Abstract
We describe the use of an auto-induction medium containing N-(phosphono-methyl)glycine (glyphosate) as a means for high-level introduction of nonstandard aromatic amino acids into a protein. We illustrate this approach by preparing maltose binding protein (MBP) wherein all eight tryptophan residues have been replaced with 6-fluorotryptophan at an incorporation level of 99.3%. Such a high level of incorporation is important for spectroscopic investigations, in particular 19F NMR, because each species’ differing amino acid sequence potentially yields a different peak pattern that complicates spectral analysis.
Keywords: protein labeling, aromatic amino acid analogs, unnatural amino acids, fluorotryptophan
Nonstandard amino acids commonly are introduced into proteins to test mechanistic hypotheses or to provide spectroscopic probes or heavy atoms used in phasing X-ray structures. The usual goals are to maximize the level of incorporation of the unusual amino acid and the overall yield of protein. Tryptophan analogs are of particular interest because of their spectral properties (1).
Incorporation of unusual amino acids into recombinant proteins produced from cells presents several obstacles. Replacement of natural residues in proteins by appropriate analogs usually is achieved by tightly controlled overproduction of the protein in an appropriate auxotrophic Escherichia coli strain growing in minimal medium with the desired unusual amino acids (1–3). Because the bacterial host is starved for the natural amino acid and supplemented with the analog—which may be toxic or may slow protein production—protein yields often are low. If a significant amount of the natural amino acid is required, its presence will lower the level of incorporation of the unusual amino acid (4,5). Furthermore, it may be necessary to identify an optimal host and then create an auxotrophic version of that strain. It may be possible to use cell-free protein production to overcome difficulties with toxicity (6), but this approach can be expensive if large amounts of protein are required. An elegant method for introducing unnatural amino acids is to make use of an engineered aminoacyl-tRNA synthetase and cognate orthogonal tRNA that recognizes the amber codon (7,8). Another method utilizes induction of an mRNA-specific endoRNase that shuts down background protein synthesis while allowing production of the target protein and introduction of toxic amino acids (9).
A convenient approach to introducing unusual aromatic amino acids into proteins produced from E. coli cells involves the use of N-(phosphonomethyl) glycine (glyphosate), commonly used as an herbicide, to inhibit the biosynthesis of aromatic amino acids (10). Cell growth and protein production in the presence of glyphosate requires the provision of all three aromatic amino acids (phenylalanine, tyrosine, and tryptophan), and the strategy is to replace one or more of these with the cognate unusual amino acid(s) to be incorporated. This approach has been used to introduce fluorotryptophan into a protein (5), but the protein yield was modest, and the level of incorporation was limited to ~80%, because unlabeled amino acid had to be supplied to support cell growth.
Recent approaches to improving protein yields include cold-shock induction with pCold vectors (11), high cell density IPTG induction method (12), and the auto-induction medium introduced by Studier (13) as an alternative to batch induction by IPTG. The auto-induction medium contains components that are metabolized differentially and support cell growth to high density, at which point protein expression is induced automatically from a lac promoter. The auto-induction approach has been adopted and refined for the production of selenomethionine and stable isotope-labeled proteins for structural and functional studies (14–16).
We describe here the use of glyphosate in an auto-induction medium as an approach for increasing both the level of incorporation of a f luorine-labeled aromatic amino acid and the overall protein yield. The approach enabled the production of maltose binding protein (MBP), wherein all eight tryptophan residues were replaced by 6-fluorotryptophan at an incorporation level of 99.3% without the use of an auxotrophic strain of E. coli.
