Abstract
The 30S subunit is composed of four structural domains, the body, platform, head, and penultimate/ultimate stems. The functional integrity of the 30S subunit is dependent upon appropriate assembly and precise orientation of all four domains. We examined 16S rRNA conformational changes during in vitro assembly using directed hydroxyl radical probing mediated by Fe(II)-derivatized ribosomal protein (r-protein) S8. R-protein S8 binds the central domain of 16S rRNA directly and independently and its iron derivatized substituents have been shown to mediate cleavage in three domains of 16S rRNA, thus making it an ideal probe to monitor multidomain orientation during assembly. Cleavages in minimal ribonucleoprotein (RNP) particles formed with Fe(II)-S8 and 16S rRNA alone were compared with that in the context of the fully assembled subunit. The minimal binding site of S8 at helix 21 exists in a structure similar to that observed in the mature subunit, in the absence of other r-proteins. However, the binding site of S8 at the junction of helices 25–26a, which is transcribed after helix 21, is cleaved with differing intensities in the presence and absence of other r-proteins. Also, assembly of the body helps establish an architecture approximating, but perhaps not identical, to the 30S subunit at helix 12 and the 5′ terminus. Moreover, the assembly or orientation of the neck is dependent upon assembly of both the head and the body. Thus, a complex interrelationship is observed between assembly events of independent domains and the incorporation of primary binding proteins during 30S subunit formation.
Keywords: small subunit, ribosome assembly, directed hydroxyl radical probing, S8, RNA folding, RNA structure, r-protein association
INTRODUCTION
The small subunit of the ribosome decodes the mRNA during translation with extremely high fidelity, hence the formation of appropriate structure in the mature subunit is crucial for cellular viability and fitness (Powers and Noller 1990; Kurland et al. 1996). The tertiary structure of the small subunit is subdivided into four structural domains: body, platform, head, and penultimate/ultimate stem, at the intersection of which lies the decoding center of the 30S subunit (Moazed and Noller 1990; Lee et al. 1997; Lodmell and Dahlberg 1997; Clemons et al. 1999; Merryman et al. 1999; Yusupov et al. 2001; Yusupova et al. 2001). The domains within the tertiary structure of the 30S subunit are formed by the association of ribosomal proteins (r-proteins) with corresponding domains of 16S rRNA within the secondary structure (5′, central, 3′ major, and 3′ minor domains) and this process can be carried out in vitro (Held et al. 1973, 1974; Cunningham et al. 1991; Culver and Noller 2000b). The aim of this study is to examine domain assembly and orientation during in vitro 30S subunit assembly using directed hydroxyl radical probing from Fe(II)-derivatized r-protein S8.
The hierarchical and cooperative principles underlying in vitro assembly of the 30S subunit have been revealed through a set of ground-breaking experiments whose results have been graphically expressed in the form of an assembly map (Fig. 1A; Mizushima and Nomura 1970; Held et al. 1974; Grondek and Culver 2004). Concisely, r-protein association leads to conformational change or altered stability of 16S rRNA tertiary structures, which in turn enhance the association of other r-proteins and subsequent folding events (Sykes and Williamson 2009). For example, r-protein S15 is an early or primary binding protein that associates directly and independently with the central domain, leading to the incorporation of mid-binding or secondary r-proteins S6, S18 and, finally, late binding or tertiary proteins S11 and S21, which together form the platform (see Fig. 1A). Each domain exhibits similar patterns of dependencies (see Fig. 1A) and can be assembled independently in vitro (Weitzmann et al. 1993; Samaha et al. 1994; Agalarov et al. 1998, 1999).
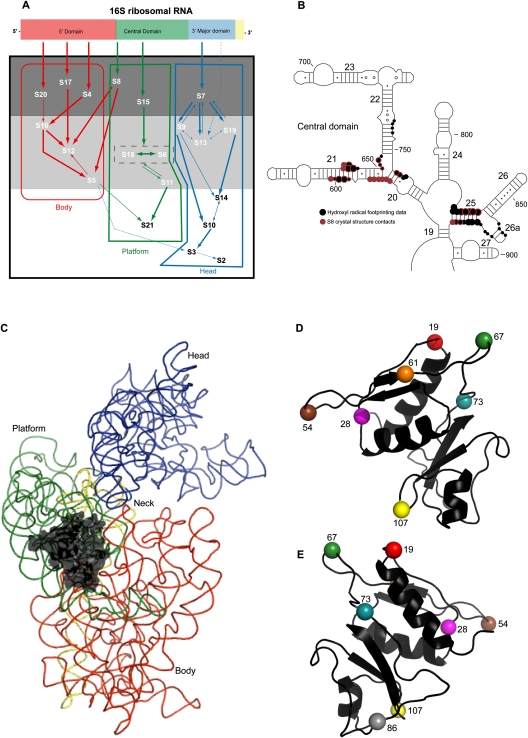
FIGURE 1.
Structure of r-protein S8 and its interactions with the 30S subunit. (A) Modified in vitro assembly map of the 30S subunit, illustrating domain organization (Mizushima and Nomura 1970; Held et al. 1974; Grondek and Culver 2004). Each domain within the secondary structure of 16S rRNA is assigned a specific color, and this scheme is extended to the r-proteins that bind 16S rRNA to form corresponding domains in the tertiary structure, delineated by boxes with the names of the domains written below. The arrows symbolize the direction of dependence of r-protein binding with the width symbolizing the strength of the dependence. R-proteins can be subdivided into early, middle, and late binding proteins contained in a dark gray, light gray, and white box, respectively. (B) Localization of S8 within the central domain of 16S rRNA. Hydroxyl radical footprinting data for S8 (Powers and Noller 1995) are represented by black circles, while S8 crystal structure contacts are represented by magenta circles. Concentric rings of color indicate sites of overlap. (C) Localization of S8 in the three-dimensional structure of the E. coli 30S subunit. S8 is depicted in black and other r-proteins were omitted for clarity. Each domain of 16S rRNA is color coded as in A. (D,E) Tethered probing sites on S8. The positions of cysteine substitutes (Lancaster et al. 2000) are indicated by number and distinctly colored. D is oriented as in C, E depicting the molecule rotated by 180° along the y-axis. All figures containing three-dimensional structures were prepared using the PyMOL and the Protein Data Bank file 2AW7 (Schuwirth et al. 2005).
Although S8 does not aid in incorporation of r-proteins that bind the central domain, the association of S8 has profound effects on central domain structure (Jagannathan and Culver 2003). Initially the minimal binding site of S8 was localized to the junction of helices 20, 21, and 22, by a number of studies, including RNase protection assays (Schaup et al. 1973; Ungewickell et al. 1975; Zimmermann et al. 1975; Muller et al. 1979), chemical cross-linking (Wower and Brimacombe 1983), and determination of binding affinity (Mougel et al. 1993; Wu et al. 1994). However, data from X-ray crystallography (see Fig. 1B; Lancaster et al. 2000; Schuwirth et al. 2005) as well as solution (see Fig. 1B; Powers and Noller 1995) and directed hydroxyl radical probing (Lancaster et al. 2000) expanded our understanding of S8 interaction with 16S rRNA to include a second site at the junction of helices 25, 26, and 26a (see Fig. 1B). The existence of two separate sites led to speculation that S8 plays a role in compacting the central domain during 5′–3′ assembly in vivo (Lancaster et al. 2000). Moreover, within the tertiary structure of the small subunit, S8 is proximal to the junction of body and platform (see Fig. 1C). This, together with its role in the incorporation of r-proteins S5 and S12 that bind near the decoding center of the 30S subunit, has led to conjectures that S8 may function in the appropriate orientation of domains during assembly via direct or indirect interactions with 16S rRNA and other r-proteins (Jagannathan and Culver 2003). In light of the data presented above, S8 appeared to be an excellent candidate r-protein for a directed hydroxyl radical probing study of interactions during 30S subunit assembly.
