Abstract
Background:
Children who snore but do not have gas exchange abnormalities or alterations of sleep architecture have primary snoring (PS). Since increasing evidence suggest that PS may be associated with morbidity, we hypothesized that assessing genome-wide gene expression in peripheral blood leukocytes (PBL) will identify a distinct signature in PS children.
Methods:
Children (aged 4–9 years) with and without habitual snoring and a normal PSG were designated as either PS or controls. Whole genome expression profiles of PBL and metabolic parameters in 30 children with PS and 30 age-, gender-, ethnicity-, and BMI-matched controls were compared. Pathway-focused gene network analysis of the PBL transcriptome was performed. Metabolic parameters were measured in an independent follow-up cohort of 98 children (64 PS and 34 controls) to evaluate the computationally derived findings.
Results:
PS was not associated with a distinct transcriptional signature in PBL. Exploratory functional network analysis of enriched gene sets identified a number of putative pathways—including those mapping to insulin signaling, adipocyte differentiation, and obesity—with significant alterations in glucose metabolism and insulin sensitivity emerging in the follow-up cohort of children with PS, but no differences in lipid profiles.
Conclusions:
PS children do not exhibit global perturbations in their PBL transcriptional response, suggesting that current normative PSG criteria are overall valid. However, subtle differences in functionally coherent pathways involved in glycemic homeostasis were detected and confirmed in a larger independent pediatric cohort indicating that PS may carry increased risk for end-organ morbidity in susceptible children.
Citation:
Khalyfa A; Gharib SA; Kim J; Capdevila OS; Kheirandish-Gozal L; Bhattacharjee R; Hegazi M; Gozal D. Peripheral blood leukocyte gene expression patterns and metabolic parameters in habitually snoring and non-snoring children with normal polysomnographic findings. SLEEP 2011;34(2):153-160.
Keywords: Snoring, sleep apnea, children, inflammation, insulin resistance
OVERNIGHT POLYSOMNOGRAPHY (PSG) OBJECTIVELY ASSESSES MOST OF THE ABNORMALITIES RELATED TO INTERMITTENT UPPER AIRWAY OBSTRUCTION during sleep including hypoxia, hypercapnia, apnea, and arousals. In recent years, the pediatric community has undertaken efforts to establish normative reference values for PSGs in order to delineate the spectrum of sleep measures in healthy children.1–5 However, we are unaware of any consensus guidelines whereby specific polysomnographic criteria have been recommended to reliably identify children requiring treatment, i.e., children suffering from the obstructive sleep apnea syndrome (OSAS), while also identifying children in whom treatment is either not indicated or can be postponed.6,7 In the absence of such guidelines, there is substantial variability in current practice among sleep physicians. However, there has been informal consensus that children who habitually snore during sleep but who exhibit an obstructive apnea-hypopnea index < 1–2/h of total sleep time in the absence of oxyhemoglobin desaturations and sleep fragmentation have a condition termed “primary snoring” (PS). PS has been traditionally perceived as being a benign condition and consequently not requiring treatment.
The previously held view on the benignity of PS has been recently challenged.8 In a number of studies assessing behavioral and cognitive functioning in school-aged children, the presence of PS was associated with an increased risk for altered behavior and reduced cognitive performance in a subset these children, suggesting the presence of individual susceptibility differences across the spectrum of pediatric sleep disordered breathing.9,10 Similarly, children with PS have been shown to have elevated systemic blood pressures as well as abnormalities in arterial wall rigidity, albeit not consistently, across various international cohorts.11–14 Furthermore, increased frequency of enuresis and somatic growth alterations have also been reported among habitually snoring children as defined by questionnaire or PSG-based definition of obstructive apnea-hypopnea index < 2 per hour.15–17
Transcriptional profiling is a well-established approach to comprehensively assess changes in expression occurring across the whole genome.18 In a recent study, we showed that a substantial number of genes were differentially expressed in peripheral blood leukocytes of non-obese children with OSA, and that inflammatory pathways were particularly over-represented, suggesting that OSA activates widespread pro-inflammatory networks that may play a role in the end-organ morbidity associated with this condition.19 Considering the uniquely high sensitivity of this approach, we hypothesized that genome-wide expression profiling may provide additional clues as to whether primary snorers fall closer to healthy non-snoring children or, have transcriptional responses that place them within the spectrum of sleep disordered breathing.
MATERIALS AND METHODS
Subjects
The study was approved by the University of Louisville Human Research Committee and the Jefferson County Public Schools Board. Parental informed consent and child assent, in the presence of a parent, were obtained. Pediatric population was voluntarily recruited from the public school system of Louisville Jefferson County Public School System. Parents of all children who enrolled were invited to complete sleep questionnaire, in which the main question of interest for the present study revolved around snoring.10,20 Responses were graded as “never,” “rarely” (once per week), “occasionally” (twice per week), “frequently” (3–4 times per week), “almost always” (4 times per week), and “always” (every night). Children with answers in the “frequently” to “always” range were invited to undergo an overnight sleep study and based upon the results of such study were defined as having primary snoring (PS; see below). Similarly, control subjects were recruited, were initially screened, and invited to participate if they had no history of snoring and no symptoms of sleep disordered breathing, and if their overnight PSG was within normal limits (see below). Children were excluded if they had any chronic medical condition, receiving medications, and if they had any genetic or craniofacial syndromes. The control children were matched for age, gender, ethnicity, and BMI z-score to those defined as having PS.
Overnight Polysomnography Evaluation
A standard overnight multichannel PSG evaluation was performed at the University of Louisville Pediatric Sleep Medicine Center. Children were studied for up to 12 h in a quiet, darkened room with an ambient temperature of 24°C in the company of one of their parents or guardian. All children were in bed with lights out between 21:00 and 21:30 and were awakened at 07:00 unless they awoke earlier. No drugs were used to induce sleep. The following parameters were measured: chest and abdominal wall movement by respiratory impedance or inductance plethysmography, heart rate by electrocardiogram, and air flow with a nasal pressure transducer and oronasal thermistor, a side-stream end-tidal capnograph, which also provided breath-by-breath assessment of end-tidal carbon dioxide levels (BCI SC-300, Menomonee Falls, WI) and an oronasal thermistor. Arterial oxygen saturation (SpO2) was assessed by pulse oximetry (Nellcor N 100; Nellcor Inc, Hayward, CA), with simultaneous recording of the pulse waveform. The bilateral electro-oculogram, 8 channels of electroencephalogram (2 frontal, 2 occipital, 2 temporal, and 2 central leads), chin and anterior tibial electromyograms, and analog output from a body position sensor (Braebon Medical Corporation, New York, NY) were also monitored. All measures were digitized using a commercially available polysomnography system (Stellate Systems, Montreal, Canada, or Medcare, Buffalo, NY). Tracheal sound was monitored with a microphone sensor (Sleepmate, Midlothian, VA), and a digital time-synchronized video recording was performed. The sleep technician followed patient behavior and confirmed sleep position by the infrared camera inside the room. Sleep architecture was assessed by standard techniques.1,21 The proportion of time spent in each sleep stage was expressed as a percentage of total sleep time (TST).
Obstructive apnea was defined as the absence of airflow with continued chest wall and abdominal movement for ≥ 2 breaths.22 Hypopnea was defined as a decrease in airflow ≥ 50% (based on nasal pressure transducer, or in case of poor transducer signal quality, the oronasal thermistor) with a corresponding decrease in SpO2 ≥ 3% and/or arousal. The obstructive apnea hypopnea index (OAHI) was defined as the number of obstructive apneas and hypopneas per hour of TST. Arousals were defined as recommended and included respiratory related (occurring immediately after an apnea, hypopnea, or snore) and spontaneous arousals—although inclusion of arousals following snore did not meet the current AASM scoring guidelines.21 Arousals were expressed as the total number of arousals per hour of sleep time. PS was defined by an OAHI ≤ 2/h TST in the presence of a positive history of snoring and documented snoring during the PSG, while controls had similar PSG criteria, but absence of documented snoring during PSG in addition to a negative history of snoring.
