs were deduced from the annotations of the top blast hits to known genes in the public databases.
Results
Construction of a Brain-Biased and PCD-Enriched cDNA Array.
Because a gene expression component has been implicated in multiple models of neuronal PCD and neurodegeneration, one aim of the present study was to optimize cDNA array technology for transcription profiling of the nervous system. We arrayed DNA elements of nonredundant ESTs selected from cDNA libraries made from the rat frontal cortex and NGF-deprived differentiated PC12 cells (20). The rationale was to enrich for genes expressed in the brain, and up-regulated during one model of neuronal PCD.
To characterize our array, which we designated Smart Chip, and to identify brain-specific genes, we hybridized cDNA prepared from ten different normal rat tissues. As expected, radiolabeled brain cDNA hybridized more intensely to most of the array elements compared with the other tissues (supplemental data, Fig. 5). In fact, 582 genes appeared to be brain-specific as defined by detection above threshold for brain but below threshold for any of the other tissues (supplemental data, Table 3). Relative tissue distribution predicted by array hybridization was confirmed for over 50 genes by Northern analysis (data not shown). Having demonstrated Smart Chip brain-bias, we expected that it would be a very useful tool for our neuronal apoptosis studies, because it was enriched for genes expressed in the brain and/or up-regulated by at least one model of PCD.
Expression Profiling of CGN Potassium and Serum-Withdrawal Reveals Several Distinct Waves of Gene Expression.
By using Smart Chip, we profiled mRNA expression during a time course of PCD in primary cultures of CGNs induced by KS-W. By profiling multiple time points we intended to group regulated genes by their temporal expression pattern and identify functional relationships between genes within each group.
We observed a large effect of KS-W on relative mRNA levels. CGNs cultured in high K+ and high serum were switched to low K+ and no serum eliciting an apoptotic mode of cell death resulting in approximately 60% cell loss by 24 h. As shown by others (6, 8), the observed PCD was completely inhibited by actD, demonstrating an obligate transcriptional component. Of 6,818 genes detected, 790 were regulated at least 3-fold over the time course of KS-W. A majority of those (556) were also regulated by medium change alone (supplemental data, Fig. 4) and were most likely induced by a residual serum component in the nonconditioned add-back medium. We decided to analyze these serum-responsive genes as part of an independent treatment, serum-add-back (SA), which did not elicit cell death.
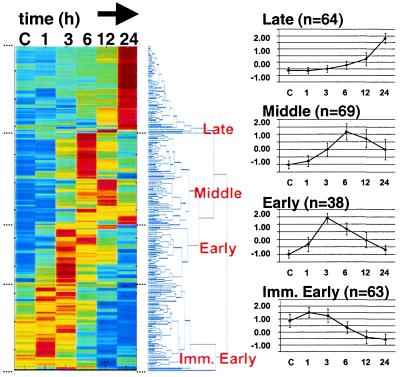
Two hundred thirty-four genes were regulated greater than 3-fold by KS-W and were not regulated by SA. By using hierarchical clustering algorithms, we ordered the regulated genes based on their expression pattern (Fig. 1). Genes segregating into four major branches of the dendrogram were assigned to temporal classes that we designated immediate early (peaking at 1 h followed by decay through 24 h), early (peaking at 3–6 h), middle (peaking at 6–12 h), and late (up-regulated at 24 h).
Figure 1.
Temporal expression clusters of genes regulated by potassium and serum-withdrawal. (Left) Hierarchical clustering was used to order 234 significantly regulated genes. Each colored row represents an expression profile for an individual gene. From left to right, control (C), 1, 3, 6, 12, and 24 h post-KS-W expression values for each gene were scaled based on the number of standard deviations from the mean intensity of each gene. Scaled expression values were color-coded such that red, yellow, and blue indicate above, at, and below mean intensity, respectively. The distance between branches in the dendrogram is proportional to the relatedness between expression patterns based on a Euclidean metric; neighboring genes are most alike. (Right) For the four major branches (Late, Middle, Early, and Immediate Early) we graphed the average profile (n = total number for each class) ± the standard deviation. Supplemental data, Table 4 provides the gene expression data.
In the immediate-early class, the majority of known genes encoded proteins involved in regulating secretion and synaptic vesicle release, including synaptotagmin, synaphin, calcium calmodulin-dependent kinase II, synapsin, and fodrin (Table 1). One possible explanation for initial up-regulation of this class of genes may have been an attempt by the neurons to maintain signaling capacity after cessation of spontaneous activity.
