Abstract
XSP10 is an abundant 10 kDa protein found in the xylem sap of tomato. The protein displays structural similarity to plant lipid transfer proteins (LTPs). LTPs are involved in various physiological processes, including disease resistance, and some are able to bind and transfer diverse lipid molecules. XSP10 abundance in xylem sap declines upon infection with Fusarium oxysporum f. sp. lycopersici (Fol), implying involvement of XSP10 in the plant–pathogen interaction. Here, the biochemical characterization of XSP10 with respect to fatty acid-binding properties is reported; a weak but significant binding to saturated fatty acids was found. Furthermore, XSP10-silenced tomato plants were engineered and it was found that these plants exhibited reduced disease symptom development upon infection with a virulent strain of Fol. Interestingly, the reduced symptoms observed did not correlate with an altered expression profile for known reporter genes of plant defence (PR-1 and WIPI). This work demonstrates that XSP10 has lipid-binding properties and is required for full susceptibility of tomato to Fusarium wilt.
Keywords: Fusarium, lipid transfer protein (LTP), tomato, xylem sap
Introduction
The interaction between Fusarium oxysporum f.sp. lycopersici (Fol) and tomato has become a model system for the study of the molecular basis of disease resistance and susceptibility (Takken and Rep, 2010). Fol is a soil-borne fungus that invades tomato roots and colonizes the xylem vessels. In early stages of infection the interface between pathogen and host is largely confined to the xylem. Analysis of the proteome of the xylem sap of Fol-infected plants revealed many fungal proteins that are secreted during colonization, including enzymes as well as small proteins (<25 kDa) with unknown functions (Houterman et al., 2007). Besides Fol-secreted proteins, many plant proteins accumulate in the xylem sap of infected plants, such as pathogenesis-related (PR) proteins (Rep et al., 2002; Houterman et al., 2007). In addition to new proteins appearing, a few were found to disappear from the xylem sap during the course of infection. One prominent low molecular weight protein that strongly decreased in abundance is XSP10. This 10 kDa protein has structural similarity to plant lipid transfer proteins (LTPs) (Rep et al., 2003).
Plant LTPs are small, often basic proteins that are characterized by their ability to bind different types of lipids and hydrophobic molecules (Cheng et al., 2004). LTPs are ubiquitous in the plant kingdom and are divided into two major subfamilies: LTP1 with molecular mass ∼9 kDa; and LTP2 with an average molecular mass of 7 kDa. Both families share several structural features, the most important being eight strictly conserved cysteine residues responsible for the formation of four disulphide bridges (Kader, 1996). The three-dimensional structure of several LTPs has been resolved, featuring four α-helices stabilized by disulphide bonds and enclosing a hydrophobic cavity (Lerche et al., 1997; Lee et al., 1998; Samuel et al., 2002; Pons et al., 2003; Hoh et al., 2005; Lascombe et al., 2008). Most LTPs are synthesized as precursors with an N-terminal signal peptide, and some of them have been shown to be secreted to the extracellular space (Thoma et al., 1993; Arondel et al., 2000).
Although several LTPs have been shown to bind lipids, including phospholipids, and can transfer them between membranes in vitro, their biochemical functions in plants remain largely obscure (Kader, 1996). Functionally, LTPs have been implicated in developmental processes such as transport of cutine monomers in cutical layer formation in carrot and tobacco (Sterk et al., 1991; Cameron et al., 2006), and in cell wall loosening (Nieuwland et al., 2005). Recently, the involvement of a root-specific LTP for establishment of a symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti was reported (Pii et al., 2009). On the other hand, many LTPs are thought to participate in defence responses against parasitic interactions. Some LTPs display direct antimicrobial activity (Cammue et al., 1995; Molina and Garcia-Olmedo, 1997; Ge et al., 2003) and their overexpression in transgenic plants leads to enhanced resistance, like overexpression of barley LTP1 in transgenic tobacco (Molina and Garcia-Olmedo, 1997). Antimicrobial activity is not always coupled to lipid transfer properties, since at least one member of the LTP family, Ace-AMP1, does not bind or transfer lipids (Tassin et al., 1998). Altogether, these observations led to classification of LTPs as pathogenesis-related proteins (PR-14) (van Loon et al., 2006).
In addition to direct antimicrobial activity, some members of the LTP family are associated with signalling pathways leading to the activation of plant defence. For instance, tobacco LTP1 has the ability to bind jasmonic acid (JA), and treatment of tobacco plants with LTP1–JA increases resistance to Phytophthora parasitica (Buhot et al., 2004). Another LTP family member, DIR1 (defective in induced resistance) from Arabidopsis thaliana, is required for systemic signal propagation during SAR (systematic acquired resistance). A dir1 mutant is unaffected in local resistance responses, but is unable to develop SAR against virulent Pseudomonas syringae (Maldonado et al., 2002). Recently, the structure of DIR1 complexed with lyso stearoylphosphatidyl choline was determined. Like other LTPs, the core of the protein consists of a left-handed superhelix formed by four α-helices that encompass the central hydrophobic cavity. This cavity binds with high affinity to two long-chain lysophospolipids. The overall fold of the protein resembles that of the LTP2 family, but it differs from these by its low pI and a characteristic PxxPxxP motif on its surface (Lascombe et al., 2008).
Although XSP10 appears to be structurally related to the LTP family, it has not been designated an LTP member because of its low level of sequence similarity and the lack of experimental data concerning lipid transfer activity. Hence it was classified as ‘a new family of secreted, plant-specific proteins with unknown function’ (Rep et al. 2003). In this study the lipid-binding properties of XSP10 are characterized and its involvement in resistance to Fol is investigated. It was found that XSP10 does have affinity for specific fatty acids and might represent an LTP1 family member. Silencing of the gene in tomato using an interfering hairpin RNA (hpRNA) approach showed that XSP10 is required for full susceptibility, as defined by reduced disease-symptom development, of tomato to Fusarium wilt.
Materials and methods
Plant material
Tomato (Solanum lycopersicon cv. Moneymaker GCR161) seedlings were grown in a greenhouse with a day/night temperature of 23–18 °C and a 16/8 h light/dark regime.
DNA isolation and sequence analysis of the XSP10 gene and its 5′- and 3′;-flanking regions
A five genome equivalent library from the breeding line Ontario 7518 (Cf18) (Lauge et al., 1998) in the pCLD04541 binary cosmid vector (Bent et al., 1994) with an average insert size of ∼20 kb was used (de Kock, 2004). The library was screened using an XSP10-specific primer set: Fxsp, 5′-GCA GGA ATG AAC TAC TTG TTG T; and Rxsp, 5′-CTG CCA CCA AAC ACA TAG GTA. Two cosmids harbouring the XSP10 sequence were identified. Detailed characterization of these cosmids by restriction mapping, DNA hybridization, and sequence analysis was performed (data not shown).
