Abstract
Tumor suppression by p53 and BRCA1 involves regulation of cell cycle, apoptosis, and DNA repair and is influenced by transcriptional coactivators and post-translational modifications. Here we show that coactivator-associated arginine methyltransferase 1 (CARM1) methylates Arg 754 in the KIX region of coactivator p300. Methylated p300 and p300 protein fragments are preferentially recognized by BRCT domains of BRCA1, identifying the BRCT domain as a novel methylarginine-binding module. CARM1 and p300 cooperate with BRCA1 and p53 to induce expression of the critical cell cycle and proliferation regulator p21WAF1/CIP1 in response to DNA damage. This induction was severely attenuated by elimination of CARM1 or its methyltransferase activity, or by mutation of Arg 754 of p300. Absence of CARM1 methyltransferase activity led to failure of cells to arrest in the G1 phase of the cell cycle in response to DNA damage. CARM1 methyltransferase activity was required for induction of some p53 target genes (p21 and Gadd45) but not others (Bax) by DNA damage. Recruitment of BRCA1 to the p53-binding region of the p21 promoter in response to DNA damage required methylation of Arg 754 of p300 by CARM1. Thus, coactivator methylation may be crucial for fine-tuning the tumor suppressor function of BRCA1 and other BRCT domain proteins.
Keywords: BRCA1, p300, CARM1, transcriptional coactivator, DNA damage, protein methylation
Protein methylation on Arg and Lys residues is an abundant protein modification that modulates many cellular functions, including protein–protein interaction, transcription, RNA processing, protein trafficking, DNA repair, and signal transduction (Stallcup 2001; Bedford and Richard 2005; DY Lee et al. 2005; Paik et al. 2007). One of the 10 currently known mammalian protein arginine methyltransferases, coactivator-associated arginine methyltransferase 1 (CARM1, also known as protein arginine methyltransferase 4), is important for embryonic development and the pluripotency of embryonic cells (Yadav et al. 2003; Torres-Padilla et al. 2007). As a coactivator, CARM1 contributes to transcriptional regulation by methylation of histone H3 and several other proteins, including p300, cyclic AMP response element-binding protein (CREB)-binding protein (CBP), and steroid receptor coactivator-3 (SRC-3) (Stallcup 2001; Bedford and Richard 2005; Lee and Stallcup 2009). p300 and CBP are related proteins with distinct functions as important coactivators for many DNA-binding transcription factors, and the severe but distinct phenotypes of heterozygous mice lacking one allele of either p300 or CBP indicate their involvement in critical physiological processes (Goodman and Smolik 2000; Vo and Goodman 2001). In addition to their histone acetyltransferase activity, p300 and CBP contain several protein–protein interaction domains, including nuclear receptor-binding domains, KIX or CREB-binding domains, bromo domains, three cysteine/histidine-rich or zinc finger-like domains, and one glutamine-rich domain; each domain interacts with many different DNA-binding transcription factors and coactivators (Goodman and Smolik 2000; Vo and Goodman 2001). p300 and CBP function synergistically with CARM1 as coactivators for several DNA-binding transcription factors, including nuclear receptors, p53, NFκB, and β-catenin/LEF1 (Koh et al. 2002; Lee et al. 2002; An et al. 2004; Covic et al. 2005). The requirement for the CARM1 methyltransferase activity in its cooperative coactivator function with p300 and CBP led to the investigation of p300 and CBP as substrates for CARM1.
Three independent groups documented different sites where p300 or CBP is methylated by CARM1 (Xu et al. 2001; Chevillard-Briet et al. 2002; YH Lee et al. 2005). Xu et al. (2001) reported methylation of multiple Arg residues within amino acids 582–672 of the KIX or CREB-binding region of CBP (and an overlapping larger region of p300), and found that methylation of the KIX domain prevented its binding to and coactivator function for CREB. Chevillard-Briet et al. (2002) reported methylation of CBP at three sites in a region (amino acids 685–774) located just C-terminal to the sites described above. They proposed that these methyl modifications may contribute to the synergistic coactivator function of CBP, CARM1, and the p160 coactivators with nuclear receptors. YH Lee et al. (2005) showed that methylation of Arg 2142 of human p300 inhibited the binding of p300 to the p160 nuclear receptor coactivator GRIP1, and thus may regulate the assembly or disassembly of the coactivator complex on the promoter.
Here we identify Arg 754 as the major arginine methylation site in the C-terminal extension of the KIX domain of p300. Arg 754 corresponds to one of the three methylation sites previously reported in CBP (Chevillard-Briet et al. 2002). We show that methylation of Arg 754 leads to enhanced interaction of the p300 KIX region with the C-terminal BRCT domain of BRCA1, an essential tumor suppressor in breast cancers and ovarian cancers, and with the BRCT domains of some other proteins. We also present evidence that CARM1 is required for DNA damage-induced expression of an important cell cycle regulator (p21WAF1/CIP1) and the DNA excision repair factor Gadd45; moreover, CARM1-mediated methylation of p300 is essential for the recruitment of BRCA1 to the p21 gene, which is a known target of BRCA1 coactivator function. These results further suggest that methylation of p300 by CARM1 is important for the tumor suppressor function of BRCA1 and possibly the functions of other BRCT family proteins.
Results
Arg 754 is the major site of methylation of p300 KIX by CARM1
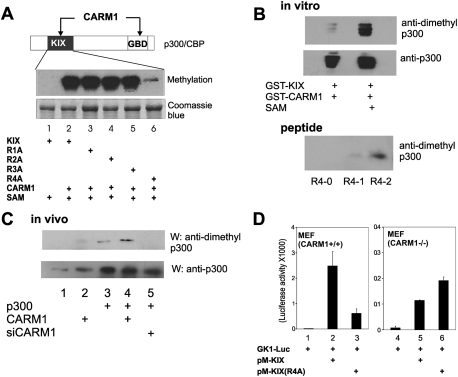
An extended KIX domain fragment of p300 (amino acids 568–828) was methylated in vitro by CARM1, and the methylated region was subsequently localized to the C-terminal part of that fragment (amino acids 669–828) (data not shown). Each of the four Arg residues (R1 = Arg 695, R2 = Arg 705, R3 = Arg 728, R4 = Arg 754) in this region (amino acids 669–828) was independently changed to Ala, and the effect of each mutation on the methylation of the p300(568–828) fragment by CARM1 was tested in vitro (Fig. 1A). The wild-type fragment and the R1, R2, and R3 mutants were methylated to similar extents, but the R4 mutation almost completely eliminated the methylation. Thus, R4 (Arg 754) is the major CARM1-mediated methylation site of the extended p300 KIX region. The low residual level of methylation of the p300 R4A mutant suggests that other arginine residues may also be methylated to a lesser extent.
Figure 1.
Arg 754 of p300 is the major CARM1 methylation site in the KIX region. (A) Protein methylation assay. The diagram shows that both the GRIP1-binding domain (GBD) (YH Lee et al. 2005) and KIX region of p300 are methylated by CARM1. GST fusion proteins of the human p300 KIX region (amino acids 568–828) were prepared with wild-type sequence (KIX) or four different Arg-to-Ala mutations (R1A, R695A; R2A, R705A; R3A, R728A; R4A, R754A). One microgram of purified GST-KIX protein was coincubated with GST-CARM1 in the presence of 3H-labeled S-adenosylmethionine (SAM). (Top panel) Methylation products were analyzed by SDS-PAGE and autoradiography. (Bottom panel) Coomassie blue staining shows that similar protein amounts of GST-KIX proteins were used. (B) Preferential recognition of methylated p300 fragments and peptides by anti-dimethyl-p300 antiserum. (Top panels) GST-KIX and GST-CARM1 were incubated in the absence or presence of unlabeled S-adenosylmethionine (SAM) in methylation reactions, as in A. Reaction products were resolved by SDS-PAGE and immunoblot, using antibodies against asymmetric dimethyl-R4 peptide or p300. (Bottom panel) R4 peptides containing unmethylated (R4-0), monomethylated (R4-1), or asymmetrically dimethylated (R4-2) Arg 754 were analyzed by SDS-PAGE and immunoblot with antiserum against asymmetric dimethyl-R4 peptide. Peptide concentrations were verified by absorbance readings of high-pressure liquid chromatography eluates (data not shown). (C) Methylation of Arg 754 of p300 by CARM1 in vivo. 293T cells in six-well plates were transfected with combinations of 200 ng of pCMV-p300, 250 ng of pSG5.HA-CARM1, and 50 pmol of duplex siRNA against CARM1, as indicated. Immunoblot was performed with antibodies against asymmetric dimethyl-R4 peptide or p300. The results shown are from a single experiment that is representative of five independent experiments. See also Supplemental Figure S1. (D) Transcriptional activation by the wild-type and mutant KIX region in MEF(CARM1+/+) and MEF(CARM1−/−) cells. GK1-Luc reporter plasmid (0.1 μg) was transfected with 0.1 μg of pM-KIX (encoding p300 KIX amino acids 568–828 fused to Gal4 DBD) or pM-KIX(R4A) into MEF(CARM1+/+) or MEF(CARM1−/−) cells, and luciferase activity in the cell extracts was determined. The results shown are from a single experiment that is representative of five independent experiments.