Assessment of Metabolic Parameters
Blood samples were drawn by venipuncture in the morning after the sleep study after an overnight fast. Blood samples were immediately centrifuged and plasma was frozen at −80°C. Plasma insulin levels were measured using a commercially available radioimmunoassay kit (Coat-A-Count Insulin, Cambridge Diagnostic Products, Inc, Fort Lauderdale, FL). Plasma glucose levels were measured using a commercial kit based on the hexokinase-glucose-6-phosphate dehydrogenase method (Flex Reagent Cartridges, Dade Behring, Newark, DE). Serum lipids, including total cholesterol, high-density lipoprotein (HDL) cholesterol, calculated low-density lipoprotein (LDL) cholesterol, and triglycerides were also assessed, as well as high sensitivity C-reactive protein (CRP) (Flex Reagent Cartridges, Dade Behring, Newark, DE). For the second cohort, insulin resistance was also assessed using the homeostasis model assessment (HOMA) equation (fasting insulin × fasting glucose ÷ 405).23
Body Mass Index
Height and weight were obtained using standard techniques from each child. BMI was then calculated (body mass/height2) and was expressed as BMI z-score using an online BMI z-score calculator (http://www.cdc.gov/epiinfo/). Children with BMI z-score values > 1.20 were classified as fulfilling the criteria for overweight/obesity.24 Children with BMI z-score values > 1.65 were considered obese and excluded from our analysis.
Statistical Analysis
For demographic and sleep measures, data are presented as mean ± SD unless otherwise indicated. All analyses were conducted using SPSS software (version 17.0; SPPS, Inc., Chicago, IL). Comparisons of demographics, sleep measures, and metabolic parameters were performed using independent t-tests or by analysis of the variance (ANOVA) followed by post hoc comparisons, with P-values adjusted for unequal variances when appropriate (Levene test for equality of variances). All P-values reported are 2-tailed with statistical significance set at < 0.05.
RNA Isolation
Following the sleep study, fasting peripheral blood samples were drawn from all children within the first hour after awakening and collected in PAXgene Blood RNA tubes (Becton Dickinson, UK). Total RNA was isolated using a PAXgene Blood RNA Kit and treated with DNase I (QIAGEN, CA), according to the manufacturer's protocol. The RNA quantity and integrity were determined using a Nanodrop Spectrophotometer and Agilent 2100 Bioanalyzer Nano 6000 LabChip assay (Agilent Technologies).
Microarray Experiments
Total RNA from 30 children with primary snoring and 30 control children was used for complementary DNA (cDNA) synthesis. Equal quantities of total RNA were labeled using Agilent's low RNA input fluorescent linear amplification kit, and hybridized to 60 independent microarrays. Briefly, total RNA (500 ng) was reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase with oligo (dT) primer. Fluorescent cRNAs were synthesized by in vitro transcription using T7 RNA polymerase and labeled with cyanine 3-dCTP (Cy3-dCTP; Perkin Elmer, Boston, MA, USA). Labeled cRNAs were further purified using RNeasy mini spin columns (Qiagen) to remove unlabeled products. Fifteen picomoles of the fluorescently labeled cRNA was used for each microarray hybridization.
Hybridizations were performed using In Situ Hybridization Kit Plus (Agilent) according to the manufacturer's protocols. Each subject's cRNA was hybridized to a human Agilent array 60-mer (G4112A) containing 44,000 human probe sequences. Following hybridization, the arrays were scanned immediately using the Agilent Microarray Scanner. The scanned microarray images were processed with Feature Extraction software v. 9.3.5 (Agilent), and captured images were analyzed and filtered by GeneSpring v. 10.0 Software (Agilent).
Five microarray experiments (4 control and 1 PS) were excluded from analysis because of poor hybridization quality that did not pass our rigorous image analysis filtering criteria. The resulting 55 experiments (26 control and 29 PS) underwent further analysis. The gene expression intensities of all 55 microarrays were normalized using the quantile method.25
Gene Expression Analysis
The presence of differential gene expression between PS and control children was assessed using: (1) a Bayesian implementation of the parametric t-test that uses a probabilistic framework with Gaussian independent modeling to obtain point estimates for parameters based on the empirical and local background variances of neighboring gene coupled with false discovery rate (FDR) analysis,26 and (2) a well-known nonparametric method, significance analysis of microarrays (SAM).27 An FDR cutoff < 0.05 was deemed significant for either statistical test. Multidimensional scaling using principal components was performed based on the covariance matrix of normalized gene expression values across all 55 subjects.28
Identification of Enriched Pathways
Enriched pathways in peripheral blood leukocytes of children with and without primary snoring were identified using gene set enrichment analysis (GSEA).29 Approximately 1890 curated and 1450 Gene Ontology gene sets were computationally assessed. A random permutation analysis (n = 2000) of gene sets was applied to determine enrichment of biological pathways in each group using an FDR cutoff < 0.05.
Gene Interaction Network Analysis
Genes mapping to gene sets enriched in the PBL of children with and without primary snoring and involved in insulin signaling, obesity and adipocyte differentiation pathways were combined. A network connecting these gene products was created based on previously published direct and indirect interactions using Ingenuity's knowledge base30 and several publicly available databases. The interaction network, or interactome, was built around genes with the highest connectivity using an iterative algorithm that systematically connected additional nodes to the initial seed.
RESULTS
Subject Characteristics and Polysomnographic Parameters
A total of 60 children were recruited for the initial study: 30 children with PS and 30 matched controls. As shown in Table 1, these children were of similar age, gender, ethnicity, and BMI. The subjects had similar sleep latency and duration, sleep architecture, mean SpO2, and periodic leg movement index. Children with PS had a trend towards a slightly higher mean obstructive AHI (P-value = 0.1). Children with PS had a small, albeit statistically significant elevation in their PETCO2 (Table 1).
Table 1.
Demographic and polysomnographic characteristics of the initial cohort
| PS (n = 30) | Controls (n = 30) | |
|---|---|---|
| Age (years) | 6.9 ± 0.6 | 7.0 ± 0.5 |
| Male (n) | 16 | 15 |
| African American (n) | 10 | 10 |
| BMI (z-score) | 0.63 ± 0.22 | 0.47 ± 0.25 |
| Sleep latency (min) | 23.8 ± 15.4 | 23.4 ± 14.2 |
| REM latency (min) | 145.5 ± 43.2 | 146.2 ± 44.4 |
| Total sleep time (h) | 8.0 ± 0.3 | 8.1 ± 0.4 |
| Sleep Efficiency (%) | 94.7 ± 3.1 | 94.6 ± 3.9 |
| Stage 1 (%) | 5.7 ± 3.7 | 5.6 ± 4.2 |
| Stage 2 (%) | 42.3 ± 6.5 | 42.0 ± 6.6 |
| Stage 3 (%) | 6.5 ± 3.2 | 5.6 ± 3.4 |
| Stage 4 (%) | 23.7 ± 6.1 | 24.9 ± 5.9 |
| REM sleep (%) | 21.8 ± 5.1 | 21.9 ± 6.0 |
| Total arousal index (/h TST) | 6.9 ± 2.3 | 6.8 ± 2.4 |
| PLM index with arousal (/h TST) | 0.2 ± 0.5 | 0.2 ± 0.6 |
| PLM index in sleep (/h TST) | 1.1 ± 1.6 | 0.7 ± 1.4 |
| OAI (/h TST) | 0.2 ± 0.5 | 0.1 ± 0.3 |
| OAHI (/h TST) | 0.6 ± 0.3 | 0.3 ± 0.2 |
| Mean SpO2 (%) | 97.9 ± 0.6 | 98.6 ± 0.4 |
| SpO2 nadir (%) | 91.3 ± 1.4 | 93.0 ± 0.8 |
| Peak PETCO2 (mm Hg) | 51.5 ± 1.6* | 48.4 ± 1.5 |
| Mean PETCO2 (mm Hg) | 47.6 ± 0.7* | 43.8 ± 2.0 |
Data shown as mean ± SD; Level of significance: *P < 0.05; SpO2, Arterial oxygen saturation measured by pulse oximetry; TST, total sleep time; PLM, periodic leg movement; OAI, obstructive apnea index; OAHI, obstructive apnea hypopnea index; PETCO2, end-tidal carbon dioxide tension measured by capnography; BMI, body mass index.
Metabolic Parameters
Table 2 summarizes the results from 18 children with PS and 24 controls in whom we had performed microarray experiments and obtained metabolic parameters. No significant differences in glucose, insulin, total cholesterol, LDL, HDL, triglyceride, or CRP levels were observed between the groups.
Table 2.