Heterologous expression of XSP10 in Pichia pastoris and affinity purification
Total RNA was isolated from roots of tomato plants using Trizol LS reagent (Invitrogen) followed by chloroform extraction and isopropanol precipitation. DNA was removed with DNase (Fermentas). Additional RNA purification was performed on RNeasy minicolumns according to the manufacturer's instructions (Qiagen). cDNA was synthesized from 1 μg of total RNA using M-MuLV Reverse Transcriptase (Fermentas) as described by the manufacturer.
The XSP10 cDNA was amplified by PCR with Fxsp and Rxsp using tomato root cDNA as template. The PCR fragment was then cloned into pGEM-T easy (Promega) and sequenced. The coding sequence was then re-amplified using oligonucleotide pairs: FxspBam (5′-CAGGATCC ATG AAC TAC TTG TTG TGT; the BamHI restriction site is underlined, and the start codon of XSP10 is highlighted in bold) and Rxsp6HNot (5′-GTGCGGCCGC TCA GTG GTG ATG GTG GTG ATG TGG CAG TGT GTA AGG TCC A; the NotI restriction site is underlined, the stop codon of XSP10 is highlighted in bold, and the six His tag is denoted by italics) for the expression of XSP10 with a native secretion signal and a six histidine tag on the C-terminus of the protein; FxspEco (5′-CAGAATTCGC CGG TGA ATG CGG GAG AA; the EcoRI restriction site is underlined, and the start codon of XSP10 is highlighted in bold) and Rxsp6HNot for the expression of XSP10 with the yeast α-factor secretion signal and a six histidine tag on the C-terminus of the protein; Fxsp6HEco (5′-CAGAATTC CAC CAT CAC CAC CAT CAT GCC GGT GAA TG CGG GAG AA; the EcoRI restriction site is underlined, the start codon of XSP10 is highlighted in bold, and the six His-tag is denoted by italics) and RxspNot (5′-GTGCGGCCGC TCA TGG CAG TGT GTA AGG T; the NotI restriction site is underlined, and the stop codon of XSP10 is highlighted in bold) for XSP10 expression with the yeast α-factor secretion signal and a six histidine tag on the N-terminus of the protein. The amplified fragment was purified and cloned into pPIC9 using the sites indicated in the primers (Invitrogen). The correct orientation of the XSP10 sequence was checked by PCR and confirmed by DNA sequencing.
Pichia pastoris transformation (strain GS115) and selection of transformants was performed according to the instructions of the manufacturer (Pichia Expression Kit, Invitrogen). The selected yeast transformants were pre-cultivated on a minimum glycerol medium [MGY: 1.34% yeast nitrogen base (YNB), 4×10−5% biotin, 1% glycerol] for 16 h, then cells were harvested by centrifugation (1500 g for 5 min at room temperature) and resuspended in minimum methanol medium (MM: 1.34% YNB, 4×10−5% biotin, 0.5% methanol) to an OD600 of 1.0. All cultures were maintained at 29 °C, in the dark, on rotary shakers at 250 rpm. After 5 d of culturing, the medium was recovered by centrifugation (10 000 g, 10 min, 4 °C), extensively dialysed against Ni-NTA loading buffer [LB: 20 mM phosphate buffer (PB) pH 7.8, 100 mM NaCl] in Spectra/Por dialysing membranes (MWCO 3500, Spectrum Laboratories) and loaded on a manually packed column containing 2 ml of Ni-NTA–agarose resin according to the manufacturer's instructions (Qiagen). To remove unbound and aspecifically bound proteins, the column was washed extensively with 10 vols of washing buffer 1 and washing buffer 2 (LB supplemented with 10 mM and 20 mM imidazole pH 7.5, respectivelly). His-tagged XSP10 bound on Ni-NTA beads was eluted in fractions of 0.5 ml with elution buffer (LB supplemented with 0.5 M imidazole). Sample, flowthrough, and wash fractions were concentrated 10-fold using acetone precipitation, and together with elution fractions analysed by 15% TRIS–Tricine SDS–PAGE (Supplementary Fig. S2 available at JXB online). The fractions containing XSP10 were combined and dialysed extensively against the buffer in which the lipid-binding assay was performed (50 mM PB pH 7.0, 50 mM NaCl). Protein concentrations were estimated using the bicinchoninic acid method (Sigma).
Mass spectrometry
Identification of the purified XSP10 protein was done with the in-gel digestion method as described (Rep et al., 2003). The eluates after the digestion of each sample were collected and washed on a ±C18 ZipTip (Millipore) and eluted in 6 μl of 60% acetonitrile (ACN) and 0.1% formic acid. The samples were identified with a nano-HPLC system (LC Packings, Dionex), which was directly coupled with a Q-Tof1 (Micromass, Waters) mass spectrometer. The separated peptides coming from the C18 PepMap column (Dionex) were selected automatically for low energy collision-induced dissociation experiments. The resulting tandem mass spectrometry (MS/MS) fragmentation spectra were used for the analysis and for identification of the protein sample.
Holomass (real molecular average mass) analysis was performed after desalting and eluting the XSP10 samples (μC4 ZipTip Millipore) in 10 μl of 50% ACN, 1% formic acid. An Econo12 emitter (New Objective, USA) was used to spray the protein solution off-line into the Q-Tof1.
The multiple charged m/z protein peaks were used to calculate the average masses within the sample by both manual calculation and after deconvolution of the raw spectra with embedded MaxEnt1 software. Calibration of the m/z range was done with a 2 pmol μl−1 in 50% ACN, 1% formic acid stock solution of horse myglobin.
Lipid binding
The ability of XSP10 to bind lipids was assayed by monitoring the displacement of the fluorescent probe 2-p-toluidinonaphthalene-6-sulphonate (TNS). Lipid-binding experiments were performed at room temperature in a FeliX32 fluorescent spectrophotometer (Photon Technology Instruments), as previously described (Buhot et al., 2004), with minor modifications. The excitation and emission wavelengths were set at 320 nm and 437 nm, respectively. TNS, with or without fatty acids or JA, was incubated for 1 min in a stirred cuvette containing 1 ml of measurement buffer before fluorescence was recorded (F0). XSP10 was then added and, after 2 min, fluorescence was recorded at equilibrium (F). Results are expressed as a percentage of XSP10–TNS complex fluorescence according to [(F–F0)/FC]×100, where FC is the fluorescence of the XSP10–TNS complex in the absence of FA or JA. All fatty acids, JA, and TNS were purchased from Sigma. FA and JA were dissolved in ethanol; TNS was dissolved in dimethylformamide and stored in aliquots at –20 °C.
