Abstract
High-conductance voltage- and Ca2+-activated K+ (BK) channels encode negative feedback regulation of membrane voltage and Ca2+ signaling, playing a central role in numerous physiological processes. We determined the x-ray structure of the human BK Ca2+ gating apparatus at a resolution of 3.0 angstroms and deduced its tetrameric assembly by solving a 6-angstrom resolution structure of a Na+-activated homolog. Two tandem C-terminal regulator of K+ conductance (RCK) domains from each of four channel subunits form a 350-kilodalton gating ring at the intracellular membrane surface. A sequence of aspartic amino acids that is known as the Ca2+ bowl, and is located within the second of the tandem RCK domains, creates four Ca2+ binding sites on the outer perimeter of the gating ring at the “assembly interface” between RCK domains. Functionally important mutations cluster near the Ca2+ bowl, near the “flexible interface” between RCK domains, and on the surface of the gating ring that faces the voltage sensors. The structure suggests that the Ca2+ gating ring, in addition to regulating the pore directly, may also modulate the voltage sensor.
High-conductance voltage- and calcium-activated K+ channels (BK or Slo1 channels) participate in numerous physiological processes, including neuronal excitability, smooth muscle contractility, and hair cell tuning (1-6). BK channels have an unusually high single-channel conductance, but their most important physiological property is dual regulation through membrane voltage and intracellular Ca2+ (7-9). Depolarization of the membrane voltage and increased intracellular Ca2+ levels both cause BK channels to open, which hyperpolarizes the membrane and closes voltage-dependent channels, including Ca2+ channels, reducing Ca2+ influx into the cell. Thus, BK channels are negative-feedback regulators of electrical excitation (membrane depolarization) as well as the numerous biochemical pathways that are stimulated through Ca2+ acting as a second messenger.
The complexity of BK channel function mirrors the complexity of its protein structure. The amino acid sequence includes the integral membrane pore shared by all K+ channels, the integral membrane voltage sensor domains present in voltage-dependent channels, and also a cytoplasmic domain (CTD) consisting of approximately 800 amino acids per subunit, which accounts for the C-terminal two thirds of the entire channel. The CTD structure confers upon the BK channel its ability to respond to changes in intracellular Ca2+. It is also the source of functional heterogeneity through alternate splicing, polymorphisms, phosphorylation, and protein interactions, which modulate BK channel activity (10-12).
The only information currently available on BK channel structure is either low resolution from cryogenic electron microscopy (cryo-EM) (13) or indirect through homology models. The pore and voltage sensors of the BK channel will undoubtedly resemble the corresponding regions of other voltage-dependent K+ channels, but the CTD structure is less certain. The BK C terminus was proposed to contain two regulator of K+ conductance (RCK) domains (14). RCK domains are found in certain prokaryotic K+ channels and K+ transport systems, but the sequence identity between BK RCK domains and prokaryotic RCK domains is so low (<20%) that homology models for the BK CTD contain a high degree of uncertainty.
Mutagenesis studies have identified the putative primary Ca2+ binding site within the BK CTD, which comprises a stretch of aspartic amino acids referred to as the “Ca2+ bowl” (15). Na+-activated (Slo2) and H+-dependent (Slo3) channels, which are homologous to the BK channel, contain a similar CTD but have different sequences of amino acids in the region corresponding to the Ca2+ bowl (9). Mutations within the Ca2+ bowl influence Ca2+ activation, whereas mutations outside, spread throughout the BK CTD primary sequence, also influence various aspects of gating. These mutational data promise rich mechanistic interpretation in the context of an atomic structure. Here, we present the structure of the human BK CTD and its quaternary organization on the cytoplasmic surface of the BK channel.
BK domain organization and structure determination
The BK channel contains four identical subunits, each comprising seven transmembrane segments and a large intracellular C terminus (Fig. 1A) (16). Transmembrane segments S1 to S4 form the voltage sensor; S5, S6, and the intervening amino acids form the pore and selectivity filter; and the C terminus forms the large CTD. The locations of disease-causing mutations, alternate splice sites, regulatory sites, and site-directed mutations that alter specific channel properties are highlighted (Fig. 1B). Blue and red secondary structure elements, on the basis of the x-ray structure presented here, indicate the locations of two RCK domains (RCK1 and RCK2). The Ca2+ bowl sequence is located within RCK2.
Fig. 1.