Metabolic parameters of the initial cohort of children with PS and matched controls
| PS (n = 18) | Control (n = 24) | P-value | |
|---|---|---|---|
| Glucose mg/dL | 86.4 ± 4.3 | 86.0 ± 6.0 | NS |
| Insulin μIU/mL | 7.9 ± 3.6 | 6.2 ± 2.5 | NS |
| Triglycerides mg/dL | 79.3 ± 64.5 | 49.6 ± 20.1 | NS |
| Cholesterol mg/dL | 151.9 ± 30.0 | 148.3 ± 23.1 | NS |
| HDL mg/dL | 44.9 ± 11.4 | 50.3 ± 8.6 | NS |
| LDL mg/dL | 91.1 ± 23.1 | 88.1 ± 23.0 | NS |
| hsCRP mg/L | 2.8 ± 2.5 | 3.2 ± 2.8 | NS |
Data shown as mean ± SD; NS, not significant; HDL, high density lipid cholesterol; LDL, low density lipid cholesterol; hsCRP, high sensitivity C-reactive protein.
Transcriptional Profiles of PBL
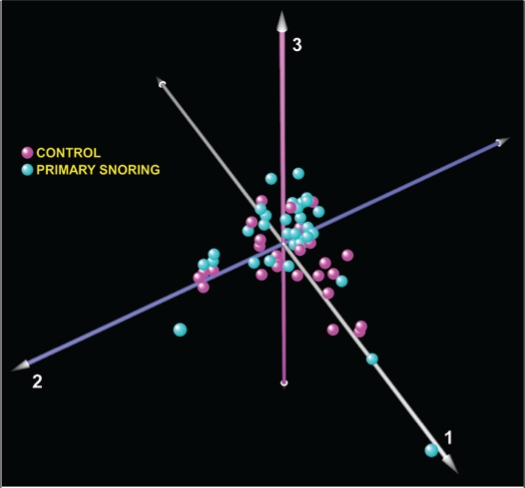
Genome-wide expression analysis on PBL of only 55 subjects with and without primary snoring was performed using 2 different statistical methods—one parametric and the other nonparametric. The other 5 samples (1 PS and 4 controls) did not pass array high stringency quality controls. No differentially expressed gene meeting our pre-designated FDR cutoff value of < 0.05 was identified using either one of the two statistical approaches. Principal components analysis of whole-genome transcriptional profiles across all subjects also failed to segregate the children with PS from matched controls (Figure 1). When this analysis was applied separately to children stratified by race (European and African ancestry), we again did not observe any segregation between the phenotypes (supplementary Figure S1). These results imply that habitual snoring in children without PSG evidence of respiratory disturbances exceeding the defined normative cut-off values does not elicit distinct global transcriptional perturbations in their PBL.
Figure 1.
Principal component analysis of peripheral blood leukocyte gene expression from 55 children with primary snoring (n = 29, cyan) and matched controls (n = 26, magenta). The absence of phenotype-specific clusters implies the lack of a significant global transcriptional perturbation.
Pathway-Focused Network Analysis
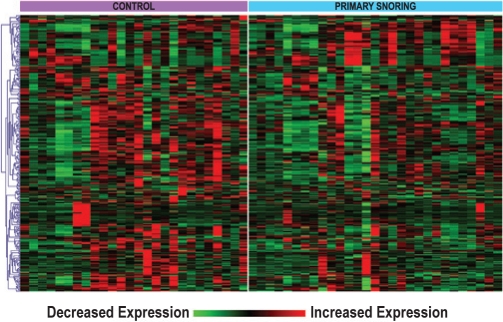
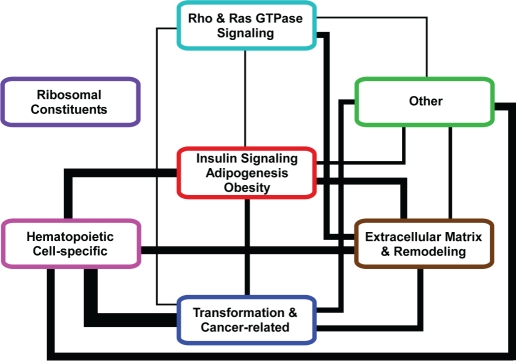
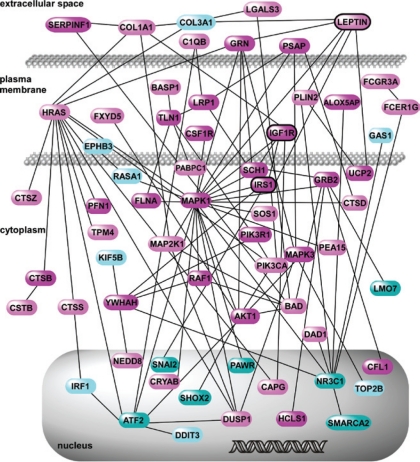
Although standard statistical methods did not detect a significant difference in PBL gene expression between children with PS and controls, these analyses were based on gene-by-gene comparisons. However, biological processes are often orchestrated by co-regulated changes in the expression of multiple genes mapping to coherent functional modules.31 We therefore performed pathway-oriented exploratory analyses, also known as gene set enrichment analysis (GSEA),29 in which over-representation of curated pathways instead of individual genes is assessed between phenotypes. Using a pre-designated FDR cutoff value of < 0.05, we identified 35 gene sets that were enriched between the 2 groups (supplementary Table S1). The enrichment of each gene set was primarily due to a subset of genes known as the “leading edge.” Figure 2 is a heatmap depiction of expression profiles of the leading edge gene set members across all 55 subjects, demonstrating distinct transcriptional patterns between the groups. Since there were overlaps between the 35 enriched gene sets, we summarized our findings by grouping them into 6 broadly distinct categories. The functional categories included among these gene sets included a module involved in adipocyte differentiation/obesity and insulin signaling (Figure 3). To gain a more detailed understanding of this metabolism-associated module in the setting of PS, we performed a network analysis based on known interactions among its gene products. This interactome was comprised of 64 nodes (Figure 4), with colors indicating whether a gene was upregulated in PS (cyan) or in control children (magenta), whereas the more intense shades of each color identify the nodes that were members of the leading edge. The resulting network highlights the complex relationships between gene products involved in insulin growth factor signaling and pathways associated with adipogenesis and obesity.
Figure 2.
Gene expression heatmap of “leading edge” members of enriched gene sets in PBL of children with PS vs. controls. One-dimensional hierarchical clustering of expression values has been performed to better depict distinct transcriptional patterns between the phenotypes.
Figure 3.
A wiring diagram depiction of the functional categories enriched between children with PS and controls. The intermodular connections reflect the fact that some genes map to multiple modules, while the line thickness is proportional to the number of shared genes.
Figure 4.
Gene product interaction network constructed from gene members mapping to the insulin signaling, adipocyte differentiation and obesity module (see Figure 3). Nodes up-regulated in PS are shown in cyan, and those up-regulated in controls are colored in magenta. Members of the leading edge are highlighted in darker shades. Among the complex interactions, note the relational links between leptin, IGF1R, and IRS1 as discussed in the text. A complete list of gene members is provided in the supplementary Table S2.
Perturbation of Glycemic Homeostasis in Children with PS
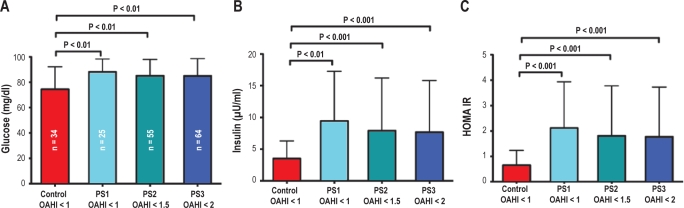
Given the subtle changes in metabolic pathways and insulin signaling identified by our bioinformatics approach, we hypothesized that the absence of any differences in serum levels of metabolic biomarkers in PS children might be due to statistical underpowering of our sample size. Thus, we assessed whether metabolic alterations were present in a large, independent cohort of children with PSG-defined PS and matched controls. A total of 98 subjects (64 PS, and 34 controls) were studied (Table 3). Since there is disagreement on normative OAHI cut-off values in children, we stratified the PS subjects into 3 groups based on their OAHI: (1) PS1 (OAHI < 1), (2) PS2 (OAHI < 1.5), and (3) PS3 (OAHI < 2). Children with PS in all 3 groups had statistically significant elevations in their fasting glucose, fasting insulin levels, and insulin resistance (HOMA-IR) compared to controls (Figure 5). There were statistical differences between the OAHI of PS2 and PS3 children relative to the non-snoring controls, but importantly, the alterations in glycemic control persisted even in subjects with OAHI < 1 (PS1), suggesting that metabolic perturbations are present in habitually snoring children with normal PSG findings. No significant differences were seen in the lipid profiles of children with and without PS (Table 3).