Table 1.
Known genes regulated by potassium and serum-withdrawal
Expression class identified based on hierarchical clustering (Fig. 1). Following KS-W, immediate early, early, middle, and late genes peaked at 1, 3–6, 6–12, and 24 h, respectively. A few down-regulated genes were not segregated from immediate-early genes because they were difficult to distinguish. Supplemental data, Table 4 provides the gene expression data.
Functional class of gene identified by blast as determined from the Medline annotation for each sequence with indicated accession no.
Accession nos.
Histones 1, 2A, and 3 fell in the early class, as did several mRNAs previously described as regulated by PCD such as mud-2 and apoptosis-associated tyrosine kinase. Interestingly, the constitutively expressed and tandemly repeated arrays of histone genes in the genome have been thought to be up-regulated 3- to 5-fold during the first 2 h following initiation of DNA synthesis during S-phase of the cell cycle (21). Up-regulation of histones early after KS-W was therefore consistent with a proposed model that neuronal PCD may be a consequence of abortive entry into the cell cycle (22). Recently, histone H1 has been shown to interact directly with and stimulate the activity of DNA-fragmentation factor (DFF), a known substrate of caspase 3 (23). Therefore, significant up-regulation of histone H1 may contribute to PCD-induced DNA damage by affecting DFF nuclease activity.
The middle temporal class included genes induced by PCD or stress, such as caspase 3 and the protein phosphatase Wip-1, as well as known death substrates such as PITSLRE. In addition, transcript levels peaked at 6–12 h for several genes identical to or similar to genes encoding mitochondrial functions such as cytochrome c oxidase and NADH dehydrogenase subunit 4. Two previous large-scale expression analyses of nonneuronal models of radiation- or p53-induced apoptosis in cancer cell lines also reported that genes encoding mitochondrial proteins and/or genes regulating oxidative stress and the generation of reactive oxygen species were targets of mRNA regulation (24, 25).
Because 40% of the cells remained alive at 24 h, genes in the late class could have been effectors of cell maturation or survival. For example, proteins involved in cell adhesion (such as SPARC, phosphacan, or GRASP) and in inhibitory neurotransmission (such as GAD, GABAA receptor, or the GABA transporter) may be markers of maturing CGNs. Other proteins involved in protein stabilization, such as the cysteine proteinase inhibitor cystatin C or elongation factor 1-gamma, may contribute to an anti-apoptotic status of the cell. Overlap between temporal regulation of pro-apoptotic and survival/cell maturation genes during CGN PCD may reflect heterogeneous population of cells committed to die or to survive, activating different gene expression programs in response to KS-W. Some of the late transcripts (such as P0, S100 β, nexin-1, or vimentin) may be markers of nonneuronal cells that were present proportionately higher in the overall cell population at later time points, when many of the neurons have died.
A Restricted Set of Known PCD Genes are Regulated by Multiple Models of CGN PCD.
We next profiled CGN PCD induced by the non-NMDA glutamate receptor agonist kainic acid. KA exposure of CGNs elicited profound cell death (80% by 24 h) that was completely insensitive to actD treatment. To minimize the effect of serum on the sham and treated samples, we also examined the effect of K-W on gene expression in the presence of low serum (0.5%). In conditioned neurobasal medium, the time points for sham K-W and KA treatment were not significantly perturbed by medium add-back (supplemental data, Fig. 4) and were, therefore, averaged and designated control.