To test for methylation in vivo, antibodies were developed against a 19-amino-acid peptide representing the R4 region and containing asymmetric dimethylarginine (the product of CARM1 methylation) at the R4 (Arg 754) position. In immunoblots, the antiserum preferentially recognized recombinant GST-KIX that had been methylated by CARM1, compared with unmethylated GST-KIX (Fig. 1B, top panels). It also interacted preferentially with the R4 peptide containing asymmetric dimethylarginine, compared with the unmethylated R4 peptide or the R4 peptide containing monomethylarginine (Fig. 1B, bottom panel). Having demonstrated appropriate specificity in vitro, the antibody was tested in immunoblots with cell extracts containing varying levels of p300 and CARM1. In extracts containing endogenous CARM1, the antiserum produced an enhanced signal when p300 was overexpressed (Fig. 1C, cf. lanes 1 and 3). Moreover, in extracts containing overexpressed p300, overexpression of CARM1 enhanced the signal (Fig. 1C, cf. lanes 3 and 4), while reduction of endogenous CARM1 by siRNA transfection decreased the signal (Fig. 1C, lanes 3,5; Supplemental Fig. S1). Since the signal from the anti-R4-dimethyl-p300 antibody varied according to the level of CARM1, while the level of total p300 remained constant, these results demonstrate that p300 is methylated in vivo by CARM1 at the R4 (Arg 754) position.
To assess the importance of R4 in transcription activation by the p300 KIX region (amino acids 568–828), the KIX region was fused to the Gal4 DNA-binding domain (DBD) and transiently transfected into cells with a Gal4-responsive reporter plasmid encoding luciferase. In wild-type mouse embryo fibroblast (MEF) cells, the wild-type KIX region activated the reporter gene much more strongly than the R4 mutant (Fig. 1D). In contrast, in CARM1−/− MEF cells, wild-type and mutant KIX fragments had approximately equal activity. Thus, CARM1 is important for transcriptional activation by the KIX region; the role of CARM1 depends on R4 and, based on the in vitro and in vivo methylation results, we proposed that it involves methylation of R4 by CARM1 and tested this further below.
R4 (Arg 754) of p300 KIX is important for interaction with the C-terminal activation domain of BRCA1
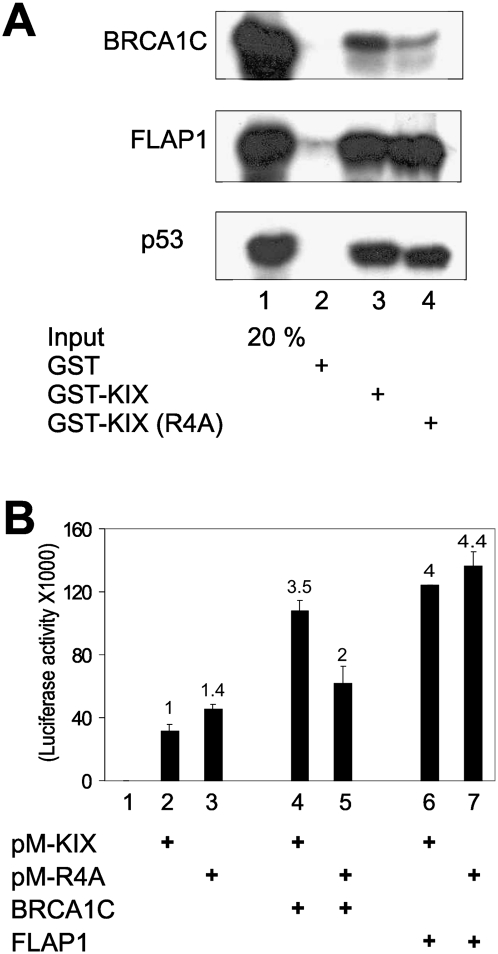
The p300 KIX region interacts with many transcription factors, including CREB, p53, FLAP1, and BRCA1 (Goodman and Smolik 2000; Vo and Goodman 2001; Lee and Stallcup 2006). The effect of the R4 mutation on these interactions was tested by GST pull-down experiments. The wild-type and R4A mutant KIX fragments bound equally to FLAP1 and p53 translated in vitro, but the weak binding of wild-type KIX to the C-terminal BRCT region of BRCA1 (BRCA1C) was rendered weaker by the R4 mutation (Fig. 2A). In an in vivo interaction assay using Gal4 DBD-KIX fusion proteins, transcriptional activation by wild-type KIX was enhanced 3.5-fold by coexpression of BRCA1C (amino acids 1528–1863), but the activity of the R4A KIX mutant was enhanced <50% by BRCA1C (Fig. 2B). In contrast, FLAP1 enhanced transcriptional activation by wild-type KIX and the R4A mutant to similar extents. Thus, R4 of p300 KIX is involved in the physical and functional interaction with the BRCA1 C terminus. Note that wild-type and mutant KIX regions had different relative activities in MEF cells (Fig. 1D) compared with 293T cells (Fig. 2B), suggesting cell type specificity in KIX action.
Figure 2.
Arg 754 (R4) of p300 KIX is important for interaction with the C-terminal (BRCT) domain of BRCA1. (A) GST pull-down assay. Sepharose bead-bound GST-KIX and GST-KIX(R4A) proteins were incubated with in vitro translated BRCA1C (BRCA1 amino acids 1599–1863), FLAP1, or p53. GST-bound proteins were analyzed by SDS-PAGE and autoradiography. Input levels were monitored by loading 20% of the in vitro translation product used in the binding assays (20% input). (B) Enhancement of p300 KIX activity by BRCA1C. GK1-Luc reporter plasmid (0.1 μg) was transfected into 293T cells along with 0.1 μg of pM-KIX or pM-KIX(R4A) and 0.1 μg of BRCA1C (amino acids 1528–1863) or FLAP1 expression vectors, and luciferase activity in the extracts was determined. The results shown are from a single experiment that is representative of four independent experiments.
Preferential interaction of the BRCT domain of BRCA1 with p300 protein and peptides containing methylated Arg 754
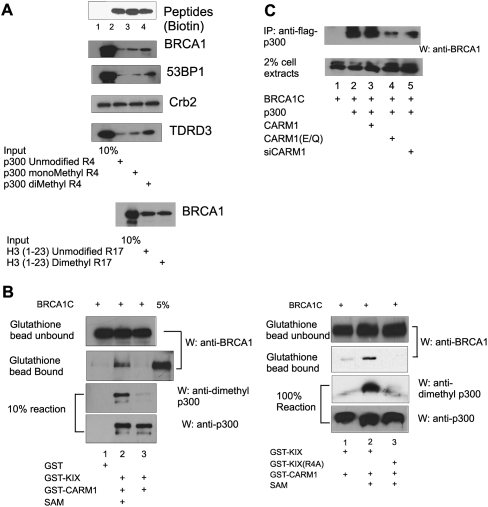
To test the effect of KIX methylation by CARM1 on binding to the BRCT domain of BRCA1, three 19-amino-acid peptides representing the Arg 754 (R4) region of KIX and containing unmethylated R4, monomethylated R4, or asymmetrically dimethylated R4 were synthesized with a biotin label at the N terminus. These peptides, captured on streptavidin-coupled beads, bound GST-BRCA1C poorly when R4 was unmodified, but bound BRCA1C more strongly when R4 of the peptide was monomethylated or asymmetrically dimethylated (Fig. 3A, top panels).
Figure 3.