The technique of directed hydroxyl radical probing has many applications, including the localization of binding sites, the determination of orientation of a protein with respect to an RNA or DNA molecule (Culver and Noller 2000a; Lancaster et al. 2000; Chen and Hahn 2003; Xu et al. 2008), the characterization of the RNA or DNA environment of the Fe(II)-derivatized protein, and monitoring conformational changes or dynamics associated with the incorporation of r-proteins with rRNA (Jagannathan and Culver 2003, 2004; Dutca and Culver 2008). In fact, a previous study had been carried out using Fe(II)-S8 to localize its binding site in the 70S ribosome (Lancaster et al. 2000). These probing experiments revealed that Fe(II)-S8 could cleave elements within the head, body, and platform of the 30S subunit, thereby suggesting that probing mediated by Fe(II)-S8 would allow assembly of these domains to be monitored.
In order to examine the architecture of these domains during assembly, iron derivatized S8 proteins were assembled into ribonucleoprotein particles (RNPs) with 16S rRNA in the presence and absence of various r-proteins and subjected to directed hydroxyl radical probing. Cleavage patterns in various complexes were compared with a view to determining a minimal RNP that recapitulated cleavage patterns observed in the 30S subunit and 70S ribosome. The association of the 30S subunit with the 50S subunit in the ribosome did not change cleavage patterns significantly as compared to free small subunits (data not shown). However, the architecture(s) of binary complexes of Fe(II)-S8 and 16S rRNA alone (Fe(II)-S8/16S) are distinct from that of the Fe(II)-S8 containing 30S subunits, as evidenced by differences in cleavage patterns. While the minimal binding site of S8 at the junction of helices 20–22 was organized in the binary complex, the cleavage pattern at the junction of helices 25-26a differed between the two sets of complexes. Thus, there is a variation in r-protein–rRNA interaction across the two S8 binding sites. Also, in both the 5′ and 3′ major domains Fe(II)-S8 mediated cleavages in 30S subunits were absent in binary complexes. Restoration of cleavage patterns required the assembly of entire domains, highlighting the significance of long-range interactions in assembly, even between independent events (Weitzmann et al. 1993; Samaha et al. 1994; Agalarov et al. 1998, 1999).
RESULTS
Comparison of Fe(II)-S8 directed cleavage data from 30S subunits and 70S ribosomes
Previous work utilized S8 proteins substituted at nine different positions by cysteine residues (Fig. 1D,E) and derivatized with iron (Fe(II)-C19-S8, Fe(II)-C28-S8, Fe(II)-C46-S8, Fe(II)-C54-S8, Fe(II)-C61-S8, Fe(II)-C67-S8, Fe(II)-C73-S8, Fe(II)-C86-S8, Fe(II)-C107-S8). These proteins and a cysteine-less control were used to determine the location of S8 within the 70S ribosome by directed hydroxyl radical probing (Lancaster et al. 2000). The present study examined 30S subunit assembly utilizing proteins derivatized at eight positions. The derivatized protein omitted in the present study was Fe(II)-C46-S8 as it demonstrated only a weak cleavage pattern at the junction of helices 25–26a in the ribosome that overlapped with that of Fe(II)-C19-S8, Fe(II)-C61-S8, and Fe(II)-C67-S8 (Lancaster et al. 2000). Cleavage patterns in both the 30S subunits and 70S ribosomes were examined to identify any possible changes due to subunit association or changes in protocol from previous work. The cleavage patterns of several derivatized proteins in 70S ribosomes were consistent with previous data (data not shown). Additionally, patterns of cleavage in 30S subunits and 70S ribosomes were extremely similar throughout 16S rRNA (data not shown), corroborating previous unpublished data (L Lancaster, unpubl.). This similarity in cleavage patterns also correlated with the estimation of probe-target distances elucidated in the study by Lancaster et al. (2000). This estimation in turn was dependent upon earlier studies establishing these correlations using a tRNA molecule as a helical ruler (Joseph et al. 1997). The data obtained in this study are therefore consistent with X-ray crystal structures of the 30S subunit (Schuwirth et al. 2005).
Examining 30S subunit assembly using S8 as a probe
Comparison of cleavage patterns between a binary complex of Fe(II)-S8 bound to 16S rRNA alone (Fe(II)-C19-S8/16S rRNA, Fe(II)-C28-S8/16S rRNA, Fe(II)-C54-S8/16S rRNA, Fe(II)-C73-S8/16S rRNA, Fe(II)-C86-S8/16S rRNA, Fe(II)-C107-S8/16S rRNA, Cysteine-less/16S rRNA) and Fe(II)-S8 in the mature subunit (Fe(II)-C19-S8/30S, Fe(II)-C28-S8/30S, Fe(II)-C54-S8/30S, Fe(II)-C73-S8/30S, Fe(II)-C86-S8/30S, Fe(II)-C107-S8/30S, Cysteine-less/30S) yielded information on the architecture of three major domains, the 5′, central, and 3′ major domains in the presence of S8 alone. Only in helix 21 within the central domain were similar patterns of cleavage between binary complexes and 30S subunits observed (Fig. 2A–C). For the other S8 binding site within the central domain as well as the 5′ and 3′ major domain, differences in cleavage patterns between the two complexes were revealed (Figs. 2, 3, 4, 5).
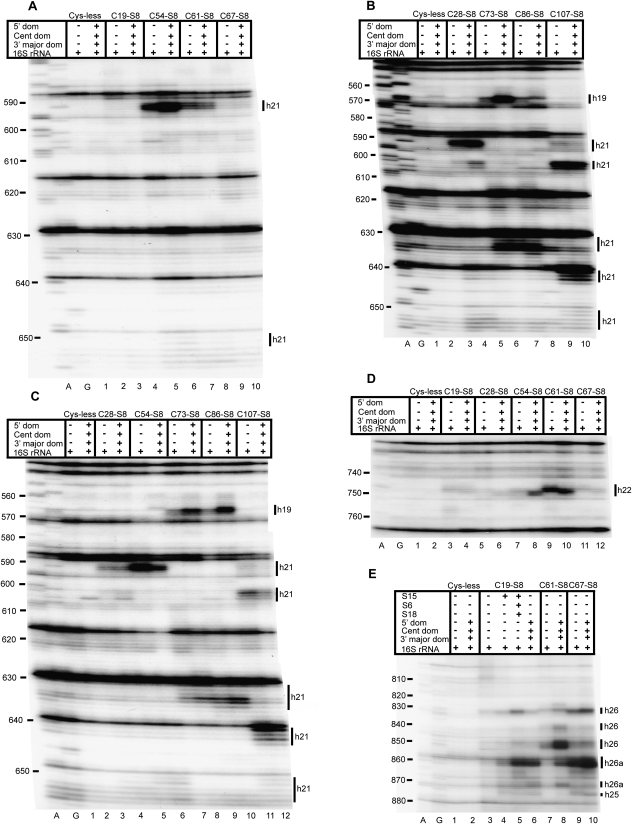
FIGURE 2.
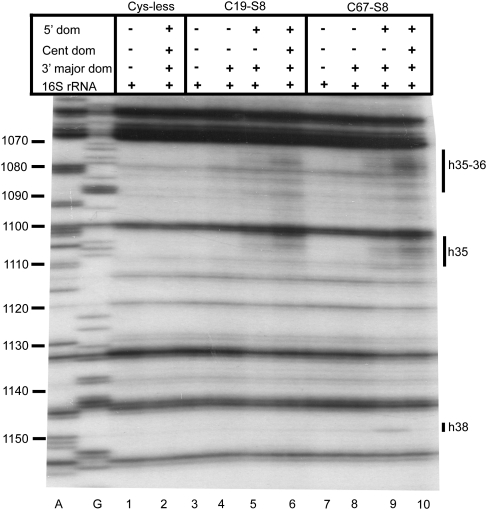
Directed hydroxyl radical cleavage of the central domain of 16S rRNA from Fe(II)-S8 in various RNPs detected by primer extension. A and G are sequencing lanes. All complexes contain natural 16S rRNA and derivatized cysteine substituents of S8, with the identity of the derivatized protein indicated at the top of each lane as Fe(II)-Cx-S8. Cys-less is the mock-treated cysteine-less control S8. The addition of individual r-proteins and mixtures is indicated by +. Composition of mixtures: 5′ dom: S4 + S5 + S12 + S16 + S17 + S20, Cent dom: S6 + S11 + S15 + S18 + S21, 3′ major dom: S2 + S3 + S7 + S9 + S10 + S13 + S14 + S19. Bars on the right indicate positions of cleavage, and the text delineates the identity of the helices cleaved. Primers utilized are 683 (A–C), 837 (D), and 1046 (E).