Table 3.
Polysomnographic and metabolic characteristics of the follow-up cohort
| PS1 (n = 25) | PS2 (n = 55) | PS3 (n = 64) | Control (n = 34) | |
|---|---|---|---|---|
| Glucose (mg/dL) | 88.1 ± 10.2* | 85.2 ± 12.8* | 85.0 ± 13.6* | 74.6 ± 17.7 |
| Insulin (μIU/mL) | 9.4 ± 7.8* | 7.9 ± 8.3** | 7.7 ± 8.1** | 3.5 ± 2.8 |
| HOMA-IR | 2.1 ± 1.8** | 1.8 ± 2.0** | 1.8 ± 2.0** | 0.7 ± 0.6 |
| Triglycerides (mg/dL) | 71.6 ± 26.9 | 74.5 ± 34.9 | 75.2 ± 33.8 | 71.3 ± 45.6 |
| Cholesterol (mg/dL) | 159.1 ± 23.7 | 160.6 ± 25.0 | 164.9 ± 26.2 | 161.8 ± 25.1 |
| HDL (mg/dL) | 53.6 ± 13.9 | 53.2 ± 13.5 | 54.1 ± 12.9 | 54.8 ± 9.6 |
| LDL (mg/dL) | 84.7 ± 28.7 | 89.6 ± 24.1 | 93.3 ± 24.7 | 92.4 ± 22.6 |
| hsCRP (mg/L) | 0.2 ± 0.1* | 0.2 ± 0.2* | 0.3 ± 0.3* | 1.1 ± 2.4 |
| SpO2 nadir (%) | 91.8 ± 4.7 | 90.4 ± 7.0 | 90.2 ± 6.7* | 92.4 ± 2.4 |
| Mean SpO2 (%) | 97.4 ± 1.7 | 97.5 ± 1.3 | 97.4 ± 1.2 | 97.5 ± 0.6 |
| Total arousal index (/h TST) | 7.9 ± 3.8* | 9.5 ± 4.9* | 10.2 ± 5.4 | 12.1 ± 6.4 |
| OAHI (/h TST) | 0.5 ± 0.3 | 0.9 ± 0.5** | 1.0 ± 0.5** | 0.5 ± 0.2 |
| BMI z-score | 0.2 ± 1.7 | 0.1 ± 1.5 | 0.1 ± 1.4 | 0.3 ± 1.0 |
| Age (years) | 6.5 ± 1.7 | 6.7 ± 1.6 | 6.8 ± 1.6 | 7.0 ± 1.1 |
Data shown as mean ± SD; *P < 0.05 compared to Control; **P < 0.001 compared to Control; PS1, primary snorers with OAHI < 1/h TST; PS2, primary snorers with OAHI < 1.5/h TST; PS3, primary snorers with OAHI < 2/h TST; HDL, high density lipid cholesterol; LDL, low density lipid cholesterol; hsCRP, high sensitivity C-reactive protein; SpO2, Arterial oxygen saturation measured by pulse oximetry; OAHI, obstructive apnea hypopnea index; BMI, body mass index.
Figure 5.
Assessment of glycemic homeostasis in an independent follow-up cohort comprised of 64 children with PS and 34 matched Controls. Habitually snoring children were stratified into three groups based on their PSG-derived OAHI (PS1: OAHI < 1/h TST, PS2: OAHI < 1.5/h TST, and PS3: OAHI < /h TST). Note that PS1 and PS2 are subsets of PS3, and PS1 is a subset of PS2. There are persistent alterations in fasting glucose, fasting insulin, and HOMA insulin resistance in children with PS compared to Controls, even in habitual snorers with OAHI < 1 /h TST (PS1). Data presented as mean ± SD, P-values are based on 2-tailed Student t-test with unequal variances.
DISCUSSION
Habitually snoring children who do not meet current guidelines for the diagnosis of obstructive sleep apnea comprise a large segment of the pediatric population. Current guidelines for clinical management of these children do not advocate the need for treatment implementation, possibly reflecting the fact that the health impact of PS is not well understood and the role of therapeutic interventions remains unexplored. In this study, we investigated the transcriptional effects of PS in peripheral blood leukocytes using a large, community-based cohort of children without polysomnographic evidence of obstructive sleep apnea. Our main finding was that PS is not associated with a distinct transcriptional signature in circulating leukocytes. This observation is in contrast to our previous report demonstrating the up-regulation of pro-inflammatory pathways in the PBL of children with OSA.19 Therefore, current PSG-based normative guidelines for pediatric sleep apnea appear to serve as a reasonable demarcation cut-off, such that children with snoring during sleep whose PSG measures remain within the normative range fail to exhibit biologically relevant transcriptional patterns in their peripheral blood leukocytes. Furthermore, PS was not associated with significant alterations in metabolic parameters or markers of inflammation in the initial cohort of children studied herein (Table 2), although the size this cohort was clearly underpowered, as subsequently shown in a larger follow-up group.
The unbiased, genome-wide interrogation of PBL gene expression on a relatively large group of subjects provided opportunities for exploratory data mining. Since many biological processes, particularly those involved in metabolism,32,33 are characterized by modest yet coordinated changes in gene expression, we employed computational methods that take advantage of this property and systematically identified enriched gene sets and networks in circulating leukocytes. Our results indicate that despite the lack of a profound global transcriptional response in PBL of children with PS, a pathway-focused approach can highlight the significant activation of a number of coherent biological processes, including those involved in insulin signaling, obesity, and adipocyte differentiation (Figures 3 and 4). Of note, selective activation and recruitment of circulating leukocytes to peripheral depots such as adipose tissue is being increasingly recognized as a key event in pathogenesis of obesity and metabolic syndrome.34–36
Several members of the gene interaction network encompassing these pathways have established roles in regulating the activity of peripheral blood leukocytes. For example, leptin has well-known immunomodulatory effects on peripheral blood mononuclear cells (PBMC)37,38 and leptin deficient mice (ob/ob) display immune dysfunction with reduced levels of peripheral T and B cells.39 Although primarily produced by adipocytes, leptin gene expression has been measured in PBMCs and is associated with blood pressure variability.40 Another network node, IRS1 (insulin receptor substrate 1) is a critical component of insulin signaling, and has been shown to undergo phosphorylation by leptin in human circulating lymphocytes.41 As depicted in Figure 4, IGF1R (insulin growth factor-1 receptor) directly interacts with IRS1 in the interactome. IGF1R is expressed on peripheral blood leukocytes,42 and serves as a primary receptor for IGF1—a polyfunctional growth hormone critical in regulating tissue growth, metabolism43,44 (e.g., insulin sensitivity), and neuroprotection.45 We recently demonstrated that plasma IGF1 levels correlate with cognitive dysfunction in children with obstructive sleep apnea.46
Given the exploratory nature of the pathway-focused network analysis and since the transcriptional effects of PS appeared to be very mild, we proceeded to confirm our findings by assessing metabolic parameters in a significantly larger cohort of children with PS. Our results, summarized in Table 3, suggest that habitual snoring in children without OSA is associated with alterations in glucose metabolism and insulin resistance, but not in their lipid profiles. These effects were present even when we limited the habitually snoring children to those with OAHI < 1. Thus, it is appears that PS may impose subtle, yet measurable changes in metabolic regulatory pathways in all children who snore, or alternatively that a sizable subset of children with PS may exhibit genetically and environmentally driven metabolic susceptibility.47–50
Several limitations in this study deserve mention, most importantly that the bioinformatics analyses conducted were exploratory in nature and intended to computationally identify putative pathways selectively enriched in circulating leukocytes of children with PS. Since the sample size used in the transcriptional profiling experiments was relatively small, some of our findings may be falsely positive due to selection of pathways by random chance. These limitations, in conjunction with the fact that we have not confirmed the activation of enriched processes using focused functional studies, imply that our results are hypothesis-generating and will require a larger study for confirmatory purposes. Additionally, the relational network created from the enriched pathway analysis is limited by our current state of knowledge, and is therefore an incomplete representation of the underlying biology. Nevertheless, the finding of perturbations in glucose metabolism and insulin resistance among children with PS in the larger, follow-up cohort is intriguing and warrants further investigation. Another limitation of this study is that we have interrogated gene expression patterns of peripheral blood leukocytes and not assessed the effects of primary snoring in the transcriptome of target end-organs. Although innate and adaptive immunity play critical roles in the pathogenesis of obesity and metabolic dysregulation, our study design only allowed for “associating” gene expression patterns of circulating immune cells with metabolic parameters. Discovering a direct link between these compartments in habitually snoring children will require organ-specific functional studies in the future.