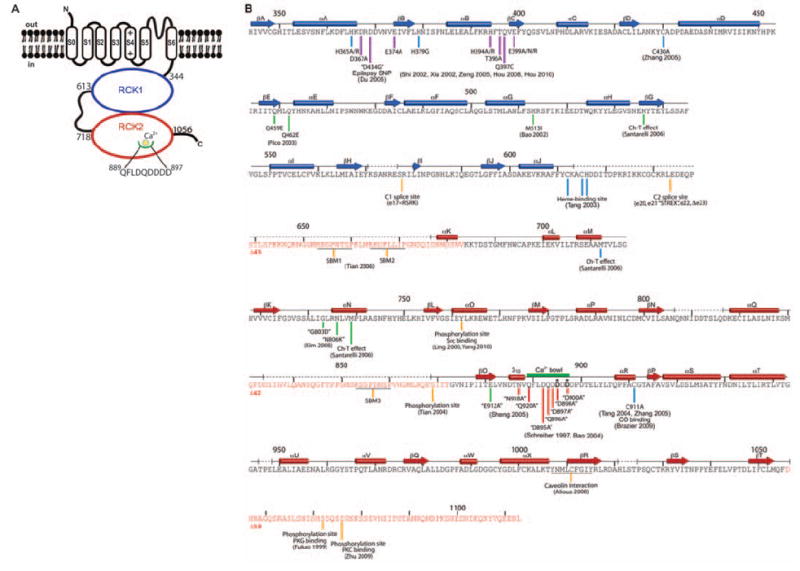
The BK channel. (A) Domain topology of BK. Domain boundaries are indicated, and the Ca2+ bowl is illustrated. (B) Secondary structure of human BK CTD. Secondary structure elements from the crystal structure are aligned to the BK sequence, showing tandem RCK domains (RCK1, blue; RCK2, red) with unresolved segments indicated by dashed lines. Deletion mutants Δ45, Δ42, and Δ58 of unresolved segments (sequences highlighted in red) were characterized functionally (Fig. 2, C and D). The Ca2+ bowl is indicated as a thick green line. Site-specific functional data are marked beneath the sequence by vertical lines with references. The complete list of corresponding references is given in table S3. The color of the lines specify the following categories: red, mutations around the Ca2+ bowl; purple, mutations facing the voltage sensor domain in our BK model (Fig. 6); green, mutations near the αF-αG/αS-αT flexible interface; yellow, splice sites, protein-protein interaction sites, and phosphorylation sites; and blue, additional sites of mutation and modification.
Residues 341 to 1056 were expressed in Spodoptera frugiperda (sf 9) insect cells and purified to homogeneity. The excluded 57 C-terminal amino acids are not essential to function (Δ58 in Fig. 2D) (17). Crystals belonging to space group P6322 with one monomer per asymmetric unit were grown in the presence of 50 mM Ca2+ and diffracted x-rays to a resolution of 3.0 Å. Experimental phases to 3.3 Å were derived from a multi-wavelength anomalous diffraction experiment with a selenomethionine-containing crystal. Experimental electron density is shown (fig. S1A) alongside electron density (2fo-fc) calculated to a resolution of 3.0 Å by using the final model refined to Rwork/Rfree = 0.25/0.28 (fig. S1B), where Rwork is the working R factor and Rfree is the free R factor.
Fig. 2.
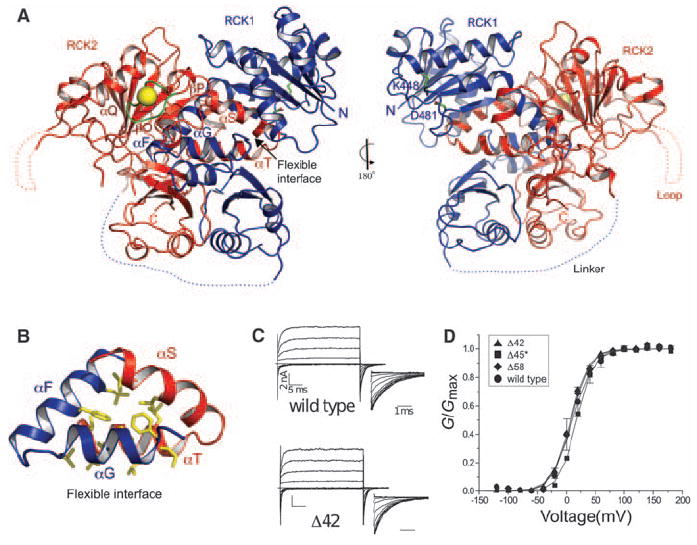
Structure of the BK CTD. (A) Ribbon representations of front and back views of the BK CTD showing RCK1 in blue and RCK2 in red. The Ca2+ bowl is colored in green, and the Ca2+ ion is shown as a yellow sphere. Large disordered segments are indicated as dashed lines. (B) Close-up view of the αF-αG/αS-αT flexible interface. Side chains of hydrophobic residues in the interface are shown as sticks. (C) Channel activation. Currents from inside-out patches are shown for wild-type BK and Δ42 deletion mutant in the presence of 10 μM Ca2+. Voltage pulses are −120 to +180 mV, ΔV = 20 mV, and a holding potential of −60 mV. (D) Normalized G-V curves for wild type and deletion mutants in 10 μM Ca2+. Δ45* was tested by means of co-expression of two separated channel segments instead of deletion from a single construct.