Preferential binding of BRCT domains to p300, p300 peptides, or p300 KIX domain containing methylated Arg 754. (A) Peptide-pull-down assay. (Top panels) GST-fused BRCT domains of BRCA1, 53BP1, and Crb2, and the tudor domain of TDRD3 were incubated with streptavidin-Sepharose beads containing 5 μg of biotinylated peptides representing p300 amino acids 745–763 and containing unmethylated, monomethylated, or asymmetrically dimethylated Arg 754. The results shown are from a single experiment that is representative of 12 independent experiments. (Bottom panel) GST-fused BRCT domain of BRCA1 was incubated with streptavidin-Sepharose beads containing 5 μg of peptides representing histone H3 amino acids 1–23 and containing unmethylated or asymmetrically dimethylated Arg 17. Bound protein fractions were analyzed by SDS-PAGE and detected by Western blot with anti-GST antibody. (Top panel) Relative peptide quantities on the beads (peptides) were assessed by Western blot with HRP-conjugated streptavidin. (B) GST pull-down assay. GST-KIX or GST-KIX(R4A) and GST-CARM1 were incubated under methylation conditions in the absence or presence of S-adenosylmethionine (SAM). These reaction mixtures or GST alone were incubated with the BRCA1 BRCT domain that had been translated in vitro. Bead-bound or unbound proteins were analyzed by SDS-PAGE and immunoblot (W) using antibodies against BRCA1, asymmetrically dimethylated p300 R4 peptide, or p300. The left and right panels are from two independent experiments. See also Supplemental Figure S2. (C) Coimmunoprecipitation from cell extracts. 293T cells in six-well plates were transfected with combinations of pcDNA.BRCA1 BRCT domain (200 ng), pCMV.Flag-p300 (200 ng), pSG5.HA-CARM1 or pSG5.HA-CARM1(E267Q) encoding a methyltransferase-deficient mutant (500 ng), or siRNA duplexes against CARM1 (80 pmol). Cell extracts were immunoprecipitated (IP) with anti-Flag antibodies, and the coprecipitated BRCA1 BRCT domain was detected by immunoblot using antibodies against BRCA1. Immunoblot of 2% of the unfractionated cell extract was also performed for comparison. The results shown are from a single experiment that is representative of three independent experiments.
Like the BRCA1 BRCT domain, the GST-fused BRCT domain from 53BP1 also bound with strong preference to the dimethylated R4 peptide and with weaker preference to the monomethylated peptide, compared with the unmethylated peptide. However, the BRCT domain of Crb2 bound equally to all three peptides. Thus, preferential binding to the methylated p300 KIX domain is a conserved property of some but not all members of the BRCT domain-containing family of proteins. The tudor domain of TDRD3 has been shown previously to bind preferentially to a wide range of Arg-methylated proteins (Cote and Richard 2005; Kim et al. 2006); as expected, TDRD3 also bound preferentially to the methylated p300 peptides (Fig. 3A, top panels). While the BRCA1 BRCT domain bound preferentially to the methylated p300 R4 peptides, it bound equally well to histone H3 N-terminal peptides that were either unmethylated or asymmetrically dimethylated at Arg 17, a well-documented site of methylation by CARM1 (Fig. 3A, bottom panel). Thus, the BRCA1 BRCT domain is not a general asymmetric dimethylarginine recognition module; instead, its recognition of asymmetric dimethylarginine depends on the amino acid sequence context of the asymmetric dimethylarginine.
Next, the effect of p300 KIX methylation on its binding to in vitro translated BRCA1C was tested by GST pull-down. Recombinant GST-KIX was first incubated with GST-CARM1 in the presence or absence of S-adenosylmethionine under methylation reaction conditions. Binding of BRCA1C to GST-KIX on glutathione-Sepharose beads was enhanced by prior methylation of GST-KIX (Fig. 3B, left panels, lane 2), compared with binding to unmethylated GST-KIX (Fig. 3B, left panels, lane 3). This result was reproducible in multiple experiments (Supplemental Fig. S2). However, the mutant GST-KIX(R4A) was not methylated by CARM1 and bound BRCA1C very weakly after incubation with CARM1 in the presence or absence of methyl donor S-adenosylmethionine under methylation conditions (Fig. 3B, right panels).
Similar tests were conducted in 293T cells overexpressing BRCA1C and Flag-tagged p300 in the presence of varying levels of CARM1. Coimmunoprecipitation of BRCA1C by the anti-Flag antibody was observed in the presence of endogenous CARM1 (Fig. 3C, top panel, lane 2). Overexpression of CARM1 did not significantly enhance the BRCA1–p300 interaction (Fig. 3C, top panel, lane 3), but overexpression of CARM1(E267Q), a methyltransferase-deficient mutant (Fig. 3C, top panel, lane 4; Lee et al. 2002), or reduction of endogenous CARM1 by siRNA transfection (Fig. 3C, top panel, lane 5) reduced coprecipitation of BRCA1C with Flag-p300. Thus, binding of BRCA1C to p300 depends on CARM1-dependent methylation both in vitro and in vivo.
Synergistic enhancement of the BRCA1 BRCT domain transcriptional activity by CARM1 and p300
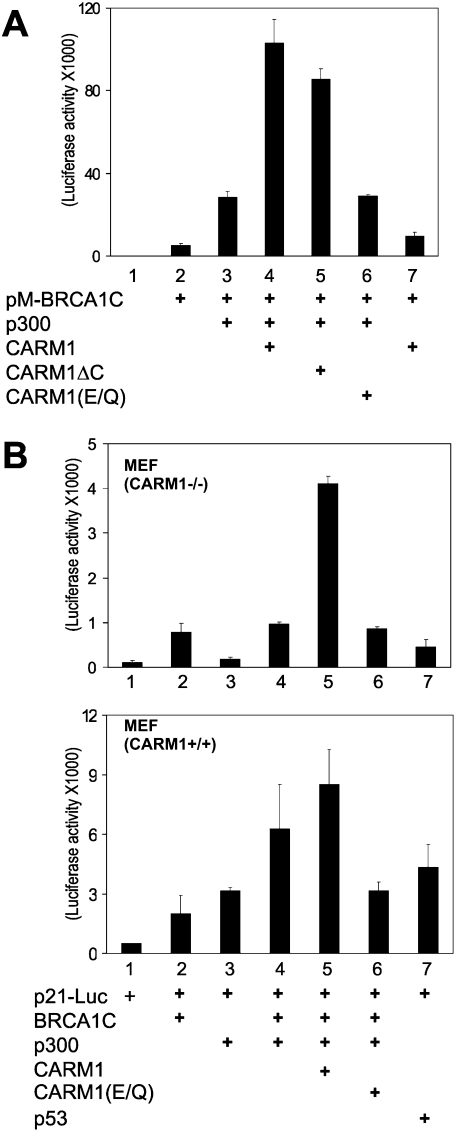
The C terminus of BRCA1 has strong transcriptional enhancement activity and interacts with many different proteins, including transcription factors, coactivators, and proteins involved in DNA repair and cell cycle checkpoint regulation (Glover et al. 2004). Transcriptional activation of a transiently transfected Gal4-responsive reporter gene in 293T cells by BRCA1C fused to Gal4 DBD was further enhanced by p300 or CARM1 and was synergistically enhanced by the combination of CARM1 and p300 (Fig. 4A). CARM1ΔC, which retains methyltransferase activity but lacks the C-terminal activation domain (Teyssier et al. 2002), also cooperated synergistically with p300, but the methyltransferase-defective E267Q mutant of CARM1 did not. Mutant and wild-type CARM1 species were expressed at similar levels (Supplemental Fig. S3B). Similarly, the activity of BRCA1C fused to Gal4 DBD was enhanced by p300 overexpression in wild-type MEF cells, but not in MEF cells lacking CARM1 (Supplemental Fig. S3A). When CARM1 was reactivated in the CARM1−/− cells by Flipase-mediated re-expression of CARM1 from its endogenous promoter (Yadav et al. 2003), the stimulation by p300 was restored (Supplemental Fig. S3A). These findings are consistent with a requirement for CARM1-mediated methylation of Arg 754 in the p300 KIX region to stabilize the interaction of p300 with BRCA1, leading to the synergistic activation of BRCA1 by CARM1 and p300.
Figure 4.
Requirement for CARM1 methyltransferase activity for synergistic transcriptional activation with p300 and BRCA1. (A) Synergistic activation of the BRCA1 BRCT domain by CARM1 and p300. GK1-Luc reporter plasmid (0.1 μg) was transfected into 293T cells along with 0.1 μg of expression vectors for BRCA1C, p300, CARM1, CARM1ΔC (lacking the C-terminal activation domain), and CARM1(E267Q) (lacking methyltransferase activity), as indicated. The luciferase activity of the cell extracts was determined. See also Supplemental Figure S3. (B) Synergistic activation of the p21 promoter by BRCA1C, CARM1, and p300. The p21-Luc reporter plasmid (0.05 μg), controlled by the p21 promoter, was transfected into MEF(CARM1−/−) (top panel) or MEF(CARM1+/+) (bottom panel) cells along with 0.1 μg each of expression vectors for BRCA1C, p300, CARM1, CARM1(E267Q), and p53, as indicated. The luciferase activity of the cell extracts was determined. The results shown are from a single experiment that is representative of four independent experiments. See also Supplemental Figure S4.