Design of the hpRNA construct
The RNA interfering hairpin construct was produced by fusing part of the XSP10 gene with a fragment of the β-glucuronidase (GUS) reporter gene (Wroblewski et al., 2007; Tomilov et al., 2008). Briefly, 309 bp covering almost the entire coding part of the XSP10 gene (TC205029, the DFCI S. lycopersicum Gene Index version 13.0, bases 7–315 from the ATG codon, Supplementary Fig. S1 at JXB online) was amplified with primers in which SfiI restriction sites were introduced: Fxsp-Sfi, 5′-ATG GCC ATG TAG GCC TAC TTG TTG TGT GTT GTA; and Rxsp-Sfi, 5′-ATG GCC AGA GAG GCC CTT ATA GCC AAC GGG ACG (15 bp adaptors bearing the SfiI cleavage site are underlined). The obtained fragment was fused to a 451 bp fragment of the GUS gene (U12639, bases 2644–3095) encoding part of the GUS protein. This chimeric fragment was used to create an inverted repeat structure in the binary vector pGSA1165 (http://www.chromdb.org). The two arms of the inverted repeat were separated by intron 3 (788 bp) of the pdk gene from Flaveria trinervia. The complete XSP10 fragment in this construct, including its borders, was sequenced using primers pGreenF2 (5′-ACT ATC CTT CGC AAG ACC C) and OCStermRev (5′-TCA TGC GAT CAT AGG CGT CT), annealing to the cauliflower mosaic virus (CaMV) 35S promoter and the OCS terminator of pGSA1165, respectively. SfiI was obtained from New England Biolabs (http://www.neb.com/). T4 DNA ligase was obtained from Fermentas (http://www.fermentas.com/).
Plant transformation and selection of transgenic plants
For tomato transformation, the hpXSP10 construct was introduced into Agrobacterium tumefaciens strain LBA4404 (Hoekema et al., 1983). Transgenic plants were produced using explants derived from cotyledons of sterile seedlings as previously reported (Cortina and Culianez-Macia, 2004).
The presence of the T-DNA insertion in primary transformants was assessed by the presence of the neomycin phosphotransferase gene (NPTII) using PCR (data not shown). DNA was isolated from leaves using the cetyltrimethylammonium bromide (CTAB) procedure (Bernatzky and Tanksley, 1986). Two primers, FNptII (CCG GTT CTT TTT GTC AAG AC) and RNptII (AGA AGA ACT CGT CAA GAA GG), were used to amplify a 661 bp fragment diagnostic of the NPTII gene. The number of T-DNA inserts was analysed by Southern blotting using the NPTII gene as probe (data not shown). Briefly, genomic DNA was digested with the restriction enzyme BglII (Fermentas), blotted on a nylon filter (Hybond-N), and hybridized with an [α-32P]NPTII radioactive probe (DecaLabel DNA labelling kit, Fermentas). Silencing in T1 progeny and T0 parents was assayed by screening for reduced GUS expression upon leaf infiltration with A. tumefaciens strain C58C1 harbouring plasmid pTFS40 (Jones et al., 1992), which allows GUS expression in planta (Wroblewski et al., 2005). Leaf infiltrations were performed as described previously (Schob et al., 1997). Histochemical GUS staining was done essentially as described before (Jefferson et al., 1987).
Fusarium bioassays
Tomato seedlings were inoculated with either a virulent race 2 isolate of Fol (Fol007) or with an avirulent race 1 isolate Fol004 (Rep et al., 2005) using the root dip method (Wellman, 1939). Briefly, conidial spores were collected from 5-day-old cultures grown in NO3 medium [3% sucrose, 5 mM KNO3, 0.17% YNB without amino acids and ammonium sulphate (Duchefa)]. After washing of the spores they were used for root inoculation at a density of 5×106 spores ml−1. Twenty 10-day-old uprooted seedlings were incubated for several minutes in the spore suspension and potted individually in a random block design (with five seedlings per block). Three weeks after inoculation, plant weight above the cotyledons was measured and disease symptoms were scored by determining the extent of browning of vessels at the height of the cotyledons. The disease index was scored on a scale of 0–4, where 0=no symptoms; 1=slightly swollen hypocotyl; 2=one or two brown vascular bundles in the hypocotyl; 3=at least two brown vascular bundles and growth distortion (strong bending of the stem and asymmetric plant development); and 4=all vascular bundles brown, plant either dead or very small and wilted. Ten seedlings of GCR161 and hpXSP10 lines were used for mock inoculation as a control. One-way analysis of variance (ANOVA) with Dunnett post-hoc test and pairwise comparison with Student's t-test for the weight measurements and the non-parametrical Mann–Whitney test for the disease index was performed using GraphPad Prism 5.0 (GraphPad Software). Leaves and roots were collected, frozen in liquid nitrogen, and stored at –70 °C for further RNA isolation.
Infection assays of 4-week-old plants were carried out by incubating the trimmed root system in a spore suspension (5×106 ml−1) for several minutes and potting the plants individually in a random design. Four weeks after mock inoculation or Fol infection, pictures of representative plants were taken and xylem sap was collected.
Xylem sap collection and SDS–PAGE analysis
To monitor XSP10 levels in hpXSP10 lines, xylem sap was collected from 8-week-old (4 weeks after mock inoculation) tomato plants as described (Satoh et al., 1992; Rep et al., 2002). Briefly, stems were cut below the second true leaf, the first droplet appearing on the cut surface was removed with blotting paper, and the plant was placed in a horizontal position. Sap dripping from the cut surface was collected in tubes placed on ice for a period of 6 h, generally yielding 7–15 ml of sap. Xylem sap was concentrated 20-fold by acetone precipitation, and the protein concentration was estimated with the bicinchoninic acid method (Sigma). Protein loading was normalized on a volume basis (Fig. 2B) or adjusted so that each sample contained 1 μg μl−1 of bovine serum albumin (BSA) equivalents (Supplementary Fig. S6 at JXB online). SDS–PAGE was performed in a Mini-Protean II electrophoresis cell (BioRad) using the TRIS–Tricine buffer system (Schagger and von Jagow, 1987). Silver staining was used to visualize proteins as described (Shevchenko et al., 1996).
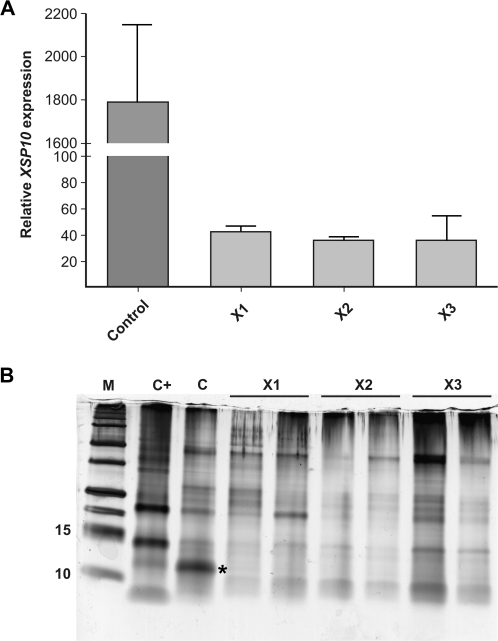
Fig. 2.
XSP10 expression is strongly reduced in hpXSP10 plants. (A) Transcript levels were determined by real-time qPCR relative to α-tubulin in roots of 5-week-old control or hpXSP10 plants. Mean transcription level of four plants and the SD are shown. (B) XSP10 levels in xylem sap of 8-week-old mock-inoculated control (C), hpXSP (X1, X2, X3 in biological duplicate), and Fol007-infected control plants (C+). Protein loading was normalized on an equal volume basis. M, protein standards, * marks XSP10 protein.