Structure of the BK CTD
The crystal structure defines two tandem RCK domains in the BK C terminus, shown in blue (RCK1) and red (RCK2) (Figs. 1B and 2A). Each RCK domain has a bi-lobed shape. The larger N-terminal lobe of each RCK domain forms a Rossmann fold, which is attached to a smaller C-terminal lobe via a helix-turn-helix connector. The tandem RCK1-RCK2 domains are folded tightly against each other, forming an interface that is reminiscent of the “flexible interface” created through the dimerization of two separate identical RCK domains in the prokaryotic MthK K+ channel (fig. S2) (18). As in the MthK RCK dimer, the flexible interface is dominated by a clasping of the helix-turn-helix connectors that attach the two lobes of each RCK domain (Fig. 2, A and B). Although the overall architecture and signature features are conserved, the BK RCK domains vary in certain details from their prokaryotic counterparts; one result of the variations is a more extensive interface between RCK domains in the BK channel.
The RCK1-RCK2 interface buries 8500 Å2 of solvent-accessible surface area and has a shape complementarity (SC) index of 0.68. The SC index is a geometric measure of a matched protein-protein interface (19). The value here is similar to that for a typical antibody-antigen interface (0.64 to 0.68) and supports the existence of a highly specific, strong interaction. This finding is compatible with experiments that show that functional channels result when two-channel fragments, separated between the RCK domains, are co-expressed in the same cell (17, 20). The extensive interface between the RCK domains supports co-assembly of the individual components, which is analogous to the assembly of separate identical RCK domains in the MthK channel (18).
Two amino acid segments are not visible in the crystal structure (Figs. 1B and 2A). The absence of electron density for the long linker connecting RCK1 to RCK2 is consistent with this region being unstructured and nonessential to function. Deletion of 45 amino acids from this loop in an experiment in which two channel fragments were co-expressed in the same cell resulted in channels with wild-type properties (Δ45* in Fig. 2D). Past studies showed that the specific amino acid sequence of this region is not important but that it has to reach at least a certain minimum length (21). The crystal structure explains the length requirement because the end of RCK1 is located a long distance away from the beginning of RCK2 (Fig. 2A, blue dotted linker). The second unstructured region connects αQ to βO in RCK2 (Figs. 1B and 2A). A 42 amino acid deletion of this loop yields normally functioning channels (Δ42 in Fig. 2, C and D). Although these two unstructured segments are not essential to basic channel function, they may play a role in higher-level channel regulation because both contain consensus sequences for binding SH3 domains (Fig. 1B) (12).
A pair of salt bridge–forming amino acids was identified in the structure of the Escherichia coli RCK domain (14). Double mutant cycle analysis of the corresponding amino acids predicted that this salt bridge is conserved in the BK channel, and this is confirmed in the structure: Amino acids K448 and D481, which are well defined in the electron density, show distances in the final model between Lys N-ζ and Asp O-δ1 and O-δ2 of 2.7 Å and 3.3 Å, respectively, which is compatible with a salt-bridge interaction (Fig. 2A) (14, 17, 22).
In the MthK channel, RCK dimers were proposed to undergo Ca2+-induced conformational changes to open the pore (18, 23). Two RCK subunits adopted distinct relative positions across the flexible interface in different crystal forms and as a function of whether or not ligand was bound (18, 23, 24). The flexible interface between two tandem RCK domains may play a similar role in Ca2+-mediated gating of the BK channel.