FIGURE 3.
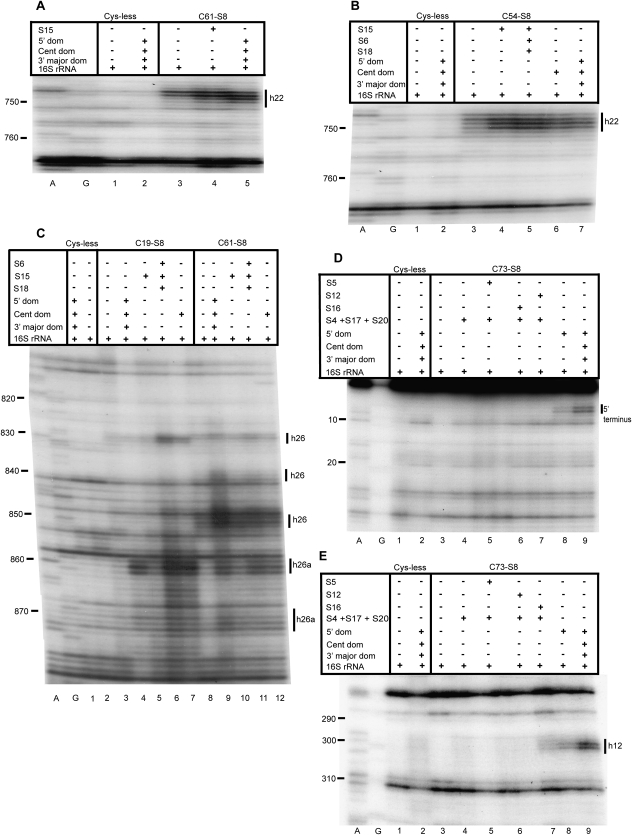
Analysis of the role of individual r-proteins in small subunit assembly. A and G are sequencing lanes. All complexes contain natural 16S rRNA and derivatized cysteine substituents of S8, with the identity of the derivatized protein indicated at the top of each lane as Fe(II)-Cx-S8. Cys-less is the mock-treated cysteine-less control S8. The addition of individual r-proteins and mixtures is indicated by +. See Figure 2 for composition of r-protein mixtures. Primers utilized are 837 (A,B), 939 (C), 161 (D), and 480 (E).
FIGURE 4.
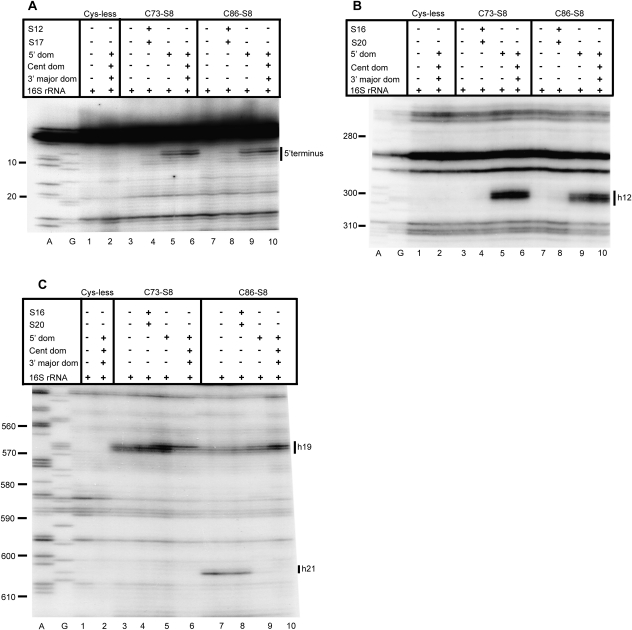
Directed hydroxyl radical cleavage of the 5′ domain of 16S rRNA from Fe(II)-S8 in various RNPs detected by primer extension. A and G are sequencing lanes. All complexes contain natural 16S rRNA and derivatized cysteine substituents of S8, with the identity of the derivatized protein indicated at the top of each lane as Fe(II)-Cx-S8. Cys-less is the mock-treated cysteine-less control S8. The addition of individual r-proteins and mixtures is indicated by +. See Figure 2 for composition of r-protein mixtures. Primers utilized are 232 (A), 480 (B), and 683 (C).
FIGURE 5.
Directed hydroxyl radical cleavage of the 3′ major domain of 16S rRNA from Fe(II)-S8 in various RNPs detected by primer extension. A and G are sequencing lanes. All complexes contain natural 16S rRNA and derivatized cysteine substituents of S8, with the identity of the derivatized protein indicated at the top of each lane as Fe(II)-Cx-S8. Cys-less is the mock-treated cysteine-less control S8. The addition of individual r-proteins and mixtures is indicated by +. See Figure 2 for composition of r-protein mixtures. Primer utilized is 1257.
Previous studies have demonstrated that differences in cleavage at specific sites between two Fe(II)-derivatized complexes assembled and probed simultaneously can be interpreted as indicating alternative conformations or dynamics of 16S rRNA in those complexes (Jagannathan and Culver 2004; Dutca and Culver 2008). Therefore, Fe(II)-S8 containing RNPs of different complexities were probed to dissect assembly events that occur subsequent to S8 association with 16S rRNA and to reprise patterns of Fe(II)-S8 cleavage observed in 30S subunits.
Assembly of the central domain
An interesting dichotomy becomes apparent upon comparison of cleavage patterns in binary complexes and 30S subunits across the central domain of 16S rRNA. All the iron derivatized S8 proteins cleave elements of the central domain of 16S rRNA in 30S subunits. Helix 21 is cleaved along its length on both faces with a similar extent and intensity in both binary complexes and 30S subunits, thus indicating that a mature structure is present in the binary complex (Figs. 2A–C, 6, 7). To elaborate, nucleotides 590–593 and 651–653 are cleaved by Fe(II)-C28-S8 (Fig. 2B, lanes 3,4; Fig. 2C, lanes 3,4), Fe(II)-C54-S8 (Fig. 2A, lanes 5,6; Fig. 2C, lanes 5,6) and Fe(II)-C61-S8 (Fig. 2A, lanes 7,8), while nucleotides 601–605 and nucleotides 630–634 are cleaved by Fe(II)-C107-S8 (Fig. 2B, lanes 9,10; Fig. 2C, lanes 11,12) and Fe(II)-C73-S8 (Fig. 2B, lanes 5,6; Fig. 2C, lanes 7,8), respectively, in binary RNPs as in 30S subunits and 70S ribosomes (Lancaster et al. 2000). Fe(II)-C107-S8 also cleaves between nucleotides 590–593 and 651–653 in both binary complexes and 30S subunits. This is consistent with previous enzymatic and mutational analyses that had been carried out on the binding site of S8 at helix 21 (Stern et al. 1989; Lancaster et al. 2000). There is an increased presence of reverse transcriptase generated stops at nucleotides 603 and 604 in complexes containing cysteine-less substituents of S8 (Fig. 2B, lane 2; Fig. 2C, lane 2); consequently, only the distinct bands generated by Fe(II)-C107-S8/16S rRNA and Fe(II)-C107-S8/30S directed cleavage (Fig. 2B, lanes 9,10; Fig. 2C, lanes 11,12) have been classified as data. Fe(II)-C86-S8/16S rRNA generates a variable cleavage pattern not observed in fully assembled particles at nucleotides 602–603 and 630–635 in half the independent probing experiments conducted (cf. Fig. 2B, lane 7, Fig. 2C, lane 9, and Fig. 4C, lane 7). Since Lysine 86 has both hydrophobic and Van der Waals interactions with C599, these cleavages may represent a disturbance of interactions between the protein and RNA at this site (Schuwirth et al. 2005). However, this variable cleavage pattern is restricted to minimal complexes, while 30S subunits (Fig. 2B, lane 8; Fig. 2C, lane 10; Fig. 4C, lane 10) and 70S ribosomes formed with Fe(II)-C86-S8 (Lancaster et al. 2000) demonstrate reproducible cleavage patterns. Overall, Fe(II)-S8 directed cleavage in helix 21 is highly similar in binary complexes and in fully assembled small subunits.