CARM1, p300, and BRCA1 synergistically activate the p21WAF1/CIP1 promoter
BRCA1 acts as a tumor suppressor partly by regulating genes involved in growth arrest. For example, the expression of p21WAF1/CIP1, an important cell cycle regulator, is controlled by BRCA1 in cooperation with tumor suppressor p53 (Somasundaram et al. 1997; MacLachlan et al. 2002). We therefore tested for synergy among BRCA1, p300, and CARM1 to enhance expression from the p21WAF1/CIP1 promoter. In CARM1−/− MEF cells, BRCA1C alone activated the p21 promoter several-fold, but p300 had little or no effect in either the presence or absence of coexpressed BRCA1C (Fig. 4B, top panel). However, coexpression of CARM1 (but not the methyltransferase-deficient CARM1 E267Q mutant) with BRCA1C and p300 caused synergistic activation of the p21WAF1/CIP1 promoter. In contrast, the requirement for exogenous CARM1 was almost eliminated in wild-type MEF cells (Fig. 4B, lower panel). In the wild-type cells, p300 alone caused several-fold enhancement of p21-luc reporter activity and several-fold enhancement of the activity achieved with BRCA1C. Coexpression of CARM1 with p300 and BRCA1 caused little if any further enhancement (presumably due to the presence of endogenous CARM1), while coexpression of the CARM1(E267Q) mutant with p300 and BRCA1C suppressed activity, presumably by competing with wild-type endogenous CARM1 for binding to p53, p300, BRCA1, or other components of the transcription machinery. As a control, overexpression of p53 caused a robust stimulation of reporter gene activity in wild-type MEF cells but not in the CARM1-deficient MEF cells, again suggesting the importance of CARM1 for activating the p21 promoter. Similar results were obtained by performing similar transient transfection experiments in wild-type MEF cells versus CARM1(KI) cells, in which a methyltransferase-eliminating point mutation was “knocked in” (KI) to the endogenous CARM1 gene (Supplemental Fig. S4). As in Figure 4B, CARM1 (either endogenous or exogenously supplied) was required for a robust coactivator activity by transiently expressed p300. Furthermore, neither CARM1 alone nor CARM1 plus BRCA1 produced a robust enhancement without transiently expressed p300, demonstrating the requirement of all three coactivators to produce synergy (Supplemental Fig. S4). Thus, CARM1 and its methyltransferase activity are important in BRCA1-mediated p21WAF1/CIP1 expression, and the coactivator activity of p300 also requires CARM1 for its function in support of BRCA1. These results are consistent with the model that CARM1 activates BRCA1-mediated transcription by methylating p300 to facilitate the binding of BRCA1 to p300.
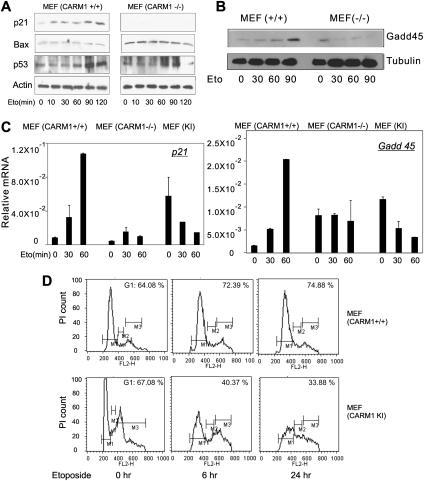
CARM1 is essential for activation of the endogenous p21WAF1/CIP1 gene after DNA damage
When cells are exposed to DNA-damaging agents, expression of p21WAF1/CIP1 is induced to arrest cell cycle progression (Somasundaram et al. 1997; MacLachlan et al. 2002). To investigate the roles of CARM1 in p21WAF1/CIP1 induction, wild-type MEF cells and MEF(CARM1−/−) cells were exposed for various time periods to etoposide, a DNA-damaging agent. In wild-type cells, the p21WAF1/CIP1 protein level increased by 10 min after etoposide treatment and was further elevated after 90 min (Fig. 5A, left panel). However, in MEF(CARM1−/−) cells, p21WAF1/CIP1 protein induction was not observed (Fig. 5A, right panel), indicating a crucial role for CARM1 in p21WAF1/CIP1 induction. When CARM1 was reactivated in the CARM1−/− cells by Flipase-mediated re-expression of CARM1 from its endogenous promoter (Yadav et al. 2003), induction of p21 expression in response to etoposide was restored (Supplemental Fig. S5A,B). Actin or tubulin served as a loading control in these experiments. The level of p53 was increased to a similar extent in both wild type and CARM1−/− cell lines in response to etoposide (Fig. 5A), and the level of BRCA1 protein was indistinguishable in the wild-type and CARM1−/− MEF cells (Supplemental Fig. 5C). Bax, another downstream target of p53 after DNA damage that does not require BRCA1 (MacLachlan et al. 2002; Aylon and Oren 2007), was not induced in either cell line in response to etoposide, perhaps due to insufficient concentration of etoposide or time of treatment. However, for unknown reasons, the level of Bax was higher in the CARM1−/− cells than in the wild-type cells (Fig. 5A). Iyer et al. (2004) also observed an elevated basal level of PUMA protein, another apoptosis regulator, in cells lacking p300. In addition to the p21 gene, we found that DNA damage-induced expression of the DNA excision repair factor Gadd45 also required CARM1 (Fig. 5B). The mRNA levels of p21 and Gadd45 were also rapidly elevated in response to etoposide in wild-type MEF cells, but not in MEF cells lacking CARM1 or containing a CARM1 mutant that lacks methyltransferase activity (Fig. 5C). In fact, mRNA levels of p21 and Gadd 45 were decreased by etoposide treatment in the cells containing methyltransferase-deficient CARM1 (KI cells), suggesting a dominant-negative effect. Although the basal levels of p21 and Gadd45 mRNA varied in the three MEF cell lines, it is inappropriate to compare basal levels between these cell lines, since each cell line was derived from a different mouse. CARM1+/+ and CARM1(KI) MEF cells express similar levels of CARM1 protein (Kim et al. 2010). It is interesting to note that DNA damage-induced expression of both p21 and Gadd45 requires BRCA1 (MacLachlan et al. 2002; Aylon and Oren 2007). Thus, CARM1 is involved in the induction of some p53-regulated genes (i.e., p21WAF1/CIP1 and Gadd45), but not others (e.g., Bax).
Figure 5.
Requirement of CARM1 for activation of endogenous p21WAF1/CIP1 and Gadd45 genes by DNA damage. (A,B) Induction of p53 target gene proteins by DNA damage in cells containing or lacking CARM1. MEF(CARM1+/+) or MEF(CARM1−/−) cells were treated with 100 μM etoposide (Eto) for the indicated times (in minutes). Cell extracts were analyzed by immunoblot with antibodies against the indicated proteins. The results shown are from a single experiment that is representative of four independent experiments. See also Supplemental Figure S5. (C) Induction of p53 target gene mRNA by DNA damage requires CARM1 methyltransferase activity. MEF(CARM1+/+) cells, MEF(CARM1−/−) cells, or MEF knock-in (KI) cells expressing methyltransferase-deficient CARM1 were treated with 100 μM etoposide for the indicated times (in minutes). Quantitative RT–PCR was performed on total cell RNA to assess p21 and Gadd45 mRNA levels, relative to β-actin mRNA. The results shown are the mean and range of variation of duplicate PCR reactions from a single experiment that is representative of five independent experiments. (D) Cell cycle analysis of etoposide-treated cells. MEF(CARM1+/+) or (CARM1 KI) cells were incubated in growth medium with 100 μM etoposide for 0–24 h and stained with propidium iodide. Cell cycle analysis was conducted by analyzing propidium iodide staining with a flow cytometer. Cell count (PI count) is plotted against propidium iodide fluorescence intensity (FL2-H), and the percent of cells in the G1 phase of the cell cycle is shown in each panel. The results shown are from a single experiment that is representative of five independent experiments.
Since CARM1 is required for induction of p21 and Gadd45 expression after etoposide treatment (Fig. 5A–C), and since p21 and Gadd45 are important for arresting the cell cycle after DNA damage, we examined the effects of etoposide treatment on the cell cycle profiles of MEF cells containing or lacking CARM1 methyltransferase activity. Etoposide treatment for 6–24 h caused an accumulation of wild-type MEF cells in the G1 phase, but caused a dramatic decrease in the percentage of G1-phase cells for MEF cells containing methyltransferase-deficient CARM1 (Fig. 5D). Thus, CARM1 methyltransferase activity plays an important role in the response to DNA damage because it is required for p21 and Gadd45 gene expression and the subsequent cell cycle arrest in G1 phase.