The Ca2+ bowl
A major focus of research on the BK channel has been aimed at defining the region of Ca2+ binding (8). Deletions and point mutations within the Ca2+ bowl sequence, DQDDDDDPD, rendered the channel less sensitive to Ca2+ and caused a shift in the conductance-voltage (G-V) curve to more depolarized membrane voltages (15). The activation process exhibited high selectivity for Ca2+ over Cd2+, even though the ionic radius of Cd2+ (0.97 Å) is only slightly smaller than that of Ca2+ (0.99 Å) (15). Other studies demonstrated a good correlation between Ca2+ binding to a C-terminal fragment of the channel using a radioactive Ca2+ assay and activation of the channel using an electrophysiological assay (25, 26). Alanine-scanning mutagenesis of the Ca2+ bowl identified the most important amino acids for Ca2+ sensing to be those corresponding to D895 and D897 in the human BK channel (26). Because of the uncertainty and disagreement in sequenced-based models of the BK channel CTD, there have been different proposals for the placement of the Ca2+ bowl sequence within the context of a model that is based on tandem RCK domains (17, 27).
Experimental electron density for the Ca2+ bowl contained less detail than most of the experimental map, but the main chain was continuous, and certain side chain features emerged with cycles of model building and refinement (Fig. 3, A and B). The model was built on the basis of crystallographic data alone, independently of information from prior mutational studies. An atom that is more electron-dense than carbon, nitrogen, or oxygen accounted for the strongest electron density at the center of the structure formed by the Ca2+ bowl sequence (Fig. 3C). This was presumably a Ca2+ ion, given its presence at 50 mM concentration in the crystallization solution.
Fig. 3.
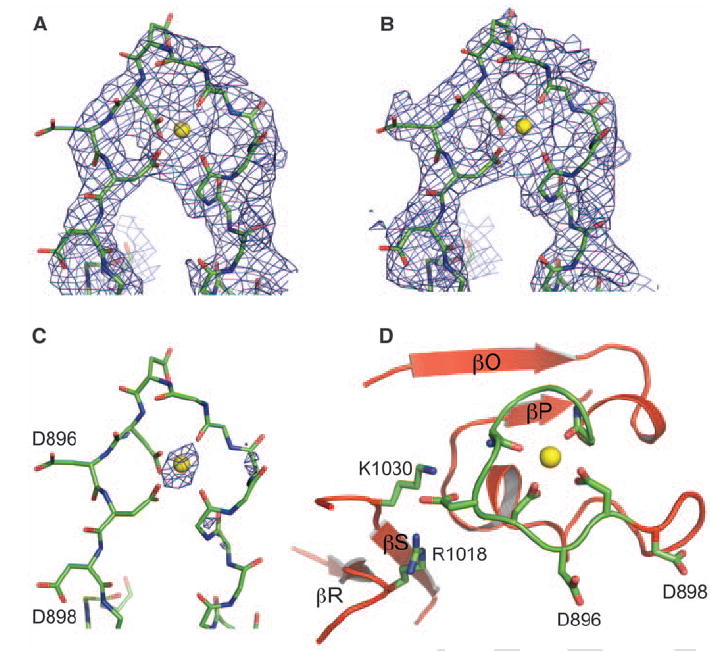
The Ca2+ bowl. (A) Experimental electron density at 3.3 Å in the Ca2+ bowl region contoured at 1.0 above the mean density, where σ is the RMSD of the density (B and C) Weighted 2fo-fc electron density at 3.0 Å after refinement contoured at 1.0 σ and 4.0 σ, respectively. The final refined model is shown as sticks, with most side chains removed for clarity. (D) Structure of the Ca2+ bowl, showing key residues coordinating the Ca2+ ion (yellow sphere).
In the context of the entire CTD, the Ca2+ bowl is located between the last two β strands, βO and βP of the RCK2 Rossmann fold (Figs. 1B, 2A, and 3D). Thus, the Ca2+ bowl does not follow but rather is an integral structural element of RCK2. Atoms making direct contact with the Ca2+ ion include two main-chain carbonyl oxygen atoms from Q889 and D892 and oxygen atoms from the side-chain carboxylate groups of D895 and D897. In searching the protein database of known structures, we observed a similar geometry of Ca2+ binding in a high-resolution crystal structure of calpain, which is an intracellular cysteine protease whose activity is regulated by Ca2+ (fig. S3).
Because of the quality of electron density, there is some uncertainty in the conformation of the Ca2+ binding site in our model. However, the model is consistent with the scanning mutagenesis studies of Cox and colleagues (26). The two most critical residues for Ca2+ activation, D895 and D897, are both directly involved in the coordination of the Ca2+ ion in the x-ray structure (Fig. 3D). The third most effective mutation involved D894, which is not in direct contact with Ca2+ but forms salt bridges simultaneously with R1018 and K1030 in the C-terminal lobe of RCK2. D894 probably plays an important role in stabilizing the conformation of the Ca2+ bowl. The mutation D896A, on the other hand, had almost no effect on the channel’s sensitivity to Ca2+ (26). In the x-ray structure, this side chain is directed out and away from the Ca2+ ion and does not make other protein contacts.