RNA isolation and analysis by real time RT-PCR measurements
For determination of relative expression of XSP10, total RNA was isolated from the three young leaves or roots of mock-inoculated or Fol007-infected plants 3 weeks after root inoculation of 10-day-old seedlings using Trizol LS reagent (Invitrogen). Additional RNA purification was performed by chloroform extraction and isopropanol precipitation. DNA was removed with DNase (Fermentas) before loading the RNA on RNeasy minicolumns according to the manufacturer's instructions (Qiagen). cDNA was synthesized from 1 μg of total RNA using M-MuLV Reverse Transcriptase (Fermentas) as described by the manufacturer in a 20 μl reaction which was diluted to 200 μl prior to using it for PCR. PCRs were performed in an ABI 7500 Real-Time PCR system (Applied Biosystems) using a Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen). PCRs of 20 μl contained 0.25 μM of each primer, 0.1 μl of ROX reference dye, and 2 μl of cDNA. The cycling program was set to 5 min 50 °C, 5 min 95 °C, 40 cycles of 15 s 95 °C and 1 min 60 °C, followed by a melting curve analysis. Amplification was tested for linearity with a standard cDNA dilution series. Primers used for XSP10 (TC205029) amplification were: qXspL (5′-AAG CAG CAT CGG ATG AGA AT) and qXspR (5′-TGG TTA TCG CAA CTT CAG GA). The expression level for XSP10 in roots was normalized to the expression of α-tubulin (TC170178) detected with primers qTubL (5′-CAG TGA AAC TGG AGC TGG AA) and qTubR (5′-TAT AGT GGC CAC GAG CAA AG). Primers used for PR-1a [AJ011520 (Van Kan et al., 1992)] amplification were: qPR1F (5′-CCC AAG ACT ATC TTG CGG TT) and qPR1R (5′-TTA CAA TCA CCC GCT CTT GA). The WIPI-II [wound-induced proteinase inhibitor II; K03291 (Graham et al., 1985)] expression level was assayed with primers: qWIPIL (5′-GAC AAG GTA CTA GTA ATC AAT TAT CC) and qWIPIR (5′-GGG CAT ATC CCG AAC CCA AGA). PR-1 and WIPI expression levels in leaves were normalized to the expression of RUB1-conjugating enzyme (RCE1; AY004247). Primers used for RCE1 amplification were: qRCE1F (5′-GAT TCT CTC TCA TCA ATC AAT TCG) and qRCE1R (5′-GAA CGT AAA TGT GCC ACC CAT A). Statistical significance was estimated by pairwise non-parametrical Mann–Whitney test with GraphPad Prism software.
Results
Sequence of XSP10
Previously, xylem sap proteomics revealed the presence of a small and relatively abundant 10 kDa protein in the xylem sap of tomato plants (Rep et al., 2002, 2003). The identity of this protein, referred to as XSP10, was determined using mass spectrometry, which allowed identification of its coding sequence in the DFCI Tomato Gene Index (TC231056). Experiments were conducted to characterize the coding sequence further and to clone the flanking DNA. Screening of a tomato cosmid library resulted in the isolation of two cosmids carrying the XSP10 gene. Restriction mapping, DNA hybridization analysis, and subsequent sequencing of part of a 4 kb EcoRI restriction fragment harbouring the XSP10 gene revealed the sequences of XSP10 and 1457 bp of the 5'-DNA flanking and 1603 bp of 3'-DNA flanking regions (Supplementary Fig. S1 at JXB online; GenBank entry HM590582). The XSP10 coding sequence contains a single 98 bp intron near the stop codon (Supplementary Fig. S1).
Heterologous expression of XSP10, and MS/MS and holomass analysis
XSP10 shows structural similarity to plant LTPs (Rep et al., 2003), a class of small globular proteins with four conserved cysteine bonds able to bind various lipid-derived molecules in vitro (Kader, 1996). This similarity prompted the assessment of the lipid-binding properties of XSP10. Production of the protein was achieved using the heterologous P. pastoris expression system. Three different expression constructs were designed. In the first one, the coding part of XSP10, encompassing its endogenous secretion signal, was fused to a C-terminal six histidine tag to allow affinity purification with nickel (Var1). In the other two constructs, the signal peptide was replaced by the yeast α-factor secretion signal, and a six histidine tag was either fused to the C-terminus of the protein or placed in between the signal peptide and the N-terminus of the protein (Var2 and Var3, respectively) (Supplementary Fig. S2 at JXB online). After affinity purification of the protein using nickel beads (Supplementary Fig. S2B shows a typical example of Var2), the identity of the expressed variants was confirmed by in-gel trypsin digestion followed by nano-HPLC combined with electrospray ionization-time of flight (ESI-Tof) MS/MS analysis (Supplementary Fig. S3A). Holomass analysis of all three purified XSP10 variants suggested the attachment of 1–3 hexose residues by P. pastoris as their masses were 1–3 times 162 Da higher than predicted for the full-length proteins (Supplementary Fig. S3B, C).
XSP10–fatty acid binding.
Purification of the XSP10 protein allowed its ability to bind fatty acids to be tested. Lipid binding was analysed using TNS, a soluble probe that is highly fluorescent when bound to a hydrophobic cavity of a protein. When a lipid is able to compete with TNS for binding to the hydrophobic pocket of a protein, quenching of the initial high level of fluorescence occurs. This method has been used successfully to study the interaction between various lipids and elicitins (Mikes et al., 1998) and with tobacco LTP1 (Buhot et al., 2004).
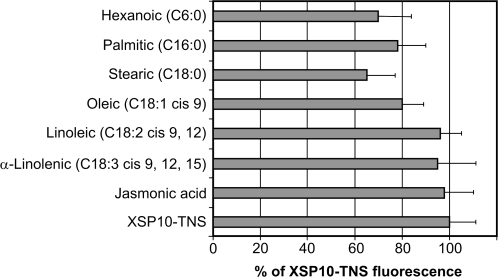
Several groups of lipids were tested in this study: saturated fatty acids (FAs) with C6, C16, and C18 chain length; C18 unsaturated FAs harbouring 1–3 double bonds; and JA. When recombinant XSP10 (Var2) was added to a mixture of TNS and FAs, the fluorescence of TNS was lower than that of the control in which XSP10 was added to TNS alone (Fig. 1; Supplementary Fig. S4 at JXB online). No quenching between FAs and TNS alone was detected, showing that these two molecules do not themselves interact directly (data not shown). Unsaturated acids and JA were poor competitors in displacing the TNS probe from XSP10, and only for oleic acid (C18:1) was a small decrease (20%) in fluorescence observed. Saturated FAs, however, were found to compete with TNS for binding to the hydrophobic pocket of XSP10, reducing the fluorescence to 65% for stearic acid (C18:0), 70% for hexanoic acid (C6:0), and 78% for palmitic acid (C16:0). Similar quenching results were recorded for the recombinant XSP10 irrespective of whether it was C- or N-terminally tagged and whether it was secreted using its endogenous or yeast α-factor secretion signal (data not shown). These data indicate that XSP10 has a weak but significant affinity towards (saturated) fatty acids.