This work has a number of notable strengths. First, to our knowledge, it is the largest study of its kind assessing metabolic disturbances in a community-based cohort of children with primary snoring and matched controls.51–60 Second, all subjects underwent meticulous demographic and polysomnographic evaluation before providing blood samples for comprehensive metabolic profiling. Third, the gene expression experiments were subjected to pathway-focused network analysis, yielding potentially novel insights into activated transcriptional programs in circulating white blood cells of children with primary snoring. Furthermore, we followed up on the relevance of the computationally identified pathways using a large independent cohort of children with PS.
Taken together, this study demonstrates that habitually snoring children without polysomnographic evidence of OSA do not have marked perturbations in their peripheral blood leukocyte transcriptome, lipid profiles, or markers of inflammation. However, there is selective enrichment of a number of metabolic pathways that are associated with subtle alterations in glucose metabolism and insulin resistance. Therefore, children with PS appear to fall closer to healthy non-snorers based on their transcriptional patterns, and yet also display evocative findings of mild dysregulation in glycemic homeostasis.
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Leila Kheirandish-Gozal has received research support from Merck. Dr. David Gozal has participated in speaking engagements for Merck and has consulted for Galleon Pharmaceuticals.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health HL065270 and HL086662 (DG), and HL074223 (SAG).
Footnotes
See commentary: Wong TK. The search on an ideal disease marker for childhood obstructive sleep apnea syndrome. SLEEP 2011;34:133–134.
Principal components analysis (PCA) on the global expression profiles of children with and without primary snoring after stratification by race (only European and African ancestry shown). Consistent with our initial PCA findings that was based on all the subjects (manuscript Figure 1) no discernable expression pattern separated the primary snoring groups from the controls in each race.
| Gene Set | Enriched Phenotype | Gene Set Size | P-value | FDR | Enrichment Score |
|---|---|---|---|---|---|
| HSA03010_RIBOSOME | PS | 69 | 0.0000 | 0.0000 | −2.51 |
| STRUCTURAL_CONSTITUENT_OF_RIBOSOME | PS | 67 | 0.0000 | 0.0000 | −2.48 |
| RIBOSOMAL_PROTEINS | PS | 75 | 0.0000 | 0.0005 | −2.29 |
| REGULATION_OF_SMALL_GTPASE_MEDIATED_SIGNAL | Control | 22 | 0.0000 | 0.0021 | 2.40 |
| REGULATION_OF_RAS_PROTEIN_SIGNAL_TRANSDUCTION | Control | 18 | 0.0000 | 0.0052 | 2.33 |
| GOLUB_ALL_VS_AML_DN | Control | 14 | 0.0000 | 0.0112 | 2.27 |
| KNUDSEN_PMNS_DN | Control | 200 | 0.0000 | 0.0113 | 2.28 |
| GNATENKO_PLATELET_UP | Control | 40 | 0.0000 | 0.0134 | 2.34 |
| GNATENKO_PLATELET | Control | 40 | 0.0000 | 0.0154 | 2.30 |
| NADLER_OBESITY_UP | Control | 50 | 0.0000 | 0.0164 | 2.21 |
| MOREAUX_TACI_HI_VS_LOW_DN | PS | 146 | 0.0000 | 0.0230 | −1.99 |
| HDACI_COLON_SUL12HRS_DN | PS | 26 | 0.0000 | 0.0243 | −2.02 |
| STEMCELL_COMMON_UP | PS | 164 | 0.0000 | 0.0248 | −1.99 |
| ADIPOCYTE DIFFERENTIATION | PS | 38 | 0.0000 | 0.0264 | −2.00 |
| BCRABL_HL60_CDNA_DN | Control | 26 | 0.0000 | 0.0286 | 2.14 |
| ET743_SARCOMA_DN | PS | 234 | 0.0000 | 0.0287 | −2.02 |
| TGFBETA_EARLY_UP | Control | 45 | 0.0000 | 0.0308 | 2.09 |
| IGF1RPATHWAY | Control | 15 | 0.0026 | 0.0311 | 2.12 |
| GRANDVAUX_IFN_NOT_IRF3_UP | Control | 13 | 0.0000 | 0.0325 | 2.09 |
| AGED_MOUSE_HYPOTH_DN | Control | 36 | 0.0000 | 0.0327 | 2.11 |
| PASSERINI_ADHESION | Control | 36 | 0.0000 | 0.0328 | 2.10 |
| SANA_IFNG_ENDOTHELIAL_UP | Control | 62 | 0.0000 | 0.0330 | 2.08 |
| ET743_SARCOMA_48HRS_DN | PS | 165 | 0.0000 | 0.0347 | −2.03 |
| HBX_HCC_DN | Control | 21 | 0.0026 | 0.0354 | 2.06 |
| IL5PATHWAY | Control | 10 | 0.0012 | 0.0378 | 2.05 |
| BRENTANI_CYTOSKELETON | Control | 19 | 0.0000 | 0.0382 | 2.04 |
| ET743_SARCOMA_24HRS_DN | PS | 97 | 0.0000 | 0.0401 | −1.94 |
| RHO_PROTEIN_SIGNAL_TRANSDUCTION | Control | 36 | 0.0000 | 0.0402 | 2.16 |
| ECMPATHWAY | Control | 21 | 0.0027 | 0.0403 | 2.04 |
| P21_MIDDLE_DN | PS | 14 | 0.0000 | 0.0438 | −1.93 |
| PLATELET_EXPRESSED | Control | 30 | 0.0014 | 0.0460 | 2.01 |
| TGFBETA_ALL_UP | Control | 78 | 0.0000 | 0.0471 | 2.00 |
| HOHENKIRK_MONOCYTE_DEND_DN | Control | 111 | 0.0000 | 0.0471 | 2.01 |
| INOS_ALL_DN | Control | 74 | 0.0000 | 0.0474 | 2.00 |
| RAY_P210_DIFF | Control | 51 | 0.0015 | 0.0476 | 1.99 |
| Gene Symbol | Gene Name | Location | Entrez Gene ID |
|---|---|---|---|
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | Cytoplasm | 207 |
| ALOX5AP | arachidonate 5-lipoxygenase-activating protein | Plasma Membrane | 241 |
| ATF2 | activating transcription factor 2 | Nucleus | 1386 |
| BAD | BCL2-associated agonist of cell death | Cytoplasm | 572 |
| BASP1 | brain abundant, membrane attached signal protein 1 | Plasma Membrane | 10409 |
| C1QB | complement component 1, q subcomponent, B chain | Extracellular Space | 713 |
| CAPG | capping protein (actin filament), gelsolin-like | Nucleus | 822 |
| CFL1 | cofilin 1 (non-muscle) | Nucleus | 1072 |
| COL1A1 | collagen, type I, alpha 1 | Extracellular Space | 1277 |
| COL3A1 | collagen, type III, alpha 1 | Extracellular Space | 1281 |
| CRYAB | crystallin, alpha B | Nucleus | 1410 |
| CSF1R | colony stimulating factor 1 receptor | Plasma Membrane | 1436 |
| CSTB | cystatin B (stefin B) | Cytoplasm | 1476 |
| CTSB | cathepsin B | Cytoplasm | 1508 |
| CTSD | cathepsin D | Cytoplasm | 1509 |
| CTSS | cathepsin S | Cytoplasm | 1520 |
| CTSZ | cathepsin Z | Cytoplasm | 1522 |
| DAD1 | defender against cell death 1 | Cytoplasm | 1603 |
| DDIT3 | DNA-damage-inducible transcript 3 | Nucleus | 1649 |
| DUSP1 | dual specificity phosphatase 1 | Nucleus | 1843 |
| EPHB3 | EPH receptor B3 | Plasma Membrane | 2049 |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | Plasma Membrane | 2207 |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | Plasma Membrane | 2214 |
| FLNA | filamin A, alpha | Cytoplasm | 2316 |
| FXYD5 | FXYD domain containing ion transport regulator 5 | Plasma Membrane | 53827 |
| GAS1 | growth arrest-specific 1 | Plasma Membrane | 2619 |
| GRB2 | growth factor receptor-bound protein 2 | Cytoplasm | 2885 |
| GRN | granulin | Extracellular Space | 2896 |
| HCLS1 | hematopoietic cell-specific Lyn substrate 1 | Nucleus | 3059 |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Plasma Membrane | 3265 |
| IGF1R | insulin-like growth factor 1 receptor | Plasma Membrane | 3480 |
| IRF1 | interferon regulatory factor 1 | Nucleus | 3659 |
| IRS1 | insulin receptor substrate 1 | Cytoplasm | 3667 |
| KIF5B | kinesin family member 5B | Cytoplasm | 3799 |
| LEP | leptin | Extracellular Space | 3952 |