When the Ca2+ bowl is deleted, the BK channel is still activated by Ca2+ but with lower affinity (15, 28, 29). We have not identified a secondary Ca2+ binding site in this structure.
Quaternary structure formed by a CTD tetramer
The BK (Slo1) Ca2+- (and voltage-) activated channel is related in sequence and structure to the Na+-activated (Slo2) and H+-dependent (Slo3) channels (fig. S4) (9). We expressed and purified the chicken Slo2.2 CTD, which included amino acids 347 to 1201. On size exclusion chromatography, the Slo2.2 CTD elutes as a tetramer, and even under the dispersive conditions of SDS–polyacrylamide gel electrophoresis (SDS-PAGE) the majority of protein migrates as a tetramer. Thus, the Slo2.2 CTD forms a very stable tetrameric complex. We crystallized this Na+-activated CTD in the presence of 500 mM NaCl. The crystals diffracted x-rays to a resolution of 6 Å and were of space group I422, with one monomer (subunit) in the asymmetric unit (table S1). Using a polyalanine model of the BK CTD, we obtained a single outstanding solution in molecular replacement against the Slo2.2 diffraction data (table S2). This solution indicates that the human BK and chicken Slo2.2 CTDs must have very similar structures. By applying the crystallographic symmetry operators, we obtained the tetramer structure of the human BK CTD (Fig. 4A). This objective solution reveals the quaternary organization of subunits but not its exact atomic details because the resolution is low and the data are from the Slo2.2 homolog.
Fig. 4.
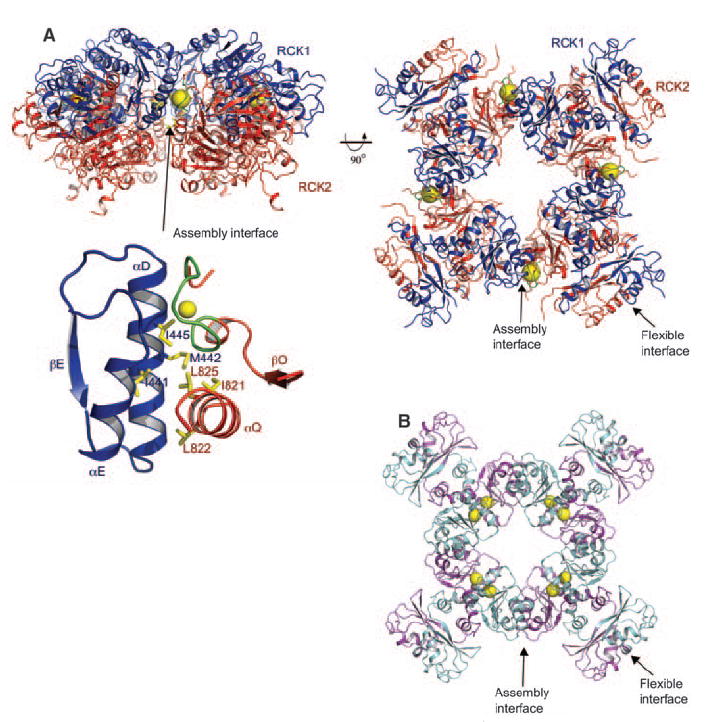
The BK gating ring solution. (A) Orthogonal views of the tetrameric gating ring structure using 6.0 Å diffraction data from the homologous chicken Slo2.2 CTD. (Right) View is down the fourfold symmetry axis, with RCK1 in blue and RCK2 in red. The Ca2+ ions are shown as yellow spheres. The flexible and assembly interfaces are labeled, and a close-up view of the assembly interface is also shown. (B) Open gating ring structure from the MthK channel (PDB accession, 1LNQ), viewed down the fourfold axis.