Fig. 1.
Some fatty acids reduce the fluorescence level of the XSP10–TNS complex. FAs or JA (10 μM) and TNS (5 μM) were incubated together for 1 min and then XSP10 (100 nM) was added. Results are expressed as the percentage of the fluorescence of the XSP10–TNS control (no FA or JA added; fluorescence level 100±11%). Experiments were performed in triplicate and results are expressed as the mean values ±SD.
Construction and selection of XSP10-silenced plant lines
The level of XSP10 in tomato xylem sap declines upon infection with Fol (Rep et al., 2002, 2003). To investigate the involvement of XSP10 in resistance/susceptibility of tomato to Fusarium wilt, transgenic lines were created in which XSP10 expression is reduced using gene silencing. Tomato cv. GCR161 was transformed using chimeric gene constructs designed to silence XSP10 and a GUS reporter gene simultaneously via the production of interfering hpRNA. Almost the entire XSP10 coding sequence was fused to a part of the coding region of the GUS gene, so as to create an inverted repeat separated by a linker fragment (see Materials and methods). This strategy was effectively used before in lettuce (Wroblewski et al., 2007) and to silence the S-adenosyl methyltransferase (SAMT) gene of tomato (Ament et al., 2010).
Ten primary hpXSP10 transformants (T0) were screened for the presence of the NPTII transgene and assayed for the number of T-DNA inserts by Southern hybridization (data not shown). T1 progeny were screened for the silencing of the GUS reporter gene using Agrobacterium-mediated transient assays, and plants showing strongly reduced GUS expression were selected (Supplementary Fig. S5 at JXB online). Of the six primary transformants with one T-DNA insert and showing strong silencing of the GUS reporter, homozygous T1 hpXSP10 lines were selected by germinating the T2 offspring on kanamycin selective medium. T1 plants yielding 100% kanamycin-resistant progeny were scored as homozygous. Finally, three homozygous hpXSP10 lines (X1, X2, and X3), each carrying a single T-DNA insert and exhibiting complete silencing of the GUS reporter gene, were selected for further study.
To analyse whether XSP10 was effectively silenced in the three selected hpXSP10 lines, the expression levels of XSP10 were measured in the roots and compared with those of a non-transgenic control plant. All three lines showed a drastic decrease in XSP10 transcription level of up to 30-fold as compared with the control (Fig. 2A). To assess whether this reduction translated into a decline in XSP10 protein abundance in the xylem sap, xylem sap was isolated from 8-week-old plants and the total protein content was visualized using SDS–PAGE. The overall protein profile for the hpXSP10 plants was comparable with that of the control plants, with the exception of XSP10, whose levels were below the detection level in hpXSP10 plants (Fig. 2B; Supplementary Fig. S6 at JXB online). The identity of the particular protein band (marked with an asterisk) was confirmed by MS/MS analysis as XSP10 (data not shown). Thus, it was concluded that expression of the XSP10 gene is drastically reduced in hpXSP10 transgenic tomato, leading to strong reduction of XSP10 protein levels in the xylem sap.
XSP10 silencing reduces susceptibility of tomato to Fusarium wilt
To investigate whether XSP10 has a role in either susceptibility or resistance of tomato to Fol, disease assays on seedlings were performed as well as on 4-week-old hpXSP10 plants. Tomato cultivar Moneymaker GCR161 contains the I gene that confers resistance to Fol races carrying Avr1 (Kroon and Elgersma, 1993; Houterman et al., 2008). Inoculations were done with either race 2 isolate Fol007 that lacks Avr1 and is virulent on GCR161 or race 1 isolate Fol004 that carries Avr1 and is avirulent on this cultivar (Rep et al., 2005).
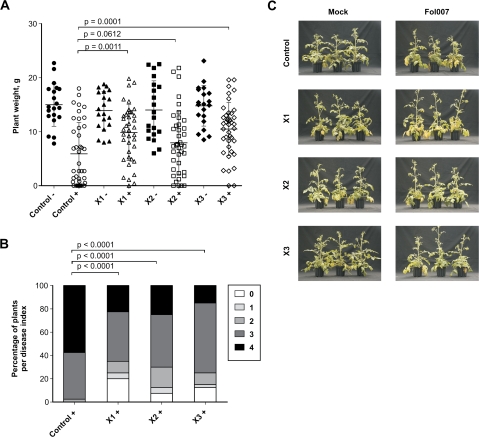
Silencing of XSP10 did not affect I-mediated resistance of GCR161 to the race 1 isolate as no differences were observed after inoculation with Fol004 as compared with the non-transgenic control (data not shown). However, compared with the non-transgenic control, the three hpXSP10 lines showed fewer disease symptoms upon race 2 infection (Fig. 3A). The percentage of plants with disease index 4 (the highest score) was 2- to 3-fold higher for the control as compared with each hpXSP10 line (Fig. 3B). Moreover, the percentage of dead plants was significantly reduced in the hpXSP10 lines as compared with the control (15% for the X1 and X2 lines, 10% for the X3 line, and 45% for the GCR161 control). The decrease in symptom development is also reflected by a significantly higher average weight 3 weeks after infection with the virulent Fol007 in the case of lines X1 and X3 (Fig. 3A). Also line X2 displayed a small, albeit non-significant, difference in plant weight compared with the control. Reduced susceptibility of hpXSP10 lines to Fol was confirmed in a separate disease assay using older (4-week-old) plants: all three hpXSP10 lines consistently showed fewer (external) disease symptoms (Fig. 3C). Taken together, these observations show that a reduced accumulation of XSP10 protein in tomato is associated with reduced disease symptom development upon infection with a virulent race of Fol.
Fig. 3.
Silencing of XSP10 decreases disease susceptibility to F. oxysporum f. sp. lycopersici. Ten-day-old seedlings of control (GCR161) and hpXSP10 lines were mock inoculated (–) or infected with Fol007 (+). Plant weight (A) and disease index (B) were determined 21 d post-inoculation. Disease index: 0=no symptoms; 1=slightly swollen hypocotyl; 2=one or two brown vascular bundles in the hypocotyl; 3=at least two brown vascular bundles and growth distortion (strong bending of the stem and asymmetric plant development); and 4=all vascular bundles brown, plant either dead or very small and wilted. Statistical significance was estimated by pairwise comparison with Student's t-test for the weight and the non-parametrical Mann–Whitney test for the disease. The graph is based on data from two independent experiments. (C) Four-week-old plants of the control and hpXSP10 lines were either mock inoculated or infected with Fol007. Pictures of three representative plants were taken 26 d post-inoculation.