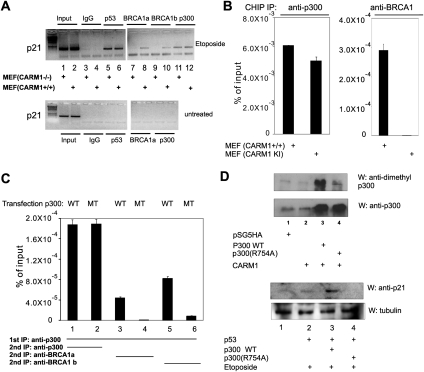
CARM1 is essential for recruitment of BRCA1 to the p21 promoter
The specific contribution of CARM1 to p21 promoter activation was investigated by chromatin immunoprecipitation (ChIP) assay. p53, BRCA1, and p300 were not detected on the p21 promoter in untreated MEF wild-type or CARM1−/− cells (Fig. 6A, bottom panel). Etoposide treatment (90 min) caused recruitment of p53 and p300 to the p21 promoter in wild-type and mutant cells, but BRCA1 recruitment, tested with two different antibodies, was observed only in wild-type cells (Fig. 6A, top panel). In a similar ChIP experiment conducted with quantitative PCR, p300 occupancy on the p21 promoter after etoposide treatment was equivalent in wild-type MEF cells and MEF(KI) cells containing enzymatically dead CARM1. In contrast, BRCA1 occupancy on the p21 promoter was observed only in the cells containing wild-type CARM1 (Fig. 6B). CARM1 was also observed binding to the p21 promoter after etoposide treatment (Supplemental Fig. S6). Thus, CARM1 is essential for recruitment of BRCA1, but not p53 and p300, to the p21 promoter.
Figure 6.
Requirement for CARM1 methylation of p300 for recruitment of BRCA1 to the p21 gene promoter in response to DNA damage. (A) Recruitment of BRCA1 to the p21 promoter. Subconfluent (CARM1+/+) and (CARM1−/−) MEF cells were treated (top panel) or not treated (bottom panel) with 100 μM etoposide for 90 min. Soluble chromatin was immunoprecipitated with the indicated antibodies (1 μg of each). Precipitated DNA was analyzed by PCR, using primers to amplify the p53-binding site region of the p21 promoter. DNA from input chromatin was also analyzed. DNA size markers are shown in the left lanes. The results shown are from a single experiment that is representative of eight independent experiments. See also Supplemental Figure S6. (B) DNA damage-induced recruitment of BRCA1 to the p21 promoter requires CARM1 methyltransferase activity. ChIP was performed as in A using (CARM1+/+) and (CARM1 KI) MEF cells, and precipitation of the p21 promoter was analyzed by quantitative PCR. The results are shown as percent of signal from the precipitated DNA compared with input chromatin. The results shown are from a single experiment that is representative of four independent experiments. (C) DNA damage-induced colocalization of BRCA1 with p300 on the p21 promoter requires Arg 754 (the CARM1 methylation site) of p300. ChIP was performed with sequential immunoprecipitations using p300 antibodies, followed by p300 antibodies again or either of two different BRCA1 antibodies. DNA from the second immunoprecipitation was analyzed for the presence of the p21 promoter by quantitative PCR. The results shown are from a single experiment that is representative of three independent experiments. (D) Essential role of Arg 754 of p300 for induction of p21 expression by DNA damage. (Top panels) 293T cells in six-well plates were transfected with combinations of 200 ng of pCMV-p300 wild type or R754A mutant and 250 ng of pSG5.HA-CARM1 (or the empty vector pSG5.HA), as indicated. Immunoblot was performed with antibodies against asymmetric dimethyl-R4 peptide or p300. (Bottom panels) Plasmids encoding wild-type or R754A mutant p300 (500 ng) and p53 or the empty vector pSG5.HA (20 ng) were transfected into 293T cells in six-well plates. After 48 h, cells were treated with 100 μM etoposide for 2 h. Levels of p21 protein in whole-cell extracts were determined by immunoblot. The results shown are from a single experiment that is representative of three independent experiments.
To test directly the role of p300 methylation in the colocalization of p300 and BRCA1 on the p21 promoter, we overexpressed full-length p300 wild-type or R754A mutant in 293T cells, treated the cells with etoposide, and then performed ChIP assays on the p21 promoter, using successive immunoprecipitations with antibodies against p300 and BRCA1. A control performed with two sequential ChIPs with antibodies against p300 demonstrated that wild-type and mutant p300 localized equally on the p21 promoter after etoposide treatment (Fig. 6C, lanes 1,2). In contrast, sequential ChIPs using antiserum against p300 followed by either of two different antisera against BRCA1 demonstrated colocalization of BRCA1 on the p21 promoter with wild-type p300 (Fig. 6C, lanes 3,5), but little or no colocalization of BRCA1 on the promoter with the mutant p300 (Fig. 6C, lanes 4,6). Similar results were obtained when the order of immunoprecipitating antibodies was reversed (data not shown). Thus, BRCA1 preferentially associates with the wild-type versus the R754A mutant p300 on the p21 promoter region, consistent with a requirement for methylation of p300 Arg 754 by CARM1 as a prerequisite for the recruitment of BRCA1 by the methylated p300.
To further explore the requirement for CARM1-mediated methylation of p300, we tested the effect of overexpressing full-length wild-type p300 and p300(R754A) on the induction of p21 expression in 293T cells in response to DNA damage. Wild-type and mutant p300 were expressed at similar levels, and the antibodies against Arg 754-dimethylated p300 recognized wild-type p300 but showed much less recognition of p300(R754A) (Fig. 6D, top panels), demonstrating further the specificity of the antibodies and the specific methylation of p300 at Arg 754. Overexpression of wild-type p300 enhanced etoposide-induced expression of p21 (Fig. 6D, bottom panels, lanes 2,3), but overexpression of p300(R754A) did not (Fig. 6D, bottom panels, lane 4). Thus, Arg 754 of p300 is essential for its ability to support induction of p21 expression by p53 in response to DNA damage.
Thus, in cells containing wild-type CARM1 and p300, p300 and BRCA1 colocalize on the p21 promoter in response to etoposide. However, elimination of the methyltransferase activity of CARM1 or the Arg 754 methylation site on p300 prevented the efficient recruitment of BRCA1 to the p21 promoter and the induction of p21 expression in response to DNA damage. The results also suggest that CARM1 and BRCA1 are key factors in regulating p21WAF1/CIP1 and Gadd45 expression, and are consistent with the known role of BRCA1 in mediating cell cycle arrest.
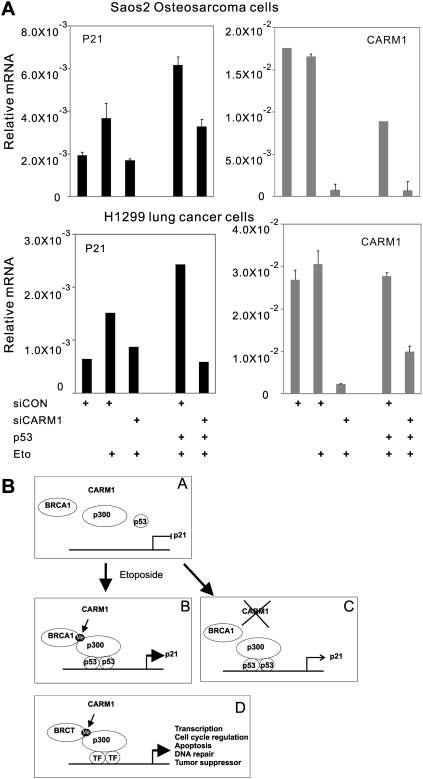
Requirement of CARM1 for induction of p21 expression by DNA damage in human cancer cell lines by p53-dependent and p53-independent mechanisms
To examine the requirement of CARM1 for induction of p21 by etoposide in other cell lines, RNAi was used to deplete CARM1 from two human cancer cell lines (Fig. 7A). Since p21 expression is regulated by both p53-dependent and p53-independent mechanisms (Macleod et al. 1995; Parker et al. 1995), the roles of CARM1 in p21 induction were investigated in two p53-negative cell lines: H1299 lung cancer cells and Saos2 osteosarcoma cells. In cells transfected with control siRNA, etoposide induced p21 mRNA levels, and transient expression of p53 enhanced p21 expression in the presence of etoposide. Depletion of CARM1 by siRNA reduced the etoposide-induced level of p21 mRNA in both the presence and absence of p53 expression. Thus, induction of p21 expression in response to DNA damage has p53-dependent and p53-independent mechanisms, both of which require CARM1.
Figure 7.
Requirement of CARM1 for induction of p21 expression by DNA damage in human cancer cell lines by p53-dependent and p53-independent mechanisms. (A) Role of CARM1 and p53 in p21 gene induction by DNA damage in human cancer cell lines. H1299 human lung cancer cells or Saos2 human osteosarcoma cells in six-well plates were transfected with 500 ng of plasmid encoding p53 and 50 pmol of duplex control siRNA (siCON) or siRNA against CARM1 (siCARM1), as indicated. Transfected cells were incubated for 120 min with or without 100 μM etoposide, and total RNA was harvested and analyzed by quantitative RT–PCR for p21 or CARM1 mRNA. Results are expressed relative to GAPDH mRNA level. (B) Model: CARM1-mediated p300 methylation induces recruitment of BRCA1. (Steps A,B) Exposure of cells to a DNA-damaging agent leads to the recruitment of p53, BRCA1, and p300 to the p21 promoter. (Step B) CARM1 stabilizes the coactivator complex by methylating p300 (Me) and thereby enhancing binding of BRCA1 to p300. (Step C) In the absence of CARM1, BRCA1 is not efficiently recruited to p21 promoter, and expression of p21WAF1/CIP1 is reduced. (Step D) Methylation of p300 by CARM1 may also induce recruitment of other BRCT family proteins to various promoters where p300 is bound. BRCT family proteins are involved in transcription, DNA repair, cell cycle regulation, tumor suppression, and other cell functions.