Two main lines of evidence support the biological relevance of this tetrameric structure. First, it is immediately apparent that the ring of subunits generated by crystallographic symmetry corresponds very well to the “gating ring” observed in the MthK channel, in which RCK domains form a ring through the alternate packing of flexible and assembly interfaces (Fig. 4, A and B) (18). In MthK, both interfaces are formed between independent identical RCK domains (Fig. 4B). In the BK channel, the flexible interface is formed between the nonidentical tandem-linked RCK domains, whereas the assembly interface joins the tandem-linked pairs together (Figs. 2A and 4A). The second line of evidence supporting a biologically relevant structure comes from prior mutational studies on the BK channel (17, 30). Pairs of amino acids were shown to interact in double mutant cycles (30). These amino acid pairs, including I441-L822, M442-L825, and I445-L825, are far away from each other in the monomer but are brought close together in space through creation of the assembly interface (Fig. 4A). Even though very low sequence identity exists between the MthK RCK domain and the C terminus of the eukaryotic Slo channels, the overall three-dimensional structure of the gating ring has been conserved across evolution. As is often the case, the protein originating from the “simpler” prokaryotic organism contains higher symmetry because it is constructed from a single repeated unit, in this case the RCK domain. Because the BK and Slo2.2 structures were determined in the absence of a transmembrane pore, we cannot conclude whether the conformations are “opened” or “closed.” However, given that both proteins were crystallized in very high concentrations of the ions that activate them, we speculate that the gating ring shown in Fig. 4A approximates an open conformation.
A notable difference exists between the MthK and BK gating rings: The ligand Ca2+ is bound on the flexible interface in MthK (Fig. 4B) and on the assembly interface in BK (Fig. 4A). On the basis of the known ligand-binding properties of Rossmann folds, we should expect ligand to be bound on or near the flexible interface, where the C-terminal edges of the central β-sheets of adjacent RCK domains come close together (Fig. 2A and fig. S2, A and B). Evolution of tandem nonidentical RCK domains in the BK channel allowed the creation of a single Ca2+ binding site at the assembly interface. Helical elements αD and αE from RCK1 pack against αQ and the Ca2+ bowl from RCK2. The Ca2+ bowl probably evolved from a helix in RCK2 that corresponded to αE in RCK1. Thus, variation through evolution has brought about a fundamental difference between the MthK and BK gating rings. In BK, Ca2+ binds at the assembly interface at a site facing the outer perimeter of the gating ring (Fig. 4A).
A model for the BK channel
Guided by amino acid sequence alignments and structural components shared by the MthK channel (18), the paddle chimera voltage-dependent K+ channel (31), and the BK gating ring, we have constructed a working structural model for the BK channel (Fig. 5 and figs. S4 and S5). The paddle chimera channel structure (residues 140 to 409) was superimposed on the MthK channel structure by aligning the main-chain atoms from the pore helix and selectivity filter (fig. S5). The pore in paddle chimera and MthK, which adopts an open conformation in both channels, superimposes with a root mean square deviation (RMSD) of 0.87 Å. The BK gating ring was then superimposed on the MthK channel by matching the secondary structural elements of the BK RCK1 domain and the MthK RCK domain. The resulting model for one subunit of the BK channel is shown with the transmembrane pore and voltage sensor in green and the gating ring in blue and red (Fig. 5A). A tetramer was generated from this model and is shown with each individual subunit in a single color (Fig. 5B). Overall, the gating ring appears to fit well against the surface of the Kv channel, with ridges from the gating ring fitting naturally into grooves that exist between the voltage sensors and the pore (Fig. 5B).
Fig. 5.
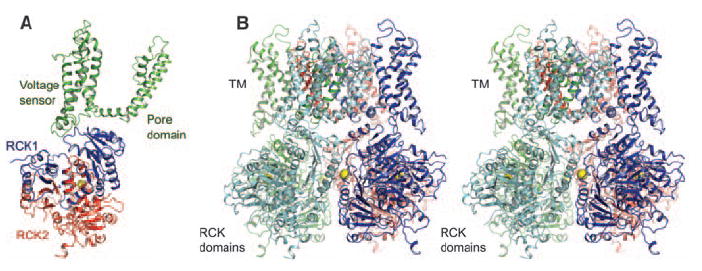
A model of the BK channel. (A) Ribbon representation of a single subunit from the BK model generated by superimposing the BK gating ring and Kv paddle chimera (PDB accession, 2R9R) onto the MthK channel (PDB accession, 1LNQ). The transmembrane domain, RCK1, and RCK2 are colored in green, blue, and red, respectively. (B) Stereoview of the BK model from the side with the transmembrane domain above. Each subunit of the tetramer is colored differently.
Mutations within the gating ring that affect BK channel function tend to occur in three distinct regions of the structure. One regional group surrounds the Ca2+ bowl (Fig. 6, red spheres) (15, 26). These mutations reduce the channel’s sensitivity to Ca2+ ions. A second group of mutations are scattered in the primary sequence but cluster together on the structure around the helix-turn-helix connector, which forms a large surface on the flexible interface (Fig. 6, green spheres) (17, 28, 32-34). These mutations probably affect conformational changes that occur in the gating ring associated with Ca2+-induced gating. A third group of mutations cluster on the N terminus of RCK1 (Fig. 6, purple spheres) (29, 35-39). These amino acids reside on the surface of the gating ring that faces the voltage sensor in the BK channel model.