XSP10 and systemic responses are not affected in hpXSP10 plants upon Fol007 infection
The reduction in disease symptom development observed for the hpXSP10 plants during Fol infection prompted the investigation of the effect of XSP10 silencing on the induction of host defence genes. Pathogen recognition generally results in the induction of host defences that are directed against the invader. Various phytohormones orchestrate the underlying defence signalling networks. The antagonistic and synergistic interactions between these plant hormones, especially salicylic acid (SA) versus JA and ethylene ET, allows fine-tuning of these immune responses (Pieterse et al., 2009). To investigate whether these pathways are affected in the silenced plants, the expression profiles of an SA and a JA/ethylene marker gene were analysed both locally and in systemic tissues. Expression profiles in roots and leaves of infected hpXSP10 plants were analysed of (i) the XSP10 gene itself; (ii) the PR-1 gene, which is often used as marker of SA-triggered defence responses (Van Kan et al., 1992; van Loon et al., 2006); and (iii) the WIPI-II gene (Graham et al., 1985), a marker for JA/ethylene-related defence responses (Farmer et al., 1992).
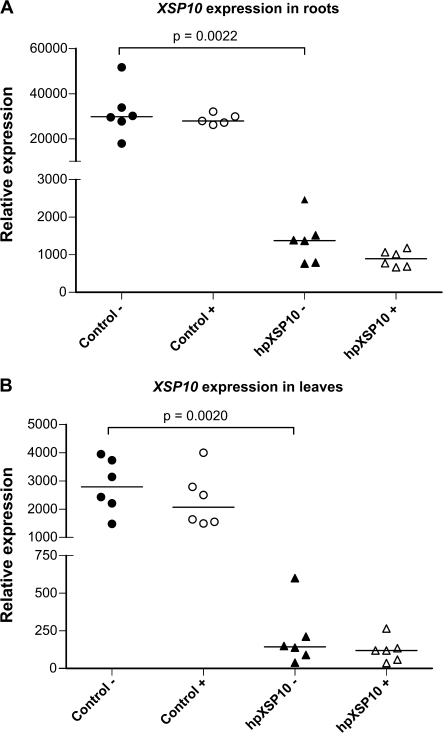
The transcription level of XSP10 in roots of tomato is one order of magnitude higher than that in leaves of the same plant (Fig. 4A, B), in agreement with earlier results using RNA blots (Rep et al., 2003). Statistically significant silencing of the XSP10 gene was observed in both roots and leaves of hpXSP10 plants as compared with the control. No changes in mRNA levels of the XSP10 gene were observed during the course of infection in either control or silenced lines.
Fig. 4.
XSP10 transcription levels are not significantly altered in roots and leaves of hpXSP10 plants upon Fol007 infection. Roots and leaves were collected 3 weeks after mock or Fol007 inoculation of 10-day-old seedlings. Transcript levels were determined by real-time qPCR relative to α-tubulin in roots and RCE in leaves of mock-inoculated (–) or Fol007-infected (+) control and hpXSP10 plants. Median and individual expression of six plants per condition is shown. Statistical significance was estimated by pairwise non-parametrical Mann–Whitney test. The experiment was repeated once with similar results.
No statistically significant differences were observed in the expression levels of the PR-1 gene, in either roots or leaves of the wild type or the silenced plants, indicating that the gene is not induced systemically upon Fol infection (Supplementary Fig. S7 at JXB online). Expression of the WIPI gene was found to be very low and unaltered after infection (Supplementary Fig. S7). These results indicate that Fol infection does not trigger systemic induction of either PR-1 or WIPI expression and that their expression profiles are not altered upon XSP10 silencing.
Discussion
XSP10 has affinity for saturated fatty acids
Xylem sap of healthy tomato plants contains various proteins of which XSP10 is the most prominent (Rep et al., 2002, 2003). Whereas during Fol infection pathogenesis-related proteins, such as PR-1, PR-2, and PR-5, accumulate, the level of XSP10 greatly decreases. This decrease suggested a possible involvement in the interaction with the invading pathogen.
XSP10 has structural similarity to plant non-specific LTPs (Rep et al., 2003). Although some plant LTPs have been shown to bind FAs and transfer phospholipids in vitro (Kader, 1996; Tassin-Moindrot et al., 2000), the in planta substrates for most LTPs are unknown. In this study, it is shown that P. pastoris-produced tomato XSP10 is able to bind particular FAs with apparently modest affinity. Heterologous production of XSP10 in yeast led to the likely attachment of one to several hexose residues to the protein (Supplementary Fig. S3 at JXB online), which might affect the affinity or specificity of XSP10 towards FAs. Yet, only particular linear FAs such as stearic, palmitic, and hexanoic were able to displace the fluorescent TNS probe from the presumed hydrophobic pocket in XSP10, indicating specific lipid binding (Fig. 1). In contrast to tobacco LTP1, neither JA nor unsaturated FAs were able to displace the TNS probe. These data imply that XSP10 has a unique, albeit weak, binding affinity towards linear saturated FAs. Further experimentation, such as measuring changes in intrinsic fluorescence as was done for DIR1 and tobacco LTP1 (Da Silva et al., 2005; Lascombe et al., 2008), is required to determine XSP10’s specificity towards various substrates and to determine dissociation constants.
XSP10 silencing compromises susceptibility of tomato to Fusarium wilt
In tomato plants the XSP10 gene is constitutively expressed at high levels in roots and the lower parts of the stem, whereas expression is low in leaves. Since XSP10 expression does not change after Fol infection (Fig. 4A, B) the decline in XSP10 protein levels in the xylem sap may reflect breakdown or modification of the protein by the pathogen (Rep et al., 2003; Fig. 2B). Possibly, a decrease in XSP10 protein levels is required for the fungus to grow vigorously in the xylem, which would fit the observation that overexpression of some LTPs leads to increased protection against microorganisms (Molina and Garcia-Olmedo, 1997; Jayaraj and Punja, 2007). Some LTPs indeed possess a direct in vitro antimicrobial activity: for instance, MtN5, a root-specific LTP from Medicago truncatula, has antimicrobial activity against Fusarium semitectum (Pii et al., 2009). The in vitro antimicrobial properties of the purified XSP10 against Fol were measured, but no direct effect of XSP10 on Fol007 spore germination or growth was found (data not shown).
Silencing of XSP10 did not affect I-mediated resistance as the XSP10-silenced lines remained fully resistant against the avirulent Avr1-carrying Fol004 isolate (data not shown). However, inoculation of seedlings with the virulent strain Fol007 revealed a significantly higher average weight, a lower disease index, and a smaller percentage of dead plants 3 weeks after infection in the XSP10-silenced plants (Fig. 3A, B). Bioassays using older plants also revealed less (external) disease symptom development in the silenced plants (Fig. 3C). These assays show that XSP10 is required for full disease symptom development of tomato upon Fusarium infection.
To explain the requirement for XSP10 for full susceptibility to Fol, two possible scenarios are proposed. One possibility is that XSP10 represents a compatibility target required for the fungus to develop disease fully. Absence of this target reduces the ability of the fungus to colonize the plant and to cause disease. In this model, XSP10 might, for instance, be involved in the trafficking of essential lipid molecules from plant membranes to the pathogen, similar to proposed sterol carrier functions of elicitins secreted by pathogenic Phytophthora or Pythium species (Blein et al., 2002). Whereas the latter produce their own lipid carriers, Fol might highjack XSP10 to serve a similar function.