Discussion
Like phosphorylation, protein methylation is used for signal transduction, by modulating protein structure and/or the ability to interact with other proteins (Stallcup 2001; Bedford and Richard 2005; DY Lee et al. 2005; Paik et al. 2007; Lee and Stallcup 2009). While there are several good examples of inhibition of protein–protein interactions by arginine-specific protein methylation, the only current example of preferential binding to proteins harboring methylarginine motifs are two tudor domain-containing proteins: SMN and TDRD3 (Cote and Richard 2005; Kim et al. 2006). The tudor domain also binds methyllysine motifs, and some of these domains have relatively relaxed requirements for the sequence context of the methylated residue (Cote and Richard 2005; Kim et al. 2006). Indeed, the tudor domain of SMN was shown recently to bind a number of CARM1 substrates, including CA150, in a CARM1-dependent fashion (Cheng et al. 2007). Here we present the BRCT domains of BRCA1 and 53BP1 as a novel type of methylarginine-binding domain; moreover, the BRCT domain of BRCA1 recognizes asymmetric dimethylarginine only within a specific amino acid sequence context. The C-terminal BRCT domain of BRCA1, composed of two BRCT repeats, has transcription activation and chromatin remodeling functions (Monteiro et al. 1996; Haile and Parvin 1999). BRCT domains are present in many proteins that have been implicated in cellular responses to DNA damage (Rosen et al. 2003). The BRCT domain of BRCA1 interacts with p300, CBP, HDAC, and p53, among other proteins (Zhang et al. 1998; Chai et al. 1999; Pao et al. 2000; Glover et al. 2004). In addition, the BRCT domains of BRCA1 and some other proteins bind preferentially to phosphoserine-containing proteins (Manke et al. 2003; Yu et al. 2003). Thus, some BRCT domains can recognize two different types of post-translational modifications.
The Arg methylation sites in CBP and p300 that were found to inhibit CREB binding are located in the originally defined KIX domain, which is highly conserved between CBP and p300 (Xu et al. 2001). The Arg 754 methylation site in p300 reported here is located in a region just on the C-terminal side of the originally defined KIX domain; this C-terminal KIX region of p300 (amino acids 669–828) shares a lower degree of homology with CBP. Of the four Arg residues located in this region of p300 (amino acids 669–828), three (R1 = Arg 695, R3 = Arg 728, and R4 = Arg 754) appear to be conserved in CBP, although the sequences surrounding some of these Arg residues have relatively low homology between p300 and CBP. It was shown previously that ablation of the p300 gene in HCT116 cells eliminated the induction of p21 mRNA in response to DNA damage (Iyer et al. 2004). Since HCT116 cells have wild-type CBP (Ozdag et al. 2002), these results indicate that CBP cannot substitute for p300 in the activation of p21 gene transcription in response to DNA damage.
The importance of coactivator methylation in transcription and coactivator synergy
CARM1 and p300 act synergistically as coactivators for several different DNA-binding transcription factors, including nuclear receptors (by binding to p160 coactivators), LEF (by binding to β-catenin), NFκB, and p53 (by binding directly to p53) (Koh et al. 2002; Lee et al. 2002; An et al. 2004; Covic et al. 2005). The synergy can be partially explained by the acetylation of histones H3 and H4 by p300 and the methylation of histone H3 by CARM1, although the exact mechanism by which these histone modifications contribute to transcriptional activation is not known. In addition, the known methylation of several coactivators by CARM1 has been proposed to regulate or contribute to the synergy (Xu et al. 2001; Chevillard-Briet et al. 2002; YH Lee et al. 2005; Feng et al. 2006; Naeem et al. 2007). For example, methylation of a C-terminal region of p300 by CARM1 inhibits binding of p300 to p160 coactivators (YH Lee et al. 2005), and methylation of the C-terminal region of the p160 coactivator SRC-3/pCIP inhibits binding of CARM1 to p160 coactivators (Feng et al. 2006; Naeem et al. 2007). These coactivator modifications could be inhibitory to transcription or could promote the disassembly of the coactivator complex to facilitate the repeated cycle of assembly and disassembly that is believed to be essential for robust transcription.
Here we show that the methylation of p300 Arg 754 by CARM1 also contributes to their synergy by regulating the recruitment of BRCA1 to the promoter of the p53 target gene, p21WAF1/CIP1 (Fig. 6). DNA damage elicits binding of p53 to the p21 promoter; p53 apparently uses its direct interactions with CARM1 and p300 to recruit them to the promoter. This presumably facilitates methylation of p300 by CARM1, which enhances binding of BRCA1 to p300 on the p21 promoter. Although the coactivator BRCA1 can bind directly to p53 (Ouchi et al. 1998; Zhang et al. 1998; Chai et al. 1999), our results indicate that stable recruitment of BRCA1 to the p21 promoter depends on methylation of p300 at Arg 754 by CARM1. This model is supported by the previously reported involvement of BRCA1 in regulating the expression of p21WAF1/CIP1 (Somasundaram et al. 1997; Randrianarison et al. 2001); the previous demonstration that endogenous p300 is required for induction of the endogenous p21 gene by DNA damage (Iyer et al. 2004); the preferential binding of the BRCA1 BRCT domain to the methylated p300 KIX peptide (Fig. 3A), to the methylated KIX domain in vitro (Fig. 3B), and to methylated p300 in vivo (Fig. 3C); the synergistic enhancement by CARM1 and the p300 KIX fragment of transcriptional activation by the BRCT domain of BRCA1, and the requirement of CARM1 methyltransferase activity for this synergy (Fig. 4A); the synergistic action of p300, CARM1, and BRCA1 on the p21 promoter, and the requirement for the methyltransferase activity of CARM1 in that synergy (Fig. 4B); the CARM1-dependent expression of two BRCA1-dependent target genes of p53 (i.e., p21WAF1/CIP1 and Gadd45) (Fig. 5A–C); the effect of eliminating CARM1 methyltransferase activity on the DNA damage-induced cell cycle profile (Fig. 5D); the requirement for Arg 754 of p300 (the CARM1 methylation site) for the ability of p300 to enhance etoposide-induced expression of p21 (Fig. 6D); and the requirement of CARM1 methyltransferase activity and Arg 754 of p300 (the CARM1 methylation site) for recruitment of BRCA1, but not p53 and p300, to the endogenous p21 promoter in response to DNA damage (Fig. 6A–C).
BRCA1 has been shown previously to play extensive and varied roles in transcription as a coregulator for p53, androgen receptor, and STAT1, among other transcription factors (Rosen et al. 2003). In addition, BRCA1 acts as a corepressor for c-Myc and estrogen receptor (Rosen et al. 2003; Glover et al. 2004). The C-terminal BRCT domain of BRCA1 contributes to the coregulator function of BRCA1 through its autonomous transcriptional enhancement activity (Monteiro et al. 1996); protein–protein interactions with various other components of the transcription machinery, including p53, p300, CBP, and histone deacetylases; and chromatin remodeling activity (Park et al. 2000; Fan et al. 2002; Glover et al. 2004). Thus, our results suggest that p300 methylation by CARM1 followed by BRCA1 binding to methylated p300 may be one of the key mechanisms for the synergy of these coactivators. In addition to methylation of p300, the role of CARM1 in transcriptional activation may also include methylation of histone H3 and other proteins in the transcription complex. However, since we observed no preferential binding of the BRCA1 BRCT domain to the histone H3 tail methylated at Arg 17 (a CARM1 methylation site) (Fig. 3A), it is unlikely that methylation of histone H3 by CARM1 is responsible for recruitment of BRCA1 to the p21 promoter in response to DNA damage.