Fig. 6.
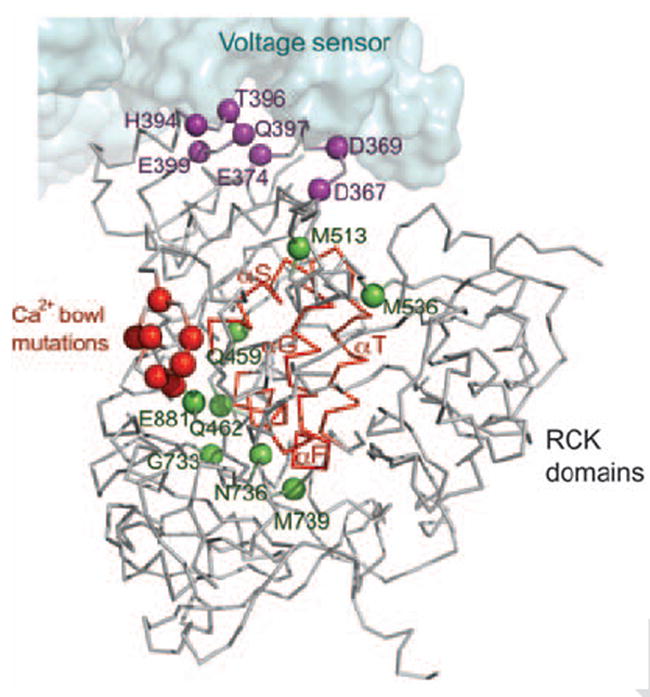
The gating ring and the voltage sensor modules. Sensitive functional mutations from the CTD (colored spheres) were mapped onto a single subunit from the BK model. Three distinct groups of mutations are distinguished by using the same colors as in Fig. 1B. The Ca2+ bowl mutations are colored in red. Mutations near the αF-αG/αS-αT flexible interface (backbone highlighted in red) are colored in green, and mutations in the RCK1 domain facing the voltage sensor domain are colored in purple. The voltage sensor domain is shown as a surface representation.
This third group of mutations support an idea concerning dual regulation by Ca2+ and voltage. The pore is controlled directly by Ca2+ through the connection of S6 to the gating ring and directly by voltage through the S4–S5 linker and voltage sensors. But the structure and this third group of mutations raise the possibility of an additional interaction between the gating ring and the voltage sensor. One interesting experiment identified a putative Mg2+ binding site bridging amino acids on the gating ring to amino acids on the voltage sensor (39). Direct interaction between the Ca2+ and voltage sensory domains would enable one input stimulus to modulate the other in the dual stimulus control of BK channel gating.
Acknowledgments
We thank K. R. Rajashankar and K. Perry at beamline 24ID-C (Advanced Photon Source, Argonne National Laboratory) and the staff at beamline X29 (National Synchrotron Light Source, Brookhaven National Laboratory) for advice at the synchrotron; members of the MacKinnon laboratory for assistance; and L. Feng and M. Whorton for comments on the manuscript. R.M. is an Investigator in the Howard Hughes Medical Institute. The research is supported by the American Asthma Foundation grant 07-0127. The x-ray crystallographic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession code 3MT5.