FIGURE 6.
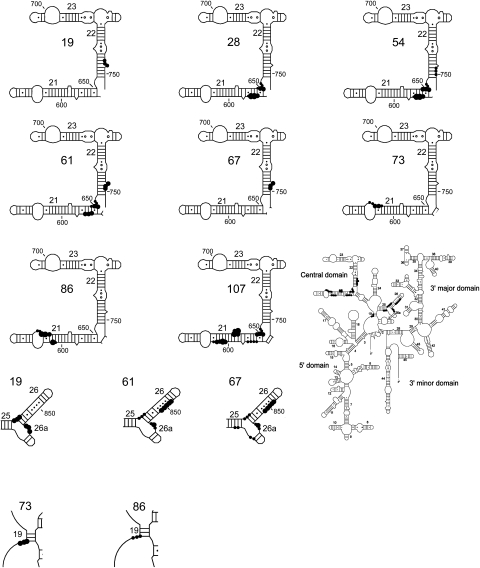
Directed hydroxyl radical cleavage from Fe(II)-S8 in binary complexes mapped on secondary structure of 16S rRNA (Cannone et al. 2002). The cleaved regions are expanded and the Fe(II)-derivatization site generating the cleavage is labeled. Cleavage strength is indicated as strong (big circle), medium (medium circle), and weak (small circle). Fe(II)-C86-S8/16S rRNA exhibits variable cleavage patterns at 601–602 and 630–635.
FIGURE 7.
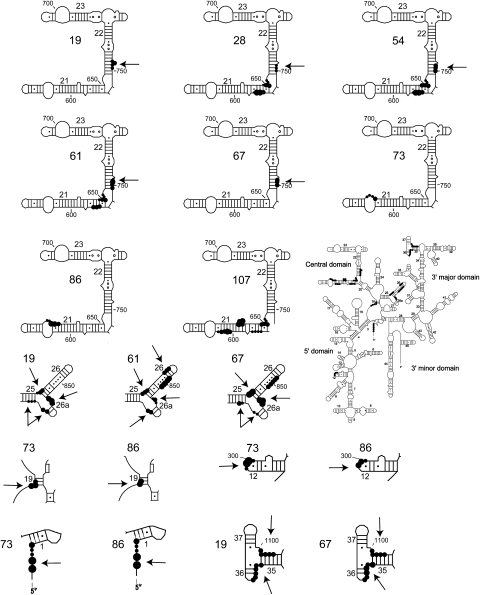
Directed hydroxyl radical cleavage sites from Fe(II)-S8 in 30S subunits mapped on the secondary structure of 16S rRNA (Cannone et al. 2002). Notations are as in Figure 5. Regions that exhibit differences in cleavage patterns or intensities from Figure 5 are distinguished by an arrow.
There are differences in cleavage patterns between binary complexes and 30S subunits at helix 22 (Figs. 2C, 6, 7). In the context of 30S subunits, Fe(II)-C19-S8 and Fe(II)-C67-S8 mediate weak cleavages extending from nucleotides 745–747 (Fig. 2D, lane 4) and 748–750 (Fig. 2D, lane 12), respectively, while Fe(II)-C54-S8 (Fig. 2D, lane 8; Fig. 3B, lane 7) and Fe(II)-C61-S8 (Fig. 2D, lane 10; Fig. 3A, lane 5) cleave strongly between 748 and 750. A 3′ offset relative to cleaved nucleotides in mature subunits occurs in binary complexes containing Fe(II)-C61-S8 (Fig. 2D, lane 9; Fig. 3A, lane 3) and Fe(II)-C67-S8 (Fig. 2D, lane 11). The complex Fe(II)-C28-S8/16S shows no cleavage in helix 22 (Fig. 2D, lane 5). Also, Fe(II)-C54-S8 cleaves with a greater intensity in 30S subunits than in binary complexes (Fig. 2D, lanes 7,8). The incorporation of r-protein S15 in a minimal RNP with Fe(II)-C19-S8, Fe(II)-C61-S8 (Fig. 3A, lane 4), and Fe(II)-C54-S8 (Fig. 3B, lane 4) with 16S rRNA leads to cleavage patterns similar to that in 30S subunits, indicating that r-protein S15 influences the architecture of helix 22. These data are consistent with earlier X-ray crystal structures and directed hydroxyl radical probing studies (Agalarov et al. 2000; Jagannathan and Culver 2003).
The binding site of S8 at the junction of helices 25, 26, and 26a is cleaved by Fe(II)-C19-S8, Fe(II)-C61-S8, and Fe(II)-C67-S8 in both binary complexes and 30S subunits (Figs. 2E, 5, 6). While cleavages at the base (nucleotides 828–830) and middle of helix 26 (nucleotides 848–852) are similar between binary complexes and 30S subunits reconstituted with Fe(II)-C19-S8 (Fig. 2E, lanes 3,6; Fig. 3C, lanes 3,4), Fe(II)-C61-S8 (Fig. 2E, lanes 7,8; Fig. 3C, lanes 8,9), and Fe(II)-C67-S8 (Fig. 2E, lanes 9,10), nucleotides 838–840 which are proximal to the loop are cleaved only in Fe(II)-C61-S8/30S subunit (Fig. 2E, lane 8; Fig. 3C, lane 9). Comparison of cleavage patterns at the base of helix 26a (nucleotides 858–860) indicates a difference in intensity between the 30S subunits (Fig. 2E, lanes 6,8,10; Fig. 3C, lanes 4,9) and binary complexes (Fig. 2E, lanes 3,7,9; Fig. 3C, lanes 3,8) irrespective of the position at which S8 is derivatized. Fe(II)-C19-S8 shows the greatest variation in cleavage intensity between complexes (Fig. 2E, lanes 3,6; Fig. 8A). Based on the aforementioned data, it appears that the more 3′ binding site for S8 is not positioned appropriately in the minimal complexes.
FIGURE 8.
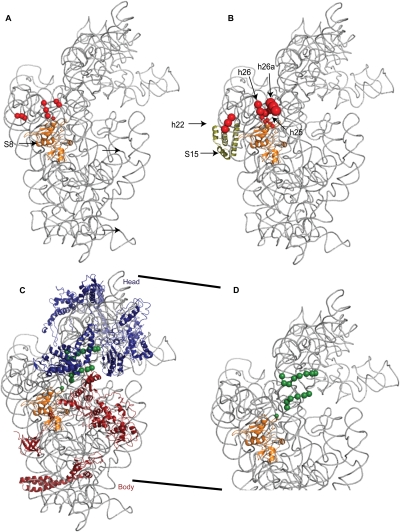
Directed hydroxyl radical cleavage sites from Fe(II)-S8 containing complexes mapped as spheres on the three-dimensional structure of the 30S ribosomal subunit. 16S rRNA is shown in gray and S8 is shown as a cartoon in orange. The other r-proteins are omitted or specifically mentioned in each subpanel. The color of the sphere indicates the probing site used (Fig. 1D), while the size corresponds to cleavage intensity. (A) Cleavages from Fe(II)-C19-S8 in the binary complex in the central domain only. (B) Cleavages from Fe(II)-C19-S8 in a minimal complex with S15, shown in olive green. (C) Cleavages from Fe(II)-C67-S8 in a minimal RNP with the head and body assembled. In blue are proteins that bind the 3′ major domain, in red are the r-proteins that bind the 5′ domain. (D) Same data as in panel C, except that the neck is expanded and r-proteins omitted.