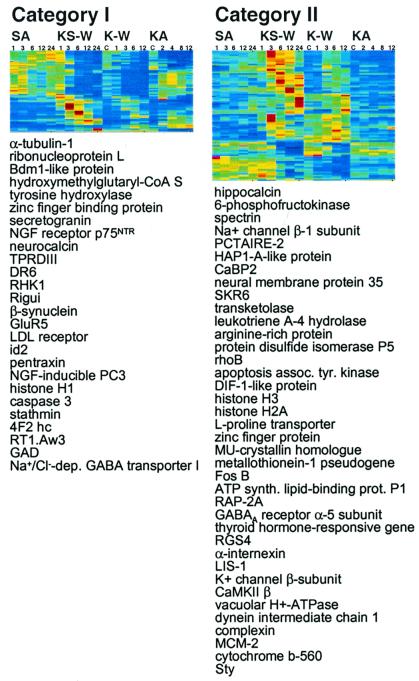
Changes in gene expression were less robust in cells cultured on neurobasal medium. Of the 6,974 genes profiled in all of the experiments, the number of genes detected above threshold was similar for K-W (6,243), KA treatment (5,828), and KS-W (5,808). However, the number of genes regulated by at least 3-fold during K-W and KA treatment was only 83 and 36, respectively, compared with 790 for KS-W. Probably because of the small number of genes regulated and the decreased magnitude of fold-differences, clustering analysis, which is adversely affected by noise in the data (26), did not reveal obvious associations between genes regulated by K-W or KA (data not shown). However, by comparing between treatments, we found that pro-apoptotic genes, such as caspase 3, death receptor DR6, and the NGF receptor p75NTR, were regulated by all three models (Fig. 2, Category I), suggesting that a feed forward mechanism may increase mRNA levels of pro-apoptotic gene products. Although the magnitude of differential expression was less during K-W, we observed that genes regulated by both KS-W and K-W exhibited similar kinetics (Fig. 2, Category II), suggesting that the overlap genes were regulated by a common mechanism. Conclusions based on direct comparisons between KS-W and K-W were difficult to make because the culturing conditions were different, but the results suggest that serum had a large additive effect on the gene regulation. These results are consistent with a previously proposed model based on the kinetics of cell death (6), that two populations of neurons coexist, one responsive to withdrawal of serum-components (likely IGF) and the other to potassium.
Figure 2.
Genes regulated by neuronal programmed cell death. 885 regulated genes were clustered (gene expression data, supplemental data, Table 5). Two classes are shown. Treatments and time points: SA 1, 3, 6, 12, and 24 h; KS-W 1, 3, 6, 12, and 24 h; K-W C, 1, 3, 6, and 12 h; KA treatment C, 2, 4, 8, and 12 h. Category I genes were regulated by all treatments eliciting PCD (KS-W, K-W, and KA). Category II genes were regulated by actD-sensitive models (KS-W and K-W), but not by actD-insensitive KA treatment.
In addition to focusing our analysis on regulated genes, we wished to determine the expression pattern of all genes on Smart Chip encoding putative PCD functions, including those that may be constitutive. One hundred twenty-four putative PCD or stress-response genes were identified by querying the top blast annotations of the ESTs for known PCD genes, or by comparing the sequence of known PCD genes to the ESTs on Smart Chip.
Most of the putative PCD genes on Smart Chip were constitutive (Table 2), which is consistent with apoptosis as a general component of the cellular repertoire. However, a subset of known PCD genes were clearly regulated (<3-fold cut-off used in initial analysis above). For example, Hsp 70 was regulated by KA treatment. In addition to caspase 3, DR6, and p75NTR, the IGF-II receptor and NGF-inducible anti-proliferative protein PC3 were regulated by neuronal PCD induced by all three models. Anti-apoptotic Bcl-2 was regulated by the actD-sensitive K+-withdrawal models. Because CGN K+-withdrawal is a model for postnatal cerebellar development, a gene expression component may integrate both cell death and survival signaling to provide the molecular machinery necessary for mediating the ultimate cell fate versus cell death decision. Different relative levels of the transcripts for caspase 3 and Bcl-2 in individual neurons would likely alter the balance of downstream PCD signaling events post KCl-withdrawal.
Table 2.
Expression classes of known cell death-related genes on Smart Chip
Expression class identified: Regulated by KS-W, by K-W, by KA treatment, by SA, by serum—i.e., up-regulated by SA and down- regulated by KS-W (Serum Up-reg.) or vice versa (Serum Down-reg.)—constitutive, constitutive with basal expression different between Eagle's and neurobasal medium as indicated, and not detected (ND) or hybridization artifact.
The programmed cell death gene identified as most similar to the array element EST by annotation or by blast to sequence with indicated accession no. Smart Chip ESTs were identified in each cluster with greater than 90–100% identity to the known rat gene, or 85–100% identity to the known human or mouse gene. Supplemental data, Table 6 provides the gene expression data.
Accession nos.
Among the constitutive PCD genes, some exhibited different basal levels of expression depending on the culture medium. For example, caspases 6 and 10, IAP-1 and -2, NAIP, XIAP, and survivin (along with other PCD-associated genes) were not regulated by PCD-induction, but their basal expression level was low in Eagle's, and high in neurobasal medium. The differential effect of culture conditions on basal expression levels of stress/PCD genes was also consistent with a model in which the susceptibility of the neuron to death or survival signals may depend on the state of global mRNA expression.