In the second scenario, XSP10 might represent a component of a signalling pathway involved in the activation of host defence after pathogen perception. Fol disease symptom development (e.g. yellowing, wilting, etc.) in tomato is a process largely controlled by the plant, as exemplified by the tomato never ripe (NR) mutant that does not develop symptoms although it is colonized to the same extent as wild-type plants (Lund et al., 1998). If XSP10 as a positive regulator is involved in a systemic signal required for symptom development, then its silencing will not lead to increased disease resistance per se, but will reduce symptom development. Alternatively, if XSP10 encodes a negative regulator, then XSP10 silencing is predicted to prime host defence, thereby restricting Fol colonization and symptom development. Recently, the protective role of such a primed defence response to Fol was exemplified by the exogenous application of SA to tomato, through either root feeding or foliar spray, that induced resistance against Fol (Mandal et al., 2009). To investigate involvement of XSP10 in induced resistance responses, the expression levels of PR-1a and WIPI-II were measured in leaves from infected plants. However, significant differences in XSP10, PR-1a and WIPI expression were observed neither locally nor systemically during the course of infection (Fig. 4). This result differs from that found by Rep and colleagues who reported an increased PR-1a expression in the lower part of the stem upon Fusarium infection, a discrepancy that might be caused by the different tissues examined, roots and leaves versus stem tissues (Rep et al., 2003). Nevertheless, since expression of both defence marker genes was unaltered in infected root tissues and in distal uninfected tissues of both wild-type and XSP10-silenced plants, the present data do not lend support to involvement of XSP10 as a negative regulator in either local or systemic signalling.
Future work to uncover the function of XSP10 should include quantification of fungal biomass in the XSP10-silenced tomato lines. These experiments can link infection to symptom development and reveal whether these plants not only show fewer symptoms, but are also more resistant to the pathogen. In addition, bioassays using other pathogens could reveal whether XSP10 is part of a general defence signalling pathway or whether it might encode a pathogenicity target unique for Fol. If XSP10 encodes a signalling component, then overexpression using a xylem-specific promoter could reveal whether it encodes a positive or a negative regulator of this pathway.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Nucleotide sequence of the XSP10 gene and its 5′- and 3′-flanking regions, and the deduced amino acid sequence of XSP10.
Figue S2. Recombinant XSP10 affinity purification.
Figure S3. MS analysis of recombinant XSP10.
Figure S4. Displacement of TNS from XSP10 by stearic, oleic, and jasmonic acids.
Figure S5. Transient GUS expression is silenced in T1 progeny of hpXSP10 lines.
Figure S6. XSP10 abundance is reduced in hpXSP10 plants.
Figure S7. PR1 and WIPI transcription levels in roots and leaves of hpXSP10 plants upon Fol007 infection.
Acknowledgments
We thank Ludek Tikovski, Harrold Lemereis, and Thijs Hendrix for taking care of our plants; Petra Houterman for helping with Fusarium bioassays; Ben Cornelissen for providing feedback on the manuscript; Marianne de Vroomen for assisting with the transformation of tomato; and Tadeusz Wroblewski for valuable suggestions and providing the vector to generate the hpSAMT silencing constructs. We are also grateful to Tadeusz for providing the C58C1 strain carrying the pTFS40 plasmid. We thank Guusje Bonnema for sharing the tomato cosmid library.
References
- Ament K, Krasikov V, Allmann S, Rep M, Takken FLW, Schuurink RC. Methyl salicylate production in tomato affects biotic interactions. The Plant Journal. 2010;62:124–134. doi: 10.1111/j.1365-313X.2010.04132.x. [DOI] [PubMed] [Google Scholar]
- Arondel VV, Vergnolle C, Cantrel C, Kader J. Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Science. 2000;157:1–12. doi: 10.1016/s0168-9452(00)00232-6. [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bernatzky R, Tanksley SD. Genetics of actin-related sequences in tomato. Theoretical and Applied Genetics. 1986;72:314–321. doi: 10.1007/BF00288567. [DOI] [PubMed] [Google Scholar]
- Blein JP, Coutos-Thevenot P, Marion D, Ponchet M. From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends in Plant Science. 2002;7:293–296. doi: 10.1016/s1360-1385(02)02284-7. [DOI] [PubMed] [Google Scholar]
- Buhot N, Gomes E, Milat ML, Ponchet M, Marion D, Lequeu J, Delrot S, Coutos-Thevenot P, Blein JP. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Molecular Biology of the Cell. 2004;15:5047–5052. doi: 10.1091/mbc.E04-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron KD, Teece MA, Smart LB. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiology. 2006;140:176–183. doi: 10.1104/pp.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammue BP, Thevissen K, Hendriks M, et al. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiology. 1995;109:445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CS, Samuel D, Liu YJ, Shyu JC, Lai SM, Lin KF, Lyu PC. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry. 2004;43:13628–13636. doi: 10.1021/bi048873j. [DOI] [PubMed] [Google Scholar]
- Cortina C, Culianez-Macia FA. Tomato transformation and transgenic plant production. Plant Cell, Tissue and Organ Culture. 2004;76:269–275. [Google Scholar]
- Da Silva P, Landon C, Industri B, Marais A, Marion D, Ponchet M, Vovelle F. Solution structure of a tobacco lipid transfer protein exhibiting new biophysical and biological features. Proteins. 2005;59:356–367. doi: 10.1002/prot.20405. [DOI] [PubMed] [Google Scholar]
- de Kock MJD. Thesis. Wageningen University; 2004. Recognition of the Cladosporium fulvum Ecp2 elicitor in tomato and non-host plants. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Johnson RR, Ryan CA. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiology. 1992;98:995–1002. doi: 10.1104/pp.98.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Chen J, Li N, Lin Y, Sun C, Cao K. Resistance function of rice lipid transfer protein LTP110. Journal of Biochemistry and Molecular Biology. 2003;36:603–607. doi: 10.5483/bmbrep.2003.36.6.603. [DOI] [PubMed] [Google Scholar]
- Graham JS, Pearce G, Merryweather J, Titani K, Ericsson LH, Ryan CA. Wound-induced proteinase inhibitors from tomato leaves. II. The cDNA-deduced primary structure of pre-inhibitor II. Journal of Biological Chemisrty. 1985;260:6561–6564. [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of Vir-region and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hoh F, Pons JL, Gautier MF, de Lamotte F, Dumas C. Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding. Acta Crystallogrographica. Section D, Biological Crystallography. 2005;61:397–406. doi: 10.1107/S0907444905000417. [DOI] [PubMed] [Google Scholar]