The addition of S15 results in a cleavage pattern similar, although not identical, to that of the fully assembled subunit (Fig. 2E, lane 4; Figs. 8B, 3C, lanes 5,10). Intriguingly, a minimal RNP containing Fe(II)-C19-S8/S15/S6/S18 and 16S rRNA (Fig. 2E, lane 5; Fig. 3C, lane 6) exhibits an increased intensity of cleavage as compared to binary complexes, 30S subunits and Fe(II)-C19-S8/S15/16S rRNA (Fig. 2E, lanes 3,6,4; Fig. 3C, lanes 3,4,5, respectively). Thus, addition of S6 and S18 to a complex of S8 and S15 appears to induce an intermediate structure at helix 26 that is resolved by the addition of all the r-proteins that bind the central domain (Fig. 3C, lane 7) and in fully formed subunits. This suggests that the assembly of the central domain may proceed through intermediate structures that are not necessarily identical to the ultimate conformation but which are then resolved by late binding proteins to assemble to the mature conformation.
Assembly of the 5′ domain
In the 5′ domain there is diminished or absent cleavage in the presence of Fe(II)-S8 and 16S rRNA as compared to 30S subunits (Figs. 4, 6, 7). The 5′ terminus is cleaved by Fe(II)-C73-S8 and Fe(II)-C86-S8 in the context of 30S subunits (Fig. 4A, lanes 6,10) but not in the binary complex (Fig. 4A, lanes 3,7). While a minimal RNP consisting of Fe(II)-C73-S8/S12/S17 (Fig. 4A, lane 4) and Fe(II)-C86-S8/S12/S17 (Fig. 4A, lane 8) is capable of establishing a distinct cleavage pattern between residues 5 and 9, only the addition of all the r-proteins that bind the 5′ domain results in the restoration of cleavage intensity to a level close to that observed in 30S subunits (Fig. 4A, lanes 5,9). A more thorough dissection of the relative significance of individual 5′ domain binding r-proteins in the assembly of this domain showed that the addition of any one of three proteins, S5 (Fig. 3D, lane 5), S16 (Fig. 3D, lane 6), or S12 (Fig. 3D, lane 7), to a minimal complex containing all the primary proteins binding the 5′ domain was not sufficient to elicit cleavage near the 5′ terminus. Cleavages were first observed only upon assembly of the 5′ domain (Fig. 3D, lane 8). This indicates that the addition of r-protein S4 to a complex containing S12, S17, and S8 results in the structural perturbation of that complex. In conclusion, the addition of all the r-proteins binding the 5′ domain is necessary in order to establish either the correct conformation or to limit the dynamicity of the 5′ terminus.
Similarly, nucleotides 298–301 within helix 12 are cleaved in the mature subunits (Fig. 4B, lanes 6,10) but not in binary complexes containing Fe(II)-C73-S8 and Fe(II)-C86-S8 (Fig. 4B, lanes 3,7). This may indicate either an alternative conformation at these residues or a greater dynamicity within the loop of helix 12 in binary complexes. Previous studies have suggested that S16 plays a significant role in stabilizing the tertiary structure of this region (Ramaswamy and Woodson 2009); however, a RNP consisting of S16 and S20 did not mediate cleavage at helix 12 (Fig. 4B, lane 4) while the addition of all the primary proteins binding the 5′ domain to this particle resulted in a weak cleavage at nucleotides 299 and 300 (Fig. 3E, lane 7). In fact, the recapitulation of cleavage patterns requires the presence of all the proteins that bind the 5′ domain (Fig. 4B, lanes 5,9), although the cleavages may be of a reduced intensity as compared to the mature subunit (Fig. 3E, lane 8). From above mentioned data, it can be deduced that the presence of all the 5′ domain binding proteins results in a conformation similar to that in the 30S subunit in helix 12.
Moreover, nucleotide 566 which pairs with nucleotide 299 via purine–purine interactions is cleaved with a decreased intensity in complexes of Fe(II)-C73-S8/16S (Fig. 4C, lane 3) and Fe(II)-C86-S8/16S (Fig. 4C, lane 7) as compared to 30S subunits (Fig. 4C, lanes 6,10). Indeed, the interdomain region between the 5′ and central domains at nucleotides 565–568 is cleaved with a differing intensity between the binary complexes and 30S subunits reconstituted with Fe(II)-C73-S8 or Fe(II)-C86-S8 (Fig. 4C, lanes 3,6,7,10). Addition of r-proteins that bind the 5′ domain results in a cleavage pattern that closely resembles that observed in 30S subunits (Fig. 4C, lanes 5,9). Therefore, assembly of the body leads to restoration of cleavage patterns within the 5′ domain to the level observed in the 30S subunits.
Assembly of the 3′ major domain
The neck (the junction of the head and body) of the 30S subunit is cleaved by Fe(II)-C19-S8 and Fe(II)-C67-S8 at nucleotides 1073–1078 and nucleotides 1101–1105 in 30S subunits (Fig. 5, lanes 6,10). In contrast, no cleavages are observed in binary complexes (Fig. 5, lanes 3,7) while within the 70S ribosome, Fe(II)-C19-S8 cleaves only nucleotides 1099–1104 (Lancaster et al. 2000). Since crystal structures (Ogle et al. 2002; Schuwirth et al. 2005) and cryo-electron microscopy techniques (Agrawal et al. 1999; Gabashvili et al. 1999; Stark et al. 2000) have reported conformational changes in the neck that enable the head to swivel with respect to the body (Ramakrishnan 2002), the change in cleavage patterns between free 30S subunits and 30S subunits associated with 50S subunits in this region of the SSU is not surprising. Reproduction of the cleavage pattern observed in 30S subunits requires the presence of r-proteins found in both the head and the body (Fig. 5, lanes 5,9; Fig. 8C,D). These data lend support to the hypothesis that there is a minor conformational change in this region and/or that this region is more dynamic in binary complexes. While the complex of Fe(II)-C67-S8 with the 5′ and 3′ major domain r-proteins and 16S rRNA recapitulates cleavage patterns at the neck, additional cleavages at nucleotide 1148 and 1149 (Fig. 5, lane 9) are also observed. These nucleotides are beyond cleaving distance from S8 in a particle with an appropriately assembled 5′ and 3′ major domain, and thus most likely represent a subpopulation with aberrant structures in these domains (Joseph et al. 1997; Lancaster et al. 2000). To summarize, assembly of both the head and the body can enable the assembly or orientation of the neck, thereby illustrating the interconnected assembly of the SSU domain structures.
DISCUSSION
Directed hydroxyl radical probing of 30S subunit assembly as carried out using Fe(II)-S8 has shed light on the role of cooperative interactions between r-proteins as well as between structural domains. Outside of the well defined S8 binding site, the cleavage patterns generated by Fe(II)-S8 are altered in binary complexes of S8 and 16S rRNA alone as compared to mature 30S subunits. Multiple equally valid interpretations are possible for this difference, such as more minimal complexes sampling numerous conformations rapidly, or existing in two or more altered conformations of varied stability. This hypothesis can apply to both the r-protein and rRNA component of the complex. At present, in common with other techniques of generating hydroxyl radicals, it is not possible to determine which of these processes result in the particular pattern of cleavage observed (Dixon et al. 1991; Nguyenle et al. 2006).