| LGALS3 | lectin, galactoside-binding, soluble, 3 | Extracellular Space | 3958 |
| LMO7 | LIM domain 7 | Cytoplasm | 4008 |
| LRP1 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | Plasma Membrane | 4035 |
| MAP2K1 | mitogen-activated protein kinase kinase 1 | Cytoplasm | 5604 |
| MAPK1 | mitogen-activated protein kinase 1 | Cytoplasm | 5594 |
| MAPK3 | mitogen-activated protein kinase 3 | Cytoplasm | 5595 |
| NEDD8 | neural precursor cell expressed, developmentally down-regulated 8 | Nucleus | 4738 |
| NR3C1 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Nucleus | 2908 |
| PABPC1 | poly(A) binding protein, cytoplasmic 1 | Cytoplasm | 26986 |
| PAWR | PRKC, apoptosis, WT1, regulator | Nucleus | 5074 |
| PEA15 | phosphoprotein enriched in astrocytes 15 | Cytoplasm | 8682 |
| PFN1 | profilin 1 | Cytoplasm | 5216 |
| PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Cytoplasm | 5290 |
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Cytoplasm | 5295 |
| PLIN2 | perilipin 2 | Plasma Membrane | 123 |
| PSAP | prosaposin | Extracellular Space | 5660 |
| RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | Cytoplasm | 5894 |
| RASA1 | RAS p21 protein activator (GTPase activating protein) 1 | Cytoplasm | 5921 |
| SERPINF1 | serpin peptidase inhibitor, clade F, member 1 | Extracellular Space | 5176 |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | Cytoplasm | 6464 |
| SHOX2 | short stature homeobox 2 | Nucleus | 6474 |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Nucleus | 6595 |
| SNAI2 | snail homolog 2 (Drosophila) | Nucleus | 6591 |
| SOS1 | son of sevenless homolog 1 (Drosophila) | Cytoplasm | 6654 |
| TLN1 | talin 1 | Plasma Membrane | 7094 |
| TOP2B | topoisomerase (DNA) II beta 180kDa | Nucleus | 7155 |
| TPM4 | tropomyosin 4 | Cytoplasm | 7171 |
| UCP2 | uncoupling protein 2 (mitochondrial, proton carrier) | Cytoplasm | 7351 |
| YWHAH | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | Cytoplasm | 7533 |
REFERENCES
- 1.Montgomery-Downs HE, O'Brien LM, Gulliver TE, Gozal D. Polysomnographic characteristics in normal preschool and early school-aged children. Pediatrics. 2006;117:741–53. doi: 10.1542/peds.2005-1067. [DOI] [PubMed] [Google Scholar]
- 2.Uliel S, Tauman R, Greenfeld M, Sivan Y. Normal polysomnographic respiratory values in children and adolescents. Chest. 2004;125:872–8. doi: 10.1378/chest.125.3.872. [DOI] [PubMed] [Google Scholar]
- 3.Redline S, Budhiraja R, Kapur V, et al. The scoring of respiratory events in sleep: reliability and validity. J Clin Sleep Med. 2007;3:169–200. [PubMed] [Google Scholar]
- 4.Grigg-Damberger M, Gozal D, Marcus CL, et al. The visual scoring of sleep and arousal in infants and children. J Clin Sleep Med. 2007;3:201–40. [PubMed] [Google Scholar]
- 5.Witmans MB, Keens TG, Davidson Ward SL, Marcus CL. Obstructive hypopneas in children and adolescents: normal values. Am J Respir Crit Care Med. 2003;168:1540. doi: 10.1164/ajrccm.168.12.954. [DOI] [PubMed] [Google Scholar]
- 6.Marcus CL. Childhood obstructive sleep apnoea: to treat or not to treat, that is the question. Thorax. 2010;65:4–5. doi: 10.1136/thx.2009.123141. [DOI] [PubMed] [Google Scholar]
- 7.Kuhle S, Urschitz MS, Eitner S, Poets CF. Interventions for obstructive sleep apnea in children: a systematic review. Sleep Med Rev. 2009;13:123–31. doi: 10.1016/j.smrv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Guilleminault C, Lee JH. Does benign “primary snoring” ever exist in children? Chest. 2004;126:1396–8. doi: 10.1378/chest.126.5.1396. [DOI] [PubMed] [Google Scholar]
- 9.Blunden S, Lushington K, Kennedy D, Martin J, Dawson D. Behavior and neurocognitive performance in children aged 5–10 years who snore compared to controls. J Clin Exp Neuropsychol. 2000;22:554–68. doi: 10.1076/1380-3395(200010)22:5;1-9;FT554. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien LM, Mervis CB, Holbrook CR, et al. Neurobehavioral implications of habitual snoring in children. Pediatrics. 2004;114:44–9. doi: 10.1542/peds.114.1.44. [DOI] [PubMed] [Google Scholar]
- 11.Kwok KL, Ng DK, Cheung YF. BP and arterial distensibility in children with primary snoring. Chest. 2003;123:1561–6. doi: 10.1378/chest.123.5.1561. [DOI] [PubMed] [Google Scholar]
- 12.Li AM, Au CT, Ho C, Fok TF, Wing YK. Blood pressure is elevated in children with primary snoring. J Pediatr. 2009;155:362–8. e1. doi: 10.1016/j.jpeds.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 13.Kaditis AG, Alexopoulos EI, Kostadima E, et al. Comparison of blood pressure measurements in children with and without habitual snoring. Pediatr Pulmonol. 2005;39:408–14. doi: 10.1002/ppul.20188. [DOI] [PubMed] [Google Scholar]
- 14.Amin R, Somers VK, McConnell K, et al. Activity-adjusted 24-hour ambulatory blood pressure and cardiac remodeling in children with sleep disordered breathing. Hypertension. 2008;51:84–91. doi: 10.1161/HYPERTENSIONAHA.107.099762. [DOI] [PubMed] [Google Scholar]
- 15.Alexopoulos EI, Kostadima E, Pagonari I, Zintzaras E, Gourgoulianis K, Kaditis AG. Association between primary nocturnal enuresis and habitual snoring in children. Urology. 2006;68:406–9. doi: 10.1016/j.urology.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Sans Capdevila O, Crabtree VM, Kheirandish-Gozal L, Gozal D. Increased morning brain natriuretic peptide levels in children with nocturnal enuresis and sleep-disordered breathing: a community-based study. Pediatrics. 2008;121:e1208–14. doi: 10.1542/peds.2007-2049. [DOI] [PubMed] [Google Scholar]
- 17.Nieminen P, Lopponen T, Tolonen U, Lanning P, Knip M, Lopponen H. Growth and biochemical markers of growth in children with snoring and obstructive sleep apnea. Pediatrics. 2002;109:e55. doi: 10.1542/peds.109.4.e55. [DOI] [PubMed] [Google Scholar]
- 18.Lander ES. Array of hope. Nat Genet. 1999;21:3–4. doi: 10.1038/4427. [DOI] [PubMed] [Google Scholar]
- 19.Khalyfa A, Capdevila OS, Buazza MO, Serpero LD, Kheirandish-Gozal L, Gozal D. Genome-wide gene expression profiling in children with non-obese obstructive sleep apnea. Sleep Med. 2009;10:75–86. doi: 10.1016/j.sleep.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Gozal D. Sleep-disordered breathing and school performance in children. Pediatrics. 1998;102:616–20. doi: 10.1542/peds.102.3.616. [DOI] [PubMed] [Google Scholar]
- 21.Iber C, Ancoli-Israel S, Chesson AL, Quan SF. Westchester IL: American Academy of Sleep Medicine; 2007. The AASM manual for the scoring of sleep and associated events. [Google Scholar]
- 22.Marcus CL, Omlin KJ, Basinki DJ, et al. Normal polysomnographic values for children and adolescents. Am Rev Respir Dis. 1992;146:1235–9. doi: 10.1164/ajrccm/146.5_Pt_1.1235. [DOI] [PubMed] [Google Scholar]
- 23.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 24.Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, et al. CDC growth charts: United States. Adv Data. 2000:1–27. [PubMed] [Google Scholar]