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/science.1190414/DC1
Materials and Methods
Figs. S1 to S5
Tables S1 to S3
References
References and Notes
- 1.Robitaille R, Garcia ML, Kaczorowski GJ, Charlton MP. Neuron. 1993;11:645. doi: 10.1016/0896-6273(93)90076-4. [DOI] [PubMed] [Google Scholar]
- 2.Fettiplace R, Fuchs PA. Annu Rev Physiol. 1999;61:809. doi: 10.1146/annurev.physiol.61.1.809. [DOI] [PubMed] [Google Scholar]
- 3.Nelson MT, et al. Science. 1995;270:633. doi: 10.1126/science.270.5236.633. [DOI] [PubMed] [Google Scholar]
- 4.Brenner R, et al. Nature. 2000;407:870. doi: 10.1038/35038011. [DOI] [PubMed] [Google Scholar]
- 5.Petkov GV, et al. J Physiol. 2001;537:443. doi: 10.1111/j.1469-7793.2001.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaczorowski GJ, Knaus HG, Leonard RJ, McManus OB, Garcia ML. J Bioenerg Biomembr. 1996;28:255. doi: 10.1007/BF02110699. [DOI] [PubMed] [Google Scholar]
- 7.Magleby KL. J Gen Physiol. 2003;121:81. doi: 10.1085/jgp.20028721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latorre R, Brauchi S. Biol Res. 2006;39:385. doi: 10.4067/s0716-97602006000300003. [DOI] [PubMed] [Google Scholar]
- 9.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. Nat Rev Neurosci. 2006;7:921. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 10.Shipston MJ. Trends Cell Biol. 2001;11:353. doi: 10.1016/s0962-8924(01)02068-2. [DOI] [PubMed] [Google Scholar]
- 11.Schubert R, Nelson MT. Trends Pharmacol Sci. 2001;22:505. doi: 10.1016/s0165-6147(00)01775-2. [DOI] [PubMed] [Google Scholar]
- 12.Tian L, et al. FASEB J. 2006;20:2588. doi: 10.1096/fj.06-6152fje. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Sigworth FJ. Nature. 2009;461:292. doi: 10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y, Pico A, Cadene M, Chait BT, MacKinnon R. Neuron. 2001;29:593. doi: 10.1016/s0896-6273(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber M, Salkoff L. Biophys J. 1997;73:1355. doi: 10.1016/S0006-3495(97)78168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meera P, Wallner M, Song M, Toro L. Proc Natl Acad Sci U S A. 1997;94:14066. doi: 10.1073/pnas.94.25.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pico AR. Ph.D. thesis, The Rockefeller University. 2003. [Google Scholar]
- 18.Jiang Y, et al. Nature. 2002;417:515. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence MC, Colman PM. J Mol Biol. 1993;234:946. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 20.Wei A, Solaro C, Lingle C, Salkoff L. Neuron. 1994;13:671. doi: 10.1016/0896-6273(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, et al. Biophys J. 2009;97:730. doi: 10.1016/j.bpj.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.In the mutants, other amino acids were substituted at certain locations; for example, R182Q indicates that arginine at position 182 was replaced by glutamine. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 23.Ye S, Li Y, Chen L, Jiang Y. Cell. 2006;126:1161. doi: 10.1016/j.cell.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 24.Dong J, Shi N, Berke I, Chen L, Jiang Y. J Biol Chem. 2005;280:41716. doi: 10.1074/jbc.M508144200. [DOI] [PubMed] [Google Scholar]
- 25.Bian S, Favre I, Moczydlowski E. Proc Natl Acad Sci U S A. 2001;98:4776. doi: 10.1073/pnas.081072398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao L, Kaldany C, Holmstrand EC, Cox DH. J Gen Physiol. 2004;123:475. doi: 10.1085/jgp.200409052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusifov T, Savalli N, Gandhi CS, Ottolia M, Olcese R. Proc Natl Acad Sci U S A. 2008;105:376. doi: 10.1073/pnas.0705261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao L, Rapin AM, Holmstrand EC, Cox DH. J Gen Physiol. 2002;120:173. doi: 10.1085/jgp.20028627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia XM, Zeng X, Lingle CJ. Nature. 2002;418:880. doi: 10.1038/nature00956. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Lim HH, Rho SH, Eom SH, Park CS. J Biol Chem. 2006;281:38573. doi: 10.1074/jbc.M604769200. [DOI] [PubMed] [Google Scholar]
- 31.Long SB, Tao X, Campbell EB, MacKinnon R. Nature. 2007;450:376. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 32.Santarelli LC, Wassef R, Heinemann SH, Hoshi T. J Physiol. 2006;571:329. doi: 10.1113/jphysiol.2005.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HJ, et al. Biophys J. 2008;94:446. doi: 10.1529/biophysj.107.108738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng JZ, et al. Biophys J. 2005;89:3079. doi: 10.1529/biophysj.105.063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi J, et al. Nature. 2002;418:876. doi: 10.1038/nature00941. [DOI] [PubMed] [Google Scholar]
- 36.Zeng XH, Xia XM, Lingle CJ. J Gen Physiol. 2005;125:273. doi: 10.1085/jgp.200409239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du W, et al. Nat Genet. 2005;37:733. doi: 10.1038/ng1585. [DOI] [PubMed] [Google Scholar]
- 38.Hou S, Xu R, Heinemann SH, Hoshi T. Nat Struct Mol Biol. 2008;15:403. doi: 10.1038/nsmb.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, et al. Nat Struct Mol Biol. 2008;15:1152. doi: 10.1038/nsmb.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]