The total sum of the data presented above, together with previous work (see Sykes and Williamson 2009), indicates that the assembly of components within the subunit, whether categorized by domain or by the binding site of individual r-proteins, is interlinked and hierarchical.
Complementary roles for r-protein S8 and S15 in central domain assembly
The data obtained by directed hydroxyl radical probing of the central domain from Fe(II)-S8 imply that only part of the central domain is appropriately structured in the presence of S8 alone (Figs. 6, 7, 8A,B). The similarity of cleavage patterns across virtually all the derivatized positions at helix 21 leads to the speculation that this helix can assume its tertiary structure in a minimal particle during biogenesis. The NMR structure of the binding site of S8 in the presence and absence of the protein also correlates this hypothesis (Kalurachchi et al. 1997).
On the other hand, the data at the junction of helices 25, 26, and 26a appears to be more complicated. While there is a change in intensity of cleavage at helix 26a and closer to the loop of helix 26, the cleavage at the base and middle of helix 26 appears unchanged between the binary complex and the 30S subunit (Figs. 6, 7, 8A,B). These data are derived from three different probing positions on S8: Fe(II)-C19-S8, Fe(II)-C61-S8, and Fe(II)-C67-S8. Although both Fe(II)-C19-S8 and Fe(II)-C67-S8 are located at similar distances from the 16S rRNA cleavage sites in the tertiary structure, only one of these, Fe(II)-C19-S8, shows a greater change in cleavage intensity at helix 26a. One possible reason for this difference is that the radicals generated from Fe(II)-C67-S8 are quenched by the segment of S8 that binds to the three-helix junction. In addition, alanine 19 of S8 interacts with U827 in the crystal structure of the 30S subunit (Schuwirth et al. 2005), thus the difference in patterns might represent a change in the interaction with 16S rRNA in the complex Fe(II)-C19-S8/16S rRNA. Yet, Fe(II)-C19-S8/16S rRNA cleaves reproducibly at nucleotides 828–830 (Figs. 6, 7, 8A,B). Also, the conformity of changes in cleavage patterns between complexes across all the derivatized positions argues against the data being artifactual in nature. The data presented above imply that the architecture of this element of 16S rRNA is influenced by other r-proteins in addition to S8.
In keeping with data from directed hydroxyl radical probing of the central domain mediated by Fe(II)-S15 (Jagannathan and Culver 2003), a complex of Fe(II)-S8/S15/16S rRNA was found to restore cleavage patterns at the three-helix junction between helices 25–26a (Fig. 8A,B). Since the cleavages are localized to the binding site of S8, three hypotheses are possible. First, S8 could bind transiently to this region in the binary complex and S15 could help directly or indirectly to stabilize the interaction of S8 with 16S rRNA. The transience of this binding could be attributed to the possibility of a conformational change in S8 before it can stably bind this region. Second, although S8 is bound to the three-helix junction, its binding site could differ in conformation from that in the 30S subunit, and S15 could aid in establishing the mature conformation. Third, the presence of a probe at the interaction site between protein and rRNA could destabilize the interaction, and S15 might complement the role of S8. While the last hypothesis is a formal possibility, data from studies on the dynamics of assembly, either by time-resolved hydroxyl radical footprinting (Adilakshmi et al. 2008) or by chemical probing (Powers et al. 1993), support the first two hypotheses. While differing in the specifics of nucleotides at which S8 dependent protections from hydroxyl radicals or chemical probes appear, both data sets demonstrate that different regions within the binding site of S8 at the junction of helices 25, 26, and 26a are protected at different stages of assembly. A study of the assembly of the central domain of Thermus thermophilus led to the proposal of a hierarchy of elements that bind S15, S6, and S18, i.e., helices 20–23 form a core RNP that is required for the association of these r-proteins while helices 24, 25, and 26 were labeled secondary binding elements that bind only the preformed core RNP (Agalarov and Williamson 2000). Thus, based on data presented in this and previous studies, it is possible to extend this theory to the assembly of the central domain in Escherichia coli, so that the junction of helices 20–22 could be considered the core element that associates with S8 and S15 while helices 25–26a are secondary binding elements that need the presence of the preformed core RNP in order to assemble. While our data are not sufficient to identify which hypothesis or combination thereof contributes to the results, it is clear that both S8 and S15, the early binding proteins that associate with the central domain, aid in the assembly of the platform. Therefore, it is possible to speculate that compaction of the central domain during biogenesis in vivo is a function of both S8 and S15 r-proteins. The function of S15 in central domain assembly appears to be redundant, as a strain bearing a deletion of rpsO is viable (Bubunenko et al. 2006). However, S15 may increase the efficiency of assembly in wild-type strains by aiding in the association of S8 with 16S rRNA and central domain compaction.
Certain elements of the 5′ domain assume a mature architecture only upon assembly to the body
The data from the 5′ domain reveal a general trend, in that the 5′ terminus, the loop of helix 12, and the stem of helix 19 are cleaved by Fe(II)-S8 to yield patterns resembling the 30S subunit only upon addition of r-proteins that bind the 5′ domain (Figs. 6, 7). The lack or decreased intensity of cleavages in binary complexes could be due to alternative conformations of these particles. Another probable explanation is that, in the absence of specific r-proteins, certain regions of 16S rRNA may be more dynamic. The appropriate orientation of the 5′ terminus is functionally significant as it is proximal to the central pseudoknot, which is a key player in translation (Poot et al. 1998), and alterations in the architecture of this region have been shown to affect fidelity (Dammel and Noller 1993; Roy-Chaudhuri et al. 2010).
While recent hydroxyl radical footprinting data suggest that S16 plays a significant role in orienting helix 12 (Ramaswamy and Woodson 2009), in our study a complex of Fe(II)-C73-S8/S16/S20/16S was unable to cleave helix 12 (Fig. 4B, lane 4). The difference in observations may be a result of the differing sensitivities of the techniques of footprinting and directed hydroxyl radical probing. Furthermore, while the study by Ramaswamy and Woodson (2009) utilized in vitro transcribed 5′ domain RNA, this study utilized full length modified 16S rRNA, consequently architectural differences may exist between the two particles.
Analysis of X-ray crystal structures reveals that among the 5′ domain proteins, S5 interacts with the 5′ terminus and G568 while S12 interacts with helix 19 and the loop of helix 12 (Allers and Shamoo 2001; Schuwirth et al. 2005). Indeed, in a previous study, the assembly of S5 caused residues at the 5′ terminus and the junction of 5′ and central domains to have altered susceptibility to reactivity with chemical probes (Stern et al. 1988). These effects were exaggerated in a complex of S5 and S12 with 16S rRNA. Moreover, Fe(II)-S5 mediates cleavage of the 5′ terminus in 30S subunits (Culver et al. 1999). However, a minimal particle of Fe(II)-C73-S8/S5/16S rRNA failed to recapitulate cleavages observed in Fe(II)-S8/30S subunit (data not shown). This conundrum may be resolved by recent studies on the dynamics of protein incorporation. Using the technique of pulse-chase quantitative mass spectrometry, it was determined that S12 and S5 are incorporated into the assembling particle at a relatively slow rate compared to the remainder of the 5′ domain binding proteins (Talkington et al. 2005). In conclusion, it is likely that these proteins are responsible for stabilizing or ordering these regions of the 5′ domain in the presence of all the other 5′ domain binding proteins.