The implication of coactivator methylation for the tumor suppressor functions of BRCA1
BRCA1 is a crucial tumor suppressor in breast and ovarian cancer (Monteiro et al. 1996; Rosen et al. 2003). Patients with familial forms of breast or ovarian cancer often have germline point or deletion mutations in BRCA1, especially in the C-terminal BRCT domain (Futreal et al. 1994; Miki et al. 1994). These mutations usually lead to deficiency in transcription activation by the BRCT domain, suggesting that transcriptional regulation by BRCA1 is one of the key mechanisms for its tumor suppressor activity. BRCA1 acts as a tumor suppressor by repressing oncogenic transcription factors like c-Myc, and by enhancing p53-dependent transcription of genes. While BRCA1 is involved in the activation of some p53 target genes, including the cell cycle regulator p21WAF1/CIP1 and DNA excision repair factor Gadd45, it is not involved in the induction of other p53 target genes, such as the apoptosis inducer Bax (MacLachlan et al. 2002; Aylon and Oren 2007). Interestingly, our study indicates that CARM1 is required for the induction of p21WAF1/CIP1 and Gadd45, but not for induction of Bax; furthermore, CARM1-mediated methylation of the coactivator p300 is required for recruitment of BRCA1 to the p21 promoter in response to DNA damage. CARM1 was implicated previously in the activation of the p53-regulated gene Gadd45 by methylating histone H3 (An et al. 2004). These findings suggest that CARM1 and its methyltransferase activity play important roles in cell cycle regulation by altering the cellular levels of BRCA1-regulated proteins p21 and Gadd45 (Fig. 5). Thus, while wild-type cells arrest in G1 in response to DNA damage, cells lacking CARM1 methyltransferase activity fail to arrest in G1 and instead accumulate in other phases of the cell cycle (Fig. 5D).
Our results suggest that the tumor suppressor activity of BRCA1 is modulated by p300 and CARM1, which control BRCA1 recruitment to specific target genes. Given the diverse roles of BRCA1 in transcriptional regulation and chromatin remodeling—and our demonstration that BRCT domains of BRCA1, 53BP1, and other proteins (Fig. 3A; data not shown) interact preferentially with methylated p300—the BRCT domain interaction with methylated p300 may be involved in recruitment of multiple BRCT domain proteins to a variety of target genes, and thus may have broader implications for many cellular processes (Fig. 7B). Thus, protein arginine methylation of coactivators may be crucial in the fine-tuning of cell cycle progression, DNA repair, and apoptosis by tumor suppressors.
Arginine methylation has been relatively understudied compared with other types of post-translational modifications. Presumably, protein arginine methylation modulates the binding of other proteins to the methylated protein, but to date very few proteins with methylarginine binding modules have been identified. Thus, our finding that the BRCT domain of BRCA1 preferentially binds to asymmetric dimethylarginine in a specific sequence context supports the idea that arginine methylation is a relatively underexplored form of signal transduction. Furthermore, our results suggest that p300 methylation by CARM1 may provide an important new mechanism for regulating the activity of the important tumor suppressor BRCA1, and this mechanism may extend to other proteins with BRCT domains. Specifically, we show that CARM1, like the tumor suppressors BRCA1 and p53, is responsible for inducing cell cycle inhibitors p21 and Gadd45 in response to DNA damage. Furthermore, we demonstrate that the methyltransferase activity of CARM1 is essential for cell cycle arrest in response to DNA damage signals, and thus for repair of DNA damage. These results suggest that CARM1 is a tumor suppressor or an important regulator for tumor suppressor pathways in cells.
Materials and methods
Plasmids
The following plasmids were described previously as indicated: plasmids encoding transcription factors and coactivators, pSG5-HA.GRIP1, pSG5-HA.CARM1, pCMV-p300, pSG5-HA.p53, pCMV-FLAP1, pSG5HA.CARM1(E267Q), pCMV-Flag-p300 (Lee et al. 2002; Lee and Stallcup 2006); pSG5HA.CARM1ΔC (Teyssier et al. 2002); pcDNA-BRCA1 (1314–1863) (Miyake et al. 2000); plasmids encoding Gal4 DBD fusion proteins, pM-BRCA1C (1528–1863) (Nadeau et al. 2000); pM-KIX containing a SmaI/SalI insert subcloned from pGEX4T-KIX; reporter plasmids, GK1-Luc, p21-Luc, described previously (Huang et al. 2001; YH Lee et al. 2005); bacterial GST-protein expression vectors, pGEX4T-KIX (p300 amino acids 669–828); pGEX4T-BRCA1(BRCT), pGEX4T-53BP1(BRCT), pGEX4T-Crb2(BRCT) (Yu et al. 2003; YH Lee et al. 2005); pGEX4T-Tudor3 (tudor domain, amino acids 495–651) (Cote and Richard 2005; Kim et al. 2006). Point mutants of the p300 KIX fragment and full-length p300 were constructed by the QuickChange mutagenesis kit according to the manufacturer's protocol.
Antiserum against R4 peptide containing asymmetric dimethylarginine at R754
Antibodies were produced against a peptide representing p300 amino acids 745–763 and containing asymmetric dimethylarginine at Arg 754 (R4). The antiserum was generated by custom order from Anaspec.
Transfection
MEF wild-type, MEF(CARM1−/−), MEF(CARM1 KI) (with a R169A mutation that eliminates methyltransferase activity), MEF(CARM1 rescued; i.e., with the knockout reversed by Flipase) (Yadav et al. 2003; Cheng et al. 2007; Kim et al. 2010), and 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum. Transfections were performed with F1 Targefect reagent (Targeting Systems) for 293T cells and with Lipofectamine 2000 (Invitrogen) for MEF cells, according to the manufacturers' protocols as described previously (Lee et al. 2004). Luciferase activity was measured in cell extracts prepared 40 h after transfection. The results shown are the mean and range of variation from two transfected cultures, and are from a single experiment that is representative of at least three independent experiments, as indicated specifically in the figure legends. Since the coactivators used enhance the expression of so-called constitutive reporter plasmids, internal controls were not used. Rather, results were tested with multiple plasmid preparations, since the major cause of differences in transfection efficiency is differences in the purity of plasmid preparations.
GST pull-down assay
The procedure for GST pull-down assays was described previously and used GST fusion proteins produced in Escherichia coli BL21 cells (Lee et al. 2004; Lee and Stallcup 2006). The transformed E. coli cells were induced by IPTG and lysed by sonication. GST-fused proteins were collected by incubation with glutathione-Sepharose beads.
Protein methylation assay
The GST-KIX protein and its mutants were prepared in E. coli, and protein methylation in vitro was performed with GST-CARM1 as described previously (YH Lee et al. 2005). One microgram of purified GST-KIX or GST-KIX mutants (R1A, R2A, R3A, and R4A) was coincubated with 1 μg of GST-CARM1 in the presence of 3H-labeled S-adenosylmethionine. Methylation was detected by using autoradiography.
Peptide pull-down assay
Three biotinylated p300 peptides containing methylated or unmethylated Arg 754 (R4) and biotinylated histone H3(1–23) peptide containing unmethylated or asymmetrically dimethylated Arg 17 were synthesized by the Yale University core facility. For equilibration of beads, 20 μL of streptavidin agarose beads (Upstate Biotechnologies) was washed three times with peptide-binding buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol, 0.5% NP-40). Five micrograms to 10 μg of biotin-labeled peptides was incubated with buffer-equilibrated streptavidin-coupled beads overnight at 4°C and washed three times with binding buffer to remove unbound peptides. To reduce nonspecific binding, ∼5 μg of GST-fused BRCT or Tudor proteins was preincubated with buffer-equilibrated agarose beads overnight. After centrifugation, the supernatant was coincubated with peptide-bound beads in 300 μL of binding buffer overnight. The beads were washed five times with 500 μL of binding buffer on a rotator for 1 min per each washing. Twenty microliters of SDS-PAGE gel loading buffer was added to the precipitates and boiled for 5 min. The samples were analyzed by SDS-PAGE and immunoblot with anti-GST antibody (SantaCruz Biotechnology). Biotinylated peptides were detected by horseradish peroxidase-conjugated streptavidin (Upstate Biotechnologies). The results shown were representative of at least three independent experiments. The synthesized p300 peptide sequences are as follows: unmodified R4 peptide, biotin-lnppmgygpRmqqpsnqgq; monomethyl R4 peptide, biotin-lnppmgygp-(monomethyl R)-mqqpsnqgq; asymetric dimethyl R4 peptide, biotin-lnppmgygp-(dimethyl R)-mqqpsnqgq.
Coimmunoprecipitation
Coimmunoprecipitation was performed as described previously (Lee et al. 2002), using antibodies against the Flag epitope (Sigma).
Immunoblot analyses
MEF(CARM1+/+), MEF(CARM1−/−), and MEF(CARM1 rescued) cells were grown in six-well plates, treated with etoposide for various time periods, and lysed in 0.25 mL of RIPA buffer (50 mM Tris-Cl at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Lysates were clarified by centrifugation for 15 min at the maximum speed of a microcentrifuge. A portion of the supernatant (40 μL) was analyzed by SDS-PAGE, and specific protein expression was detected by immunoblot analysis (Lee et al. 2002) with antibodies against p21, Bax, actin, Gadd45, and p53 (SantaCruz Biotechnology).