- 25.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 26.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 27.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 31.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 32.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 33.Patti ME, Butte AJ, Crunkhorn S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kintscher U, Hartge M, Hess K, et al. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol. 2008;28:1304–10. doi: 10.1161/ATVBAHA.108.165100. [DOI] [PubMed] [Google Scholar]
- 36.Odegaard JI, Chawla A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab. 2008;4:619–26. doi: 10.1038/ncpendmet0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Cava A, Matarese G. The weight of leptin in immunity. Nat Rev Immunol. 2004;4:371–9. doi: 10.1038/nri1350. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Margalet V, Martin-Romero C, Santos-Alvarez J, Goberna R, Najib S, Gonzalez-Yanes C. Role of leptin as an immunomodulator of blood mononuclear cells: mechanisms of action. Clin Exp Immunol. 2003;133:11–9. doi: 10.1046/j.1365-2249.2003.02190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 40.Samara A, Marie B, Pfister M, Visvikis-Siest S. Leptin expression in peripheral blood mononuclear cells (PBMCs) is related with blood pressure variability. Clin Chim Acta. 2008;395:47–50. doi: 10.1016/j.cca.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 41.Hennige AM, Stefan N, Kapp K, et al. Leptin down-regulates insulin action through phosphorylation of serine-318 in insulin receptor substrate 1. FASEB J. 2006;20:1206–8. doi: 10.1096/fj.05-4635fje. [DOI] [PubMed] [Google Scholar]
- 42.Baudler S, Baumgartl J, Hampel B, et al. Insulin-like growth factor-1 controls type 2 T cell-independent B cell response. J Immunol. 2005;174:5516–25. doi: 10.4049/jimmunol.174.9.5516. [DOI] [PubMed] [Google Scholar]
- 43.Sandhu MS, Heald AH, Gibson JM, Cruickshank JK, Dunger DB, Wareham NJ. Circulating concentrations of insulin-like growth factor-I and development of glucose intolerance: a prospective observational study. Lancet. 2002;359:1740–5. doi: 10.1016/S0140-6736(02)08655-5. [DOI] [PubMed] [Google Scholar]
- 44.Sesti G, Sciacqua A, Cardellini M, et al. Plasma concentration of IGF-I is independently associated with insulin sensitivity in subjects with different degrees of glucose tolerance. Diabetes Care. 2005;28:120–5. doi: 10.2337/diacare.28.1.120. [DOI] [PubMed] [Google Scholar]
- 45.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–35. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gozal D, Sans Capdevila O, McLaughlin Crabtree V, Serpero LD, Witcher LA, Kheirandish-Gozal L. Plasma IGF-1 levels and cognitive dysfunction in children with obstructive sleep apnea. Sleep Med. 2009;10:167–73. doi: 10.1016/j.sleep.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Gozal D, Kheirandish-Gozal L. The multiple challenges of obstructive sleep apnea in children: morbidity and treatment. Curr Opin Pediatr. 2008;20:654–8. doi: 10.1097/MOP.0b013e328316ec2d. [DOI] [PubMed] [Google Scholar]
- 48.Gozal D. Sleep, sleep disorders and inflammation in children. Sleep Med. 2009;10(Suppl 1):S12–6. doi: 10.1016/j.sleep.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Goldbart AD, Row BW, Kheirandish-Gozal L, Cheng Y, Brittian KR, Gozal D. High fat/refined carbohydrate diet enhances the susceptibility to spatial learning deficits in rats exposed to intermittent hypoxia. Brain Res. 2006;1090:190–6. doi: 10.1016/j.brainres.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 50.Gozal D, Nair D, Goldbart AD. Physical activity attenuates intermittent hypoxia-induced spatial learning deficits and oxidative stress. Am J Respir Crit Care Med. 2010;182:104–12. doi: 10.1164/rccm.201001-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsaoussoglou M, Bixler EO, Calhoun S, Chrousos GP, Sauder K, Vgontzas AN. Sleep-disordered breathing in obese children is associated with prevalent excessive daytime sleepiness, inflammation, and metabolic abnormalities. J Clin Endocrinol Metab. 2010;95:143–50. doi: 10.1210/jc.2009-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apostolidou MT, Alexopoulos EI, Damani E, et al. Absence of blood pressure, metabolic, and inflammatory marker changes after adenotonsillectomy for sleep apnea in Greek children. Pediatr Pulmonol. 2008;43:550–60. doi: 10.1002/ppul.20808. [DOI] [PubMed] [Google Scholar]
- 53.Gozal D, Capdevila OS, Kheirandish-Gozal L. Metabolic alterations and systemic inflammation in obstructive sleep apnea among nonobese and obese prepubertal children. Am J Respir Crit Care Med. 2008;177:1142–9. doi: 10.1164/rccm.200711-1670OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redline S, Storfer-Isser A, Rosen CL, et al. Association between metabolic syndrome and sleep-disordered breathing in adolescents. Am J Respir Crit Care Med. 2007;176:401–8. doi: 10.1164/rccm.200703-375OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tauman R, Serpero LD, Capdevila OS, et al. Adipokines in children with sleep disordered breathing. Sleep. 2007;30:443–9. doi: 10.1093/sleep/30.4.443. [DOI] [PubMed] [Google Scholar]
- 56.Li AM, Chan MH, Chan DF, et al. Insulin and obstructive sleep apnea in obese Chinese children. Pediatr Pulmonol. 2006;41:1175–81. doi: 10.1002/ppul.20508. [DOI] [PubMed] [Google Scholar]
- 57.Korner A, Kratzsch J, Gausche R, et al. Metabolic syndrome in children and adolescents--risk for sleep-disordered breathing and obstructive sleep-apnoea syndrome? Arch Physiol Biochem. 2008;114:237–43. doi: 10.1080/13813450802306685. [DOI] [PubMed] [Google Scholar]
- 58.Nakra N, Bhargava S, Dzuira J, Caprio S, Bazzy-Asaad A. Sleep-disordered breathing in children with metabolic syndrome: the role of leptin and sympathetic nervous system activity and the effect of continuous positive airway pressure. Pediatrics. 2008;122:e634–42. doi: 10.1542/peds.2008-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tauman R, O'Brien LM, Ivanenko A, Gozal D. Obesity rather than severity of sleep-disordered breathing as the major determinant of insulin resistance and altered lipidemia in snoring children. Pediatrics. 2005;116:e66–73. doi: 10.1542/peds.2004-2527. [DOI] [PubMed] [Google Scholar]
- 60.Verhulst SL, Schrauwen N, Haentjens D, et al. Sleep-disordered breathing and the metabolic syndrome in overweight and obese children and adolescents. J Pediatr. 2007;150:608–12. doi: 10.1016/j.jpeds.2007.01.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal components analysis (PCA) on the global expression profiles of children with and without primary snoring after stratification by race (only European and African ancestry shown). Consistent with our initial PCA findings that was based on all the subjects (manuscript Figure 1) no discernable expression pattern separated the primary snoring groups from the controls in each race.