Assembly of the 3′ major domain alone is not sufficient for appropriate orientation of the neck
It was found that the head is capable of rotating at the neck (Schuwirth et al. 2005), thus the difference in cleavage patterns between 70S ribosomes (Lancaster et al. 2000), free 30S subunits, and binary particles within this region is not surprising (Figs. 6, 7, 8C,D). The quenching of radicals from Fe(II)-C19-S8 or changes in conformation of this region upon subunit association could explain the diminished cleavages in 70S ribosomes. The lack of cleavages in the binary complex could be indicative of conformational changes in the neck, or due to increased motion of the neck, or an altered alignment of the 3′ domain with respect to the 5′ domain. Yet, addition of r-proteins that bind the 3′ domain to form the head did not result in the restoration of Fe(II)-S8 cleavage patterns. Previously, a fragment of 16S rRNA corresponding to the 3′ major domain had been assembled with r-proteins to form the head, and was demonstrated to possess reactivity to chemical probes and spectinomycin binding affinity similar to the head domain in intact 30S subunits (Samaha et al. 1994). Taken together, these studies suggest that assembly of the head in the presence of S8 is not sufficient to organize the neck. The recapitulation of cleavage patterns requires the presence of all the r-proteins binding the head and body. While it is highly probable that the interaction of S5 with the head and body plays a significant role in the restoration of cleavage patterns (Schuwirth et al. 2005), a minimal RNP of Fe(II)-C19-S8/S5/16S and Fe(II)-C67-S8/S5/16S does not cleave the neck (data not shown). One explanation for this is that S5 is dependent upon multiple proteins for incorporation (see Fig. 1A). Also, as mentioned above, S5 binds with slower kinetics than other 5′ domain binding proteins with the exception of S12. It is also possible that S8 is not appropriately aligned in particles containing only the r-proteins binding the 3′ major domain, and addition of r-proteins that bind the body aids in appropriate organization. Since the cleavages occur close to the functional center of the subunit, the necessity for the appropriate assembly of multiple domains in order to achieve mature structure may be a quality control mechanism during in vivo biogenesis. Moreover, previous studies have noted that the movement of the head relative to the remainder of the subunit may play a significant role in increasing the fidelity of translation (see Zaher and Green 2009).
A common principle that appears to underlie the results presented above is the interdependence of assembly between different regions of 16S rRNA, both within a domain and across domains. The assembly of the junction of helices 25–26a appears to be dependent upon the establishment of appropriate structure at the junction of helices 20–23. Similarly, the orientation of the neck is dependent upon the assembly of the head and body. Indeed, the 30S subunit can be considered a collection of ribonucleoprotein complexes that fold through modular pathways that influence one another. This hierarchy of folding behavior has been compared to another large, multidomain RNA, the Tetrahymena ribozyme (Laggerbauer et al. 1994; Zarrinkar and Williamson 1994; Agalarov and Williamson 2000), and has also been observed in the group II intron ribozyme ai5γD135 (Su et al. 2005). Thus, multidomain RNAs may fold through pathways that contain intermediates characterized by segmented folding, with the folding of certain key segments playing a significant role in the architecture of the entire molecule.
MATERIALS AND METHODS
Mutagenesis, expression, purification, and derivatization of S8
Previously, a cysteine-less mutant of rpsH had been constructed in pET 21b vector by the replacement of C126 with Alanine, and eight positions had been substituted to cysteine residues in this background. All nine constructs were expressed and the cells were collected as described (Lancaster et al. 2000). The cells were lysed in Buffer E (20 mM K+-HEPES [pH 7.6], 20 mM KCl, and 6 mM βME) in an Avestin cell lyser. Inclusion bodies containing S8 were pelleted by spinning at 7000 rpm for 20 min at 4°C and resuspended in Buffer C (20 mM sodium acetate [pH 5.6], 1 M KCl, 6 M Urea and 6 mM βME) followed by overnight dialysis into Buffer B (20 mM sodium acetate [pH 5.6], 20 mM KCl, 6 M Urea and 6 mM βME) with three changes. The purification protocol was also similar to Lancaster et al. (2000) except that elution was carried out over a 125 mL salt gradient from 20 to 130 mM KCl in Buffer B and the proteins eluted at ∼22 mM KCl. Purified proteins were dialyzed into Buffer D (80 mM K+-HEPES [pH 7.6], 1 M KCl, and 6 mM βME), aliquoted and frozen in liquid nitrogen. The derivatization reaction was carried out as in Lancaster et al. (2000).
Reconstitution and purification of RNPs containing derivatized protein
16S rRNA was isolated as described (Moazed et al. 1986), heat-activated by incubating at 42°C for 15 min in Buffer A (80 mM K+-Hepes [pH 7.6], 330 mM KCl, 20 mM MgCl2, and 0.01% Nikkol), and then incubated on ice for 10 min. The binary complexes were formed by mixing 240 pmol of Fe(II)-derivatized proteins with 40 pmol of 16S rRNA in a final volume of 100 μL with a final KCl concentration of 330 mM. If a more complex RNP was desired, the derivatized proteins were mixed with either 360 pmol of individual proteins or 120 pmol of domain mixes prior to the addition of RNA. The 5′ domain mix is composed of r-proteins S4, S5, S12, S16, S17, and S20, the central domain mix of r-proteins S6, S11, S15, S18, and S21 while the 3′ major domain mix is composed of r-proteins S2, S3, S7, S9, S10, S13, S14, S19. In order to form the RNPs, the reactions were incubated at 42°C for 20 min. These were then cooled on ice for 10 min and purified by passing them through spin columns with a speed of 2500 rpm for 3.5 min as described (Dutca and Culver 2008). In order to minimize non-directed probing, 1.6 μL of 125 mM EDTA was added to yield a final concentration of 2 mM EDTA per reaction except to the complexes depicted in Figure 2C. The RNPs were incubated on ice for 10 min prior to directed hydroxyl radical probing.
Directed hydroxyl radical probing
Directed hydroxyl radical probing and primer extension analysis were carried out on various RNPs as in Culver and Noller (2000a). Each probing experiment and the subsequent analysis were performed at least two times and intensity of cleavage was determined visually.
Structural analysis
All analysis was carried out on the PDB file 2AW7 (Schuwirth et al. 2005). The structure was visualized by PyMOL (The PyMOL Molecular Graphics System). The program ENTANGLE was utilized to examine interactions within the structure (Allers and Shamoo 2001).
ACKNOWLEDGMENTS
The S8 cysteine substituents were a gift from the laboratory of Dr. H.F. Noller (University of California Santa Cruz). We thank all members of the Culver lab. This work was supported by NIH grant GM62432 to G.M.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2332511.