Overall, the regulation of multiple genes including caspase 3, c-fos, c-jun, mud-2, Wip-1, and mitochondrial proteins was remarkably consistent with previous reports in the literature of transcription regulation during CGN PCD and/or other models. This coincidence suggests that our unselected approach of arraying genes from two relevant EST libraries successfully enriched for transcripts involved in neuronal apoptosis. Notably missing from our library screen were a few genes, such as cyclin D1 and Fas ligand, previously shown to be regulated by CGN PCD (10, 27).
Discussion
RNA synthesis has been shown to be required in many models of neuronal apoptosis and neurodegeneration. However, limited progress has been made in identifying the constituents and understanding the mechanism of action of this transcription requirement. In this study, comprehensive profiling with a brain-biased array delineated an orchestrated transcription component of CGN PCD consisting of immediate early, early, middle, and late temporal classes that correlated with functional aspects of survival or cell death. These included synaptic vesicle release, histone biosynthesis, mitochondrial functions, PCD/stress-response signaling, and cell maturation. Simultaneous regulation of both survival and cell-death genes suggested that subsets of neurons in the population were tuned to respond divergently to KS-W treatment.
As with known protein signaling mechanisms, RNA expression appeared to be specifically and coordinately regulated, implicating the importance of gene expression during PCD. The overall rate of RNA synthesis has been shown to decrease after CGN KS-W (6). However, most of the differential expression in our study appeared to consist mainly of genes induced rather than repressed relative to overall RNA levels, underscoring the importance of these transcripts to the physiology of cell death and survival.
Among the discreet gene expression waves observed in vitro, an unexpected observation was that an immediate early response encoded for many synaptic vesicle proteins, perhaps as a response to reduced neuronal signaling capacity. The transcription factor CREB, implicated in both plasticity and cell-death signaling (28), is an attractive candidate for the coordinate regulation of this group of genes. Other candidate factors likely involved in the observed orchestrated transcription include Fos and Jun (29). Both Fos and Jun are members of a family of AP-1 transcription factors that, in addition to other target genes, regulate their own transcripts. c-fos and c-jun regulation diverged in our experiments. c-fos was up-regulated by all treatments, including sham medium replacements, whereas the c-jun response coincided with serum presence in the medium change (Table 2). Other transcription factors (such as Forkhead, NFκB, and Brn-3a) required for neuronal PCD (27, 30, 31) might also be expected to activate subsets of the expression waves observed.
Given the tractability and accumulated physiological data for the CGN PCD model system, we will be able to investigate the mechanism of action of identified target genes in CGN PCD and survival pathways. In fact, several ESTs identified in this study as coregulated with pro-apoptotic genes encode for proteins of unknown function (data not shown). The focus of ongoing research includes full-length cloning and expression studies to determine the potential function of these genes in neuronal PCD. The mechanistic role of transcription and signaling is being investigated by overexpressing and/or down-regulating candidate transcription factors and, then, comparing the overlap and kinetics of downstream target expression to the patterns observed in this study. Experimentally observed expression waves that functionally segregate with specific transcription factors may be reconciled by regulatory sequences present within the target genes. Intervention with pharmacological reagents will identify the signal transduction pathways required to activate transcription factors mediating specific expression waves. As our database of Smart Chip profiles increases, signature profiles for different pathways will be defined and compared with other apoptosis models. Ultimately, they may serve as markers of the predominant cellular mechanisms of survival and PCD that may be operational in vivo during neurodegeneration.
Supplementary Material
Acknowledgments
We thank Keith Robison for blast analysis; Stan Smith for RNA preparations; and Millennium core groups, cDNA libraries, sequencing, process technology, informatics, and TRACE for library construction, EST sequencing, data acquisition, software, and array production. We are grateful to John Bertin, Anjen Chenn, Brad Geddes, Michael Jacobson, and Tom Novak for helpful comments on the manuscript.