- Houterman PM, Cornelissen BJ, Rep M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathogens. 2008 doi: 10.1371/journal.ppat.1000061. 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman PM, Speijer D, Dekker H, de Koster CG, Cornelissen BJC, Rep M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Molecular Plant Pathology. 2007;8:215–221. doi: 10.1111/j.1364-3703.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- Jayaraj J, Punja ZK. Combined expression of chitinase and lipid transfer protein genes in transgenic carrot plants enhances resistance to foliar fungal pathogens. Plant Cell Reports. 2007;26:1539–1546. doi: 10.1007/s00299-007-0368-x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Resrarch. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- Kader JC. Lipid-transfer proteins in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- Kroon BAM, Elgersma DM. Interactions between race-2 of Fusarium oxysporum f sp lycopersici and near-isogenic resistant and susceptible lines of intact plants or callus of tomato. Journal of Phytopathology-Phytopathologische Zeitschrift. 1993;137:1–9. [Google Scholar]
- Lascombe MB, Bakan B, Buhot N, Marion D, Blein JP, Larue V, Lamb C, Prangé T. The structure of ‘defective in induced resistance’ protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Science. 2008;17:1522–1530. doi: 10.1110/ps.035972.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauge R, Joosten MHAJ, Haanstra JP, Goodwin PH, Lindhout P, De Wit PJGM. Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proceedings of the National Academy of Sciences, USA. 1998;95:9014–9018. doi: 10.1073/pnas.95.15.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Min K, Cha H, Shin DH, Hwang KY, Suh SW. Rice non-specific lipid transfer protein: the 1.6 Å crystal structure in the unliganded state reveals a small hydrophobic cavity. Journal of Molecular Biology. 1998;276:437–448. doi: 10.1006/jmbi.1997.1550. [DOI] [PubMed] [Google Scholar]
- Lerche MH, Kragelund BB, Bech LM, Poulsen FM. Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure. 1997;5:291–306. doi: 10.1016/s0969-2126(97)00186-x. [DOI] [PubMed] [Google Scholar]
- Lund ST, Stall RE, Klee HJ. Ethylene regulates the susceptible response to pathogen infection in tomato. The Plant Cell. 1998;10:371–382. doi: 10.1105/tpc.10.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- Mandal S, Mallick N, Mitra A. Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiology and Biochemistry. 2009;47:642–649. doi: 10.1016/j.plaphy.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Mikes V, Milat ML, Ponchet M, Panabieres F, Ricci P, Blein JP. Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochemical and Biophysical Research Communications. 1998;245:133–139. doi: 10.1006/bbrc.1998.8341. [DOI] [PubMed] [Google Scholar]
- Molina A, Garcia-Olmedo F. Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. The Plant Journal. 1997;12:669–675. doi: 10.1046/j.1365-313x.1997.00669.x. [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BA, Fasolino A, Hilbers CW, Derksen J, Mariani C. Lipid transfer proteins enhance cell wall extension in tobacco. The Plant Cell. 2005;17:2009–2019. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5:308–16. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Pii Y, Astegno A, Peroni E, Zaccardelli M, Pandolfini T, Crimi M. The Medicago truncatula N5 gene encoding a root-specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti. Molecular Plant-Microbe Interactions. 2009;22:1577–1587. doi: 10.1094/MPMI-22-12-1577. [DOI] [PubMed] [Google Scholar]
- Pons JL, de Lamotte F, Gautier MF, Delsuc MA. Refined solution structure of a liganded type 2 wheat nonspecific lipid transfer protein. Journal of Biological Chemistry. 2003;278:14249–14256. doi: 10.1074/jbc.M211683200. [DOI] [PubMed] [Google Scholar]
- Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, de Koster CG, Cornelissen BJC. A tomato xylem sap protein represents a new family of small cysteine-rich proteins with structural similarity to lipid transfer proteins. FEBS Letters. 2003;534:82–86. doi: 10.1016/s0014-5793(02)03788-2. [DOI] [PubMed] [Google Scholar]
- Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, Speijer D, Back JW, de Koster CG, Cornelissen BJC. Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiology. 2002;130:904–917. doi: 10.1104/pp.007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Meijer M, Houterman PM, van der Does HC, Cornelissen BJC. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Molecular Plant-Microbe Interactions. 2005;18:15–23. doi: 10.1094/MPMI-18-0015. [DOI] [PubMed] [Google Scholar]
- Samuel D, Liu YJ, Cheng CS, Lyu PC. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa) Journal of Biological Chemistry. 2002;277:35267–35273. doi: 10.1074/jbc.M203113200. [DOI] [PubMed] [Google Scholar]
- Satoh S, Iizuka C, Kikuchi A, Nakamura N, Fujii T. Proteins and carbohydrates in xylem sap from squash root. Plant and Cell Physiology. 1992;33:841–847. [Google Scholar]
- Schagger H, von Jagow G. Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schob H, Kunz C, Meins F., Jr. Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigating sequence requirements for gene silencing. Molecular and General Genetics. 1997;256:581–585. doi: 10.1007/s004380050604. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins on silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC. Cell-specific expression of the carrot EP2 lipid transfer protein gene. The Plant Cell. 1991;3:907–921. doi: 10.1105/tpc.3.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Rep M. The arms race between tomato and Fusarium oxysporum. Molecular Plant Pathology. 2010;11:309–314. doi: 10.1111/j.1364-3703.2009.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin S, Broekaert WF, Marion D, Acland DP, Ptak M, Vovelle F, Sodano P. Solution structure of Ace-AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry. 1998;37:3623–3637. doi: 10.1021/bi9723515. [DOI] [PubMed] [Google Scholar]
- Tassin-Moindrot S, Caille A, Douliez JP, Marion D, Vovelle F. The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. European Journal of Biochemistry. 2000;267:1117–1124. doi: 10.1046/j.1432-1327.2000.01109.x. [DOI] [PubMed] [Google Scholar]
- Thoma S, Kaneko Y, Somerville C. A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. The Plant Journal. 1993;3:427–436. doi: 10.1046/j.1365-313x.1993.t01-25-00999.x. [DOI] [PubMed] [Google Scholar]
- Tomilov AA, Tomilova NB, Wroblewski T, Michelmore R, Yoder JI. Trans-specific gene silencing between host and parasitic plants. The Plant Journal. 2008;56:389–397. doi: 10.1111/j.1365-313X.2008.03613.x. [DOI] [PubMed] [Google Scholar]
- Van Kan JAL, Joosten MHAJ, Wagemakers CA, Van den Berg-Velthuis GC, De Wit PJGM. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Molecular Biology. 1992;20:513–527. doi: 10.1007/BF00040610. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Wellman FL. A technique for studying host resistance and pathogenicity in tomato Fusarium wilt. Phytopathology. 1939;29:945–956. [Google Scholar]
- Wroblewski T, Piskurewicz U, Tomczak A, Ochoa O, Michelmore RW. Silencing of the major family of NBS-LRR-encoding genes in lettuce results in the loss of multiple resistance specificities. The Plant Journal. 2007;51:803–818. doi: 10.1111/j.1365-313X.2007.03182.x. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczakt A, Michelmore R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnology Journal. 2005;3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.