Quantitative RT–PCR
RNA was isolated from cells, and quantitative RT–PCR was performed as described previously (Jeong et al. 2009) using the following primers: p21, 5′-CCTGGTGATGTCCGACCTG-3′ and 5′-TTATCGGGGTCTACGTTGAGC-3′; Gadd45, 5′-CCGAAAGGATGGACACGGTG-3′ and 5′-TTATCGGGGTCTACGTTGAGC-3′; β-actin, 5′-GGCTGTATTCCCCTCCATCG-3′ and 5′-CCAGTTGGTAACAATGCCATGT-3′. The results shown are normalized to β-actin mRNA.
RNAi
Duplex siRNA against CARM1 were transfected into cultured cells using Lipofectamine 2000 as described previously (Lee et al. 2004).
Cell cycle analysis
Distribution of cells in various stages of the cell cycle was determined by propidium iodide staining. CARM1+/+ and CARM1(KI) MEF cells were treated with etoposide for 6–24 h. Collected cells were fixed by 70% ethanol for 30 min. After incubation with 2 N HCl/1% Triton X-100 for 30 min, cells were washed with PBS. The fixed cells were centrifuged, resuspended in propidium iodide solution (5 μg/mL), and analyzed by flow cytometry at 488 nm.
ChIP assay
This procedure was described previously (Lee et al. 2004). MEF(CARM1−/−, CARM1 KI, and CARM1+/+) cells were cultured for 90 min with or without etoposide, and soluble chromatin was prepared by sonication. Antibodies against p300, BRCA1, and p53 were purchased from SantaCruz Biotechnology, and antibodies against CARM1 were from Upstate Biotechnologies (currently Millipore). The primers for detecting the p21 promoter were as follows: p21 forward primer, CCTTTCTATCAGCCCCAGAGGATA; p21 reverse primer, GGGACATCCTTAATTATCTGGGGT (Chao et al. 2003). PCR products were analyzed by agarose gel electrophoresis or quantitative PCR. The results of the quantitative ChIP assays are shown as the mean and range of variation of duplicate PCR reactions from a single experiment. The results shown are representative of multiple independent experiments, as indicated in the figure legends.
Sequential ChIP with two different antibodies was performed as described previously (Lee et al. 2004), and the precipitated DNA fragments were analyzed by quantitative PCR, using primers against the p21 promoter.
Acknowledgments
We thank Dr. Rong Li (University of Texas Health Science Center at San Antonio), Dr. Wafik S. El-Deiry (University of Pennsylvania School of Medicine), and Dr. Luc Gaudreau (Université de Sherbrooke) for providing plasmids encoding BRCA1, p53, and p21-Luc reporter; and D. Gerke (University of Southern California) for his technical assistance. This work was supported by United States Public Health Service grants DK55274 (to M.R.S.) and DK62248 (to M.T.B.) from the National Institutes of Health.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1975811.
Supplemental material is available for this article.
References
- An W, Kim J, Roeder RG 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Oren M 2007. Living with p53, dying of p53. Cell 130: 597–600 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S 2005. Arginine methylation: An emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Chai YL, Cui J, Shao N, Shyam E, Reddy P, Rao VN 1999. The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene 18: 263–268 [DOI] [PubMed] [Google Scholar]
- Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, Hollstein M, Appella E, Xu Y 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem 278: 41028–41033 [DOI] [PubMed] [Google Scholar]
- Cheng D, Cote J, Shaaban S, Bedford MT 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25: 71–83 [DOI] [PubMed] [Google Scholar]
- Chevillard-Briet M, Trouche D, Vandel L 2002. Control of CBP co-activating activity by arginine methylation. EMBO J 21: 5457–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Richard S 2005. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem 280: 28476–28483 [DOI] [PubMed] [Google Scholar]
- Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO 2005. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO J 24: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Ma YX, Wang C, Yuan RQ, Meng Q, Wang JA, Erdos M, Goldberg ID, Webb P, Kushner PJ, et al. 2002. p300 Modulates the BRCA1 inhibition of estrogen receptor activity. Cancer Res 62: 141–151 [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O'Malley BW 2006. Signaling within a coactivator complex: Methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 26: 7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. 1994. BRCA1 mutations in primary breast and ovarian carcinomas. Science 266: 120–122 [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS 2004. Interactions between BRCT repeats and phosphoproteins: Tangled up in two. Trends Biochem Sci 29: 579–585 [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev 14: 1553–1577 [PubMed] [Google Scholar]
- Haile DT, Parvin JD 1999. Activation of transcription in vitro by the BRCA1 carboxyl-terminal domain. J Biol Chem 274: 2113–2117 [DOI] [PubMed] [Google Scholar]
- Huang SM, Schonthal AH, Stallcup MR 2001. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene 20: 2134–2143 [DOI] [PubMed] [Google Scholar]
- Iyer NG, Chin SF, Ozdag H, Daigo Y, Hu DE, Cariati M, Brindle K, Aparicio S, Caldas C 2004. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc Natl Acad Sci 101: 7386–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KW, Lee YH, Stallcup MR 2009. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J Biol Chem 284: 29298–29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, Bedford MT 2010. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem 285: 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR 2002. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and β-catenin with two different classes of DNA-binding transcriptional activators. J Biol Chem 277: 26031–26035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR 2006. Interplay of Fli-I and FLAP1 for regulation of β-catenin dependent transcription. Nucleic Acids Res 34: 5052–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR 2009. Minireview: Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol 23: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR 2002. Synergy among nuclear receptor coactivators: Selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol 22: 3621–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Campbell HD, Stallcup MR 2004. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol 24: 2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR 2005. Role of protein methylation in regulation of transcription. Endocr Rev 26: 147–170 [DOI] [PubMed] [Google Scholar]
- Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci 102: 3611–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan TK, Takimoto R, El-Deiry WS 2002. BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets. Mol Cell Biol 22: 4280–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T 1995. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev 9: 935–944 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71 [DOI] [PubMed] [Google Scholar]
- Miyake T, Hu YF, Yu DS, Li R 2000. A functional comparison of BRCA1 C-terminal domains in transcription activation and chromatin remodeling. J Biol Chem 275: 40169–40173 [DOI] [PubMed] [Google Scholar]
- Monteiro AN, August A, Hanafusa H 1996. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci 93: 13595–13599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau G, Boufaied N, Moisan A, Lemieux KM, Cayanan C, Monteiro AN, Gaudreau L 2000. BRCA1 can stimulate gene transcription by a unique mechanism. EMBO Rep 1: 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J 2007. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol 27: 120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H 1998. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci 95: 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdag H, Batley SJ, Forsti A, Iyer NG, Daigo Y, Boutell J, Arends MJ, Ponder BA, Kouzarides T, Caldas C 2002. Mutation analysis of CBP and PCAF reveals rare inactivating mutations in cancer cell lines but not in primary tumours. Br J Cancer 87: 1162–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik WK, Paik DC, Kim S 2007. Historical review: The field of protein methylation. Trends Biochem Sci 32: 146–152 [DOI] [PubMed] [Google Scholar]
- Pao GM, Janknecht R, Ruffner H, Hunter T, Verma IM 2000. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci 97: 1020–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Irvine RA, Buchanan G, Koh SS, Park JM, Tilley WD, Stallcup MR, Press MF, Coetzee GA 2000. Breast cancer susceptibility gene 1 (BRCAI) is a coactivator of the androgen receptor. Cancer Res 60: 5946–5949 [PubMed] [Google Scholar]
- Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267: 1024–1027 [DOI] [PubMed] [Google Scholar]
- Randrianarison V, Marot D, Foray N, Cabannes J, Meret V, Connault E, Vitrat N, Opolon P, Perricaudet M, Feunteun J 2001. BRCA1 carries tumor suppressor activity distinct from that of p53 and p21. Cancer Gene Ther 8: 759–770 [DOI] [PubMed] [Google Scholar]
- Rosen EM, Fan S, Pestell RG, Goldberg ID 2003. BRCA1 gene in breast cancer. J Cell Physiol 196: 19–41 [DOI] [PubMed] [Google Scholar]
- Somasundaram K, Zhang H, Zeng YX, Houvras Y, Peng Y, Wu GS, Licht JD, Weber BL, El-Deiry WS 1997. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature 389: 187–190 [DOI] [PubMed] [Google Scholar]
- Stallcup MR 2001. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20: 3014–3020 [DOI] [PubMed] [Google Scholar]
- Teyssier C, Chen D, Stallcup MR 2002. Requirement for multiple domains of the protein arginine methyltransferase CARM1 in its transcriptional coactivator function. J Biol Chem 277: 46066–46072 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH 2001. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem 276: 13505–13508 [DOI] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM 2001. A transcriptional switch mediated by cofactor methylation. Science 294: 2507–2511 [DOI] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci 100: 6464–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber BL, El-Deiry WS 1998. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 16: 1713–1721 [DOI] [PubMed] [Google Scholar]