Abbreviations
- PCD
programmed cell death
- CGN
cerebellar granule neuron
- KS-W
potassium and serum withdrawal
- K-W
potassium-withdrawal alone
- KA
kainic acid
- SA
serum-add-back
- actD
actinomycin D
- NGF
nerve growth factor
- EST
expressed sequence tag
References
- 1.Pettmann B, Henderson C E. Neuron. 1998;20:633–747. doi: 10.1016/s0896-6273(00)81004-1. [DOI] [PubMed] [Google Scholar]
- 2.Hetts S W. J Am Med Assoc. 1998;279:300–307. doi: 10.1001/jama.279.4.300. [DOI] [PubMed] [Google Scholar]
- 3.Raff M C, Barres B A, Burne J F, Coles H S, Ishizaki Y, Jacobson M D. Science. 1993;262:695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 4.Dragunow M, Preston K. Brain Res Rev. 1995;21:1–28. doi: 10.1016/0165-0173(95)00003-l. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman R J. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 6.Miller T M, Johnson E M., Jr J Neurosci. 1996;16:7487–7495. doi: 10.1523/JNEUROSCI.16-23-07487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Mello S R, Galli C, Ciotti T, Calissano P. Proc Natl Acad Sci USA. 1993;90:10989–10993. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz J B, Weller M, Klockgether T. J Neurosci. 1996;16:4696–4706. doi: 10.1523/JNEUROSCI.16-15-04696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galli C, Meucci O, Scorziello A, Werge T M, Calissano P, Schettini G. J Neurosci. 1995;15:1172–1179. doi: 10.1523/JNEUROSCI.15-02-01172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller T M, Moulder K L, Knudson C M, Creedon D J, Deshmukh M, Korsmeyer S J, Johnson E M., Jr J Cell Biol. 1997;139:205–217. doi: 10.1083/jcb.139.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;95:14863–14868. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato K, Puttfarcken P S, Lyons W E, Coyle J T. J Pharmacol Exp Ther. 1991;256:402–411. [PubMed] [Google Scholar]
- 13.Le-Niculescu H, Bonfoco E, Kasuya Y, Claret F-X, Green D R, Karin M. Mol Cell Biol. 1999;19:751–763. doi: 10.1128/mcb.19.1.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Mello S R, Borodezt K, Soltoff S P. J Neurosci. 1997;17:1548–1560. doi: 10.1523/JNEUROSCI.17-05-01548.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Britten R J, Graham D E, Neufeld B R. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- 17.Miller T M, Johnson E M., Jr J Neurosci. 1996;16:7487–7495. doi: 10.1523/JNEUROSCI.16-23-07487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonian N A, Getz R L, Leveque J C, Konradi C, Coyle J T. Neuroscience. 1996;74:675–683. doi: 10.1016/0306-4522(96)00141-8. [DOI] [PubMed] [Google Scholar]
- 19.Everitt B S. Cluster Analysis. New York: Oxford Univ. Press; 1993. [Google Scholar]
- 20.Rukenstein A, Rydel R E, Greene L A. J Neurosci. 1991;11:2552–2563. doi: 10.1523/JNEUROSCI.11-08-02552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein G S, Stein J L, Lian J B, van Wijnen A J, Wright K L, Pauli U. Cell Biophys. 1989;15:210–223. doi: 10.1007/BF02989684. [DOI] [PubMed] [Google Scholar]
- 22.Evan G I, Brown L, Whyte M, Harrington E. Curr Opin Cell Biol. 1995;7:825–834. doi: 10.1016/0955-0674(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Zou H, Widlak P, Garrard W, Wang X. J Biol Chem. 1999;274:13836–13840. doi: 10.1074/jbc.274.20.13836. [DOI] [PubMed] [Google Scholar]
- 24.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 25.Voehringer D W, Hirschberg D L, Xiao J, Lu Q, Roederer M, Lock C B, Herzenberg L A, Steinman L, Herzenberg L A. Proc Natl Acad Sci USA. 2000;97:2680–2685. doi: 10.1073/pnas.97.6.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milligan G W. In: Clustering and Classification. Arabie P, Hubert L J, De Soete G, editors. Singapore: World Scientific Publishing Co.; 1996. pp. 341–375. [Google Scholar]
- 27.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 28.Riccio A, Ahn S, Davenpoart C M, Blendy J A, Ginty D D. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 29.Watson A, Eilers A, Lallemand D, Kyriakis J, Rubin L L, Ham J. J Neurosci. 1998;18:751–762. doi: 10.1523/JNEUROSCI.18-02-00751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamatani M, Che Y H, Matsuzaki H, Ogawa S, Okado H, Miyake S-i, Mizuon T, Tohyama M. J Biol Chem. 1999;274:8531–8538. doi: 10.1074/jbc.274.13.8531. [DOI] [PubMed] [Google Scholar]
- 31.Latchman D S. Int J Biochem Cell Biol. 1998;30:1153–1157. doi: 10.1016/s1357-2725(98)00090-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.