| Gene Set | Enriched Phenotype | Gene Set Size | P-value | FDR | Enrichment Score |
|---|---|---|---|---|---|
| HSA03010_RIBOSOME | PS | 69 | 0.0000 | 0.0000 | −2.51 |
| STRUCTURAL_CONSTITUENT_OF_RIBOSOME | PS | 67 | 0.0000 | 0.0000 | −2.48 |
| RIBOSOMAL_PROTEINS | PS | 75 | 0.0000 | 0.0005 | −2.29 |
| REGULATION_OF_SMALL_GTPASE_MEDIATED_SIGNAL | Control | 22 | 0.0000 | 0.0021 | 2.40 |
| REGULATION_OF_RAS_PROTEIN_SIGNAL_TRANSDUCTION | Control | 18 | 0.0000 | 0.0052 | 2.33 |
| GOLUB_ALL_VS_AML_DN | Control | 14 | 0.0000 | 0.0112 | 2.27 |
| KNUDSEN_PMNS_DN | Control | 200 | 0.0000 | 0.0113 | 2.28 |
| GNATENKO_PLATELET_UP | Control | 40 | 0.0000 | 0.0134 | 2.34 |
| GNATENKO_PLATELET | Control | 40 | 0.0000 | 0.0154 | 2.30 |
| NADLER_OBESITY_UP | Control | 50 | 0.0000 | 0.0164 | 2.21 |
| MOREAUX_TACI_HI_VS_LOW_DN | PS | 146 | 0.0000 | 0.0230 | −1.99 |
| HDACI_COLON_SUL12HRS_DN | PS | 26 | 0.0000 | 0.0243 | −2.02 |
| STEMCELL_COMMON_UP | PS | 164 | 0.0000 | 0.0248 | −1.99 |
| ADIPOCYTE DIFFERENTIATION | PS | 38 | 0.0000 | 0.0264 | −2.00 |
| BCRABL_HL60_CDNA_DN | Control | 26 | 0.0000 | 0.0286 | 2.14 |
| ET743_SARCOMA_DN | PS | 234 | 0.0000 | 0.0287 | −2.02 |
| TGFBETA_EARLY_UP | Control | 45 | 0.0000 | 0.0308 | 2.09 |
| IGF1RPATHWAY | Control | 15 | 0.0026 | 0.0311 | 2.12 |
| GRANDVAUX_IFN_NOT_IRF3_UP | Control | 13 | 0.0000 | 0.0325 | 2.09 |
| AGED_MOUSE_HYPOTH_DN | Control | 36 | 0.0000 | 0.0327 | 2.11 |
| PASSERINI_ADHESION | Control | 36 | 0.0000 | 0.0328 | 2.10 |
| SANA_IFNG_ENDOTHELIAL_UP | Control | 62 | 0.0000 | 0.0330 | 2.08 |
| ET743_SARCOMA_48HRS_DN | PS | 165 | 0.0000 | 0.0347 | −2.03 |
| HBX_HCC_DN | Control | 21 | 0.0026 | 0.0354 | 2.06 |
| IL5PATHWAY | Control | 10 | 0.0012 | 0.0378 | 2.05 |
| BRENTANI_CYTOSKELETON | Control | 19 | 0.0000 | 0.0382 | 2.04 |
| ET743_SARCOMA_24HRS_DN | PS | 97 | 0.0000 | 0.0401 | −1.94 |
| RHO_PROTEIN_SIGNAL_TRANSDUCTION | Control | 36 | 0.0000 | 0.0402 | 2.16 |
| ECMPATHWAY | Control | 21 | 0.0027 | 0.0403 | 2.04 |
| P21_MIDDLE_DN | PS | 14 | 0.0000 | 0.0438 | −1.93 |
| PLATELET_EXPRESSED | Control | 30 | 0.0014 | 0.0460 | 2.01 |
| TGFBETA_ALL_UP | Control | 78 | 0.0000 | 0.0471 | 2.00 |
| HOHENKIRK_MONOCYTE_DEND_DN | Control | 111 | 0.0000 | 0.0471 | 2.01 |
| INOS_ALL_DN | Control | 74 | 0.0000 | 0.0474 | 2.00 |
| RAY_P210_DIFF | Control | 51 | 0.0015 | 0.0476 | 1.99 |
| Gene Symbol | Gene Name | Location | Entrez Gene ID |
|---|---|---|---|
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | Cytoplasm | 207 |
| ALOX5AP | arachidonate 5-lipoxygenase-activating protein | Plasma Membrane | 241 |
| ATF2 | activating transcription factor 2 | Nucleus | 1386 |
| BAD | BCL2-associated agonist of cell death | Cytoplasm | 572 |
| BASP1 | brain abundant, membrane attached signal protein 1 | Plasma Membrane | 10409 |
| C1QB | complement component 1, q subcomponent, B chain | Extracellular Space | 713 |
| CAPG | capping protein (actin filament), gelsolin-like | Nucleus | 822 |
| CFL1 | cofilin 1 (non-muscle) | Nucleus | 1072 |
| COL1A1 | collagen, type I, alpha 1 | Extracellular Space | 1277 |
| COL3A1 | collagen, type III, alpha 1 | Extracellular Space | 1281 |
| CRYAB | crystallin, alpha B | Nucleus | 1410 |
| CSF1R | colony stimulating factor 1 receptor | Plasma Membrane | 1436 |
| CSTB | cystatin B (stefin B) | Cytoplasm | 1476 |
| CTSB | cathepsin B | Cytoplasm | 1508 |
| CTSD | cathepsin D | Cytoplasm | 1509 |
| CTSS | cathepsin S | Cytoplasm | 1520 |
| CTSZ | cathepsin Z | Cytoplasm | 1522 |
| DAD1 | defender against cell death 1 | Cytoplasm | 1603 |
| DDIT3 | DNA-damage-inducible transcript 3 | Nucleus | 1649 |
| DUSP1 | dual specificity phosphatase 1 | Nucleus | 1843 |
| EPHB3 | EPH receptor B3 | Plasma Membrane | 2049 |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | Plasma Membrane | 2207 |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | Plasma Membrane | 2214 |
| FLNA | filamin A, alpha | Cytoplasm | 2316 |
| FXYD5 | FXYD domain containing ion transport regulator 5 | Plasma Membrane | 53827 |
| GAS1 | growth arrest-specific 1 | Plasma Membrane | 2619 |
| GRB2 | growth factor receptor-bound protein 2 | Cytoplasm | 2885 |
| GRN | granulin | Extracellular Space | 2896 |
| HCLS1 | hematopoietic cell-specific Lyn substrate 1 | Nucleus | 3059 |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Plasma Membrane | 3265 |
| IGF1R | insulin-like growth factor 1 receptor | Plasma Membrane | 3480 |
| IRF1 | interferon regulatory factor 1 | Nucleus | 3659 |
| IRS1 | insulin receptor substrate 1 | Cytoplasm | 3667 |
| KIF5B | kinesin family member 5B | Cytoplasm | 3799 |
| LEP | leptin | Extracellular Space | 3952 |
| LGALS3 | lectin, galactoside-binding, soluble, 3 | Extracellular Space | 3958 |
| LMO7 | LIM domain 7 | Cytoplasm | 4008 |
| LRP1 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | Plasma Membrane | 4035 |
| MAP2K1 | mitogen-activated protein kinase kinase 1 | Cytoplasm | 5604 |
| MAPK1 | mitogen-activated protein kinase 1 | Cytoplasm | 5594 |
| MAPK3 | mitogen-activated protein kinase 3 | Cytoplasm | 5595 |
| NEDD8 | neural precursor cell expressed, developmentally down-regulated 8 | Nucleus | 4738 |
| NR3C1 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Nucleus | 2908 |
| PABPC1 | poly(A) binding protein, cytoplasmic 1 | Cytoplasm | 26986 |
| PAWR | PRKC, apoptosis, WT1, regulator | Nucleus | 5074 |
| PEA15 | phosphoprotein enriched in astrocytes 15 | Cytoplasm | 8682 |
| PFN1 | profilin 1 | Cytoplasm | 5216 |
| PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Cytoplasm | 5290 |
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Cytoplasm | 5295 |
| PLIN2 | perilipin 2 | Plasma Membrane | 123 |
| PSAP | prosaposin | Extracellular Space | 5660 |
| RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | Cytoplasm | 5894 |
| RASA1 | RAS p21 protein activator (GTPase activating protein) 1 | Cytoplasm | 5921 |
| SERPINF1 | serpin peptidase inhibitor, clade F, member 1 | Extracellular Space | 5176 |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | Cytoplasm | 6464 |
| SHOX2 | short stature homeobox 2 | Nucleus | 6474 |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Nucleus | 6595 |
| SNAI2 | snail homolog 2 (Drosophila) | Nucleus | 6591 |
| SOS1 | son of sevenless homolog 1 (Drosophila) | Cytoplasm | 6654 |
| TLN1 | talin 1 | Plasma Membrane | 7094 |
| TOP2B | topoisomerase (DNA) II beta 180kDa | Nucleus | 7155 |
| TPM4 | tropomyosin 4 | Cytoplasm | 7171 |
| UCP2 | uncoupling protein 2 (mitochondrial, proton carrier) | Cytoplasm | 7351 |
| YWHAH | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | Cytoplasm | 7533 |