REFERENCES
- Adilakshmi T, Bellur DL, Woodson SA 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalarov SC, Williamson JR 2000. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA 6: 402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalarov SC, Zheleznyakova EN, Selivanova OM, Zheleznaya LA, Matvienko NI, Vasiliev VD, Spirin AS 1998. In vitro assembly of a ribonucleoprotein particle corresponding to the platform domain of the 30S ribosomal subunit. Proc Natl Acad Sci 95: 999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalarov SC, Selivanova OM, Zheleznyakova EN, Zheleznaya LA, Matvienko NI, Spirin AS 1999. Independent in vitro assembly of all three major morphological parts of the 30S ribosomal subunit of Thermus thermophilus. Eur J Biochem 266: 533–537 [DOI] [PubMed] [Google Scholar]
- Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR 2000. Structure of the S15,S6,S18-rRNA complex: Assembly of the 30S ribosome central domain. Science 288: 107–113 [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6: 643–647 [DOI] [PubMed] [Google Scholar]
- Allers J, Shamoo Y 2001. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol 311: 75–86 [DOI] [PubMed] [Google Scholar]
- Bubunenko M, Korepanov A, Court DL, Jagannathan I, Dickinson D, Chaudhuri BR, Garber MB, Culver GM 2006. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA 12: 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 doi: 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HT, Hahn S 2003. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol Cell 12: 437–447 [DOI] [PubMed] [Google Scholar]
- Clemons WM Jr, May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V 1999. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature 400: 833–840 [DOI] [PubMed] [Google Scholar]
- Culver GM, Noller HF 2000a. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol 318: 461–475 [DOI] [PubMed] [Google Scholar]
- Culver GM, Noller HF 2000b. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol 318: 446–460 [DOI] [PubMed] [Google Scholar]
- Culver GM, Heilek GM, Noller HF 1999. Probing the rRNA environment of ribosomal protein S5 across the subunit interface and inside the 30 S subunit using tethered Fe(II). J Mol Biol 286: 355–364 [DOI] [PubMed] [Google Scholar]
- Cunningham PR, Richard RB, Weitzmann CJ, Nurse K, Ofengand J 1991. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S ribosomal subunit of Escherichia coli. Biochimie 73: 789–796 [DOI] [PubMed] [Google Scholar]
- Dammel CS, Noller HF 1993. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev 7: 660–670 [DOI] [PubMed] [Google Scholar]
- Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD 1991. Hydroxyl radical footprinting. Methods Enzymol 208: 380–413 [DOI] [PubMed] [Google Scholar]
- Dutca LM, Culver GM 2008. Assembly of the 5′ and 3′ minor domains of 16S ribosomal RNA as monitored by tethered probing from ribosomal protein S20. J Mol Biol 376: 92–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Grassucci R, Squires CL, Dahlberg AE, Frank J 1999. Major rearrangements in the 70S ribosomal 3D structure caused by a conformational switch in 16S ribosomal RNA. EMBO J 18: 6501–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondek JF, Culver GM 2004. Assembly of the 30S ribosomal subunit: Positioning ribosomal protein S13 in the S7 assembly branch. RNA 10: 1861–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held WA, Mizushima S, Nomura M 1973. Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J Biol Chem 248: 5720–5730 [PubMed] [Google Scholar]
- Held WA, Ballou B, Mizushima S, Nomura M 1974. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem 249: 3103–3111 [PubMed] [Google Scholar]
- Jagannathan I, Culver GM 2003. Assembly of the central domain of the 30S ribosomal subunit: Roles for the primary binding ribosomal proteins S15 and S8. J Mol Biol 330: 373–383 [DOI] [PubMed] [Google Scholar]
- Jagannathan I, Culver GM 2004. Ribosomal protein-dependent orientation of the 16 S rRNA environment of S15. J Mol Biol 335: 1173–1185 [DOI] [PubMed] [Google Scholar]
- Joseph S, Weiser B, Noller HF 1997. Mapping the inside of the ribosome with an RNA helical ruler. Science 278: 1093–1098 [DOI] [PubMed] [Google Scholar]
- Kalurachchi K, Uma K, Zimmermann RA, Nikonowicz EP 1997. Structural features of the binding site for ribosomal protein S8 in Escherichia coli 16S rRNA defined using NMR spectroscopy. Proc Natl Acad Sci 94: 2139–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Hughes D, Ehrenberg M 1996. Limitations of translational accuracy. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology (ed. Neidhardt FC et al. ), Vol. 1, pp. 979–1004 ASM Press, Washington DC [Google Scholar]
- Laggerbauer B, Murphy FL, Cech TR 1994. Two major tertiary folding transitions of the Tetrahymena catalytic RNA. EMBO J 13: 2669–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Culver GM, Yusupova GZ, Cate JH, Yusupov MM, Noller HF 2000. The location of protein S8 and surrounding elements of 16S rRNA in the 70S ribosome from combined use of directed hydroxyl radical probing and X-ray crystallography. RNA 6: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Varma S, SantaLucia J Jr, Cunningham PR 1997. In vivo determination of RNA structure-function relationships: Analysis of the 790 loop in ribosomal RNA. J Mol Biol 269: 732–743 [DOI] [PubMed] [Google Scholar]
- Lodmell JS, Dahlberg AE 1997. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277: 1262–1267 [DOI] [PubMed] [Google Scholar]
- Merryman C, Moazed D, McWhirter J, Noller HF 1999. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J Mol Biol 285: 97–105 [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nomura M 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226: 1214–1218 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1990. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol 211: 135–145 [DOI] [PubMed] [Google Scholar]
- Moazed D, Stern S, Noller HF 1986. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol 187: 399–416 [DOI] [PubMed] [Google Scholar]
- Mougel M, Allmang C, Eyermann F, Cachia C, Ehresmann B, Ehresmann C 1993. Minimal 16S rRNA binding site and role of conserved nucleotides in Escherichia coli ribosomal protein S8 recognition. Eur J Biochem 215: 787–792 [DOI] [PubMed] [Google Scholar]
- Muller R, Garrett RA, Noller HF 1979. The structure of the RNA binding site of ribosomal proteins S8 and S15. J Biol Chem 254: 3873–3878 [PubMed] [Google Scholar]
- Nguyenle T, Laurberg M, Brenowitz M, Noller HF 2006. Following the dynamics of changes in solvent accessibility of 16 S and 23 S rRNA during ribosomal subunit association using synchrotron-generated hydroxyl radicals. J Mol Biol 359: 1235–1248 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Poot RA, van den Worm SH, Pleij CW, van Duin J 1998. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res 26: 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF 1990. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci 87: 1042–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF 1995. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA 1: 194–209 [PMC free article] [PubMed] [Google Scholar]
- Powers T, Daubresse G, Noller HF 1993. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol 232: 362–374 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V 2002. Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Ramaswamy P, Woodson SA 2009. S16 throws a conformational switch during assembly of 30S 5′ domain. Nat Struct Mol Biol 16: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Kirthi N, Culver GM 2010. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci 107: 4567–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha RR, O'Brien B, O'Brien TW, Noller HF 1994. Independent in vitro assembly of a ribonucleoprotein particle containing the 3′ domain of 16S rRNA. Proc Natl Acad Sci 91: 7884–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaup HW, Sogin ML, Kurland CG, Woese CR 1973. Localization of a binding site for ribosomal protein S8 within the 16S ribosomal ribonucleic acid of Escherichia coli. J Bacteriol 115: 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W 2000. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100: 301–309 [DOI] [PubMed] [Google Scholar]
- Stern S, Powers T, Changchien LM, Noller HF 1988. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16 S rRNA. J Mol Biol 201: 683–695 [DOI] [PubMed] [Google Scholar]
- Stern S, Powers T, Changchien LM, Noller HF 1989. RNA-protein interactions in 30S ribosomal subunits: Folding and function of 16S rRNA. Science 244: 783–790 [DOI] [PubMed] [Google Scholar]
- Su LJ, Waldsich C, Pyle AM 2005. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res 33: 6674–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes MT, Williamson JR 2009. A complex assembly landscape for the 30S ribosomal subunit. Annu Rev Biophys 38: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Garrett R, Ehresmann C, Stiegler P, Fellner P 1975. An investigation of the 16-S RNA binding sites of ribosomal proteins S4, S8, S15, and S20 from Escherichia coli. Eur J Biochem 51: 165–180 [DOI] [PubMed] [Google Scholar]
- Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J 1993. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J 7: 177–180 [DOI] [PubMed] [Google Scholar]
- Wower I, Brimacombe R 1983. The localization of multiple sites on 16S RNA which are cross-linked to proteins S7 and S8 in Escherichia coli 30S ribosomal subunits by treatment with 2-iminothiolane. Nucleic Acids Res 11: 1419–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Jiang L, Zimmermann RA 1994. The binding site for ribosomal protein S8 in 16S rRNA and spc mRNA from Escherichia coli: Minimum structural requirements and the effects of single bulged bases on S8-RNA interaction. Nucleic Acids Res 22: 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, O'Farrell HC, Rife JP, Culver GM 2008. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol 15: 534–536 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009. Fidelity at the molecular level: Lessons from protein synthesis. Cell 136: 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR 1994. Kinetic intermediates in RNA folding. Science 265: 918–924 [DOI] [PubMed] [Google Scholar]
- Zimmermann RA, Mackie GA, Muto A, Garrett RA, Ungewickell E, Ehresmann C, Stiegler P, Ebel JP, Fellner P 1975. Location and characteristics of ribosomal protein binding sites in the 16S RNA of Escherichia coli. Nucleic Acids Res 2: 279–302 [DOI] [PMC free article] [PubMed] [Google Scholar]