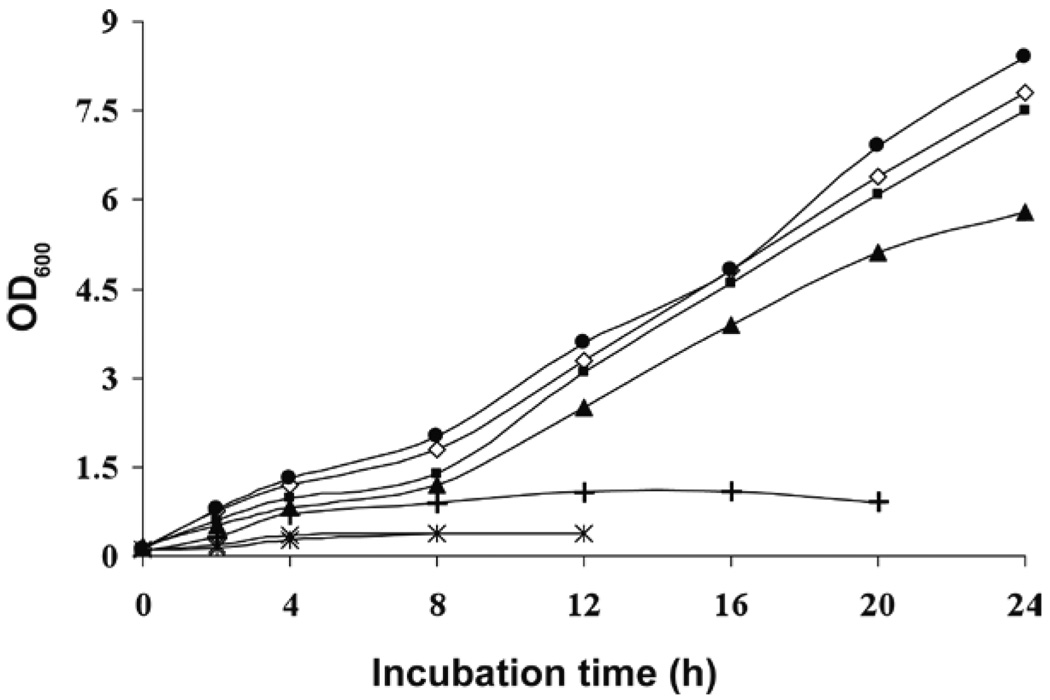
To evaluate and optimize the approach, we investigated the production of recombinant MBP in the presence of the unnatural aromatic amino acid 6-fluoro-D,L-tryptophan (6F-Trp). We compared the glyphosate-containing medium developed by Kim et al. (5), which utilizes IPTG induction, with a glyphosate-containing auto-induction medium [Studier MDA-5052 (13)]. We found that E. coli cells [BL21-Codon Plus(DE3)-RIL carrying pMAL-p2E] grew to high density (OD600 ≥ 8.0 after 24 h) on the auto-induction medium and also in the presence of glyphosate (OD600 ≈ 7.5 after 24 h), provided that aromatic amino acids were supplied (Figure 1). The rate of cell growth was nearly as high when 86% of the Trp was replaced by 6F-Trp, but with 100% 6F-Trp in the medium, the rate of cell growth fell significantly (OD600 ≈ 6.0 after 24 h). As expected, the cells failed to grow in the presence of glyphosate without added tyrosine (Tyr) and Trp. The level of cell growth on the glyphosate-containing medium described by Kim et al. (5) was poor (OD600 ≈ 1 after 20 h).
Figure 1. Time course of E. coli [BL21-Codon Plus(DE3)-RIL carrying pMAL-p2E] cells grown in 250-mL conical shaker flasks (225 rpm) at 30°C on 50 mL media of different composition.
(●) Auto-induction medium with amino acid mixture I [all 20 amino acids except cysteine (Cys) and Trp] without added glyphosate. (◊) Auto-induction medium with glyphosate (1 g/L), amino acid mixture I, and Trp (200 µg/L). (■) Auto-induction medium with glyphosate (1 g/L), amino acid mixture I, 6F-Trp (316 mg/L), and Trp (50 µg/L). (▲) Auto-induction medium with glyphosate (1 g/L), amino acid mixture I, and 6F-Trp (316 mg/L). (×) Auto-induction medium with glyphosate (1 g/L), amino acid mixture I, and Tyr (200 µg/L). (∗) Auto-induction medium with glyphosate (1 g/L), amino acid mixture II (all 20 amino acids except for Cys, Trp, and Tyr), and Trp (200 µg/L). (+) Kim et al. (5) medium (with IPTG induction) containing glyphosate and 6F-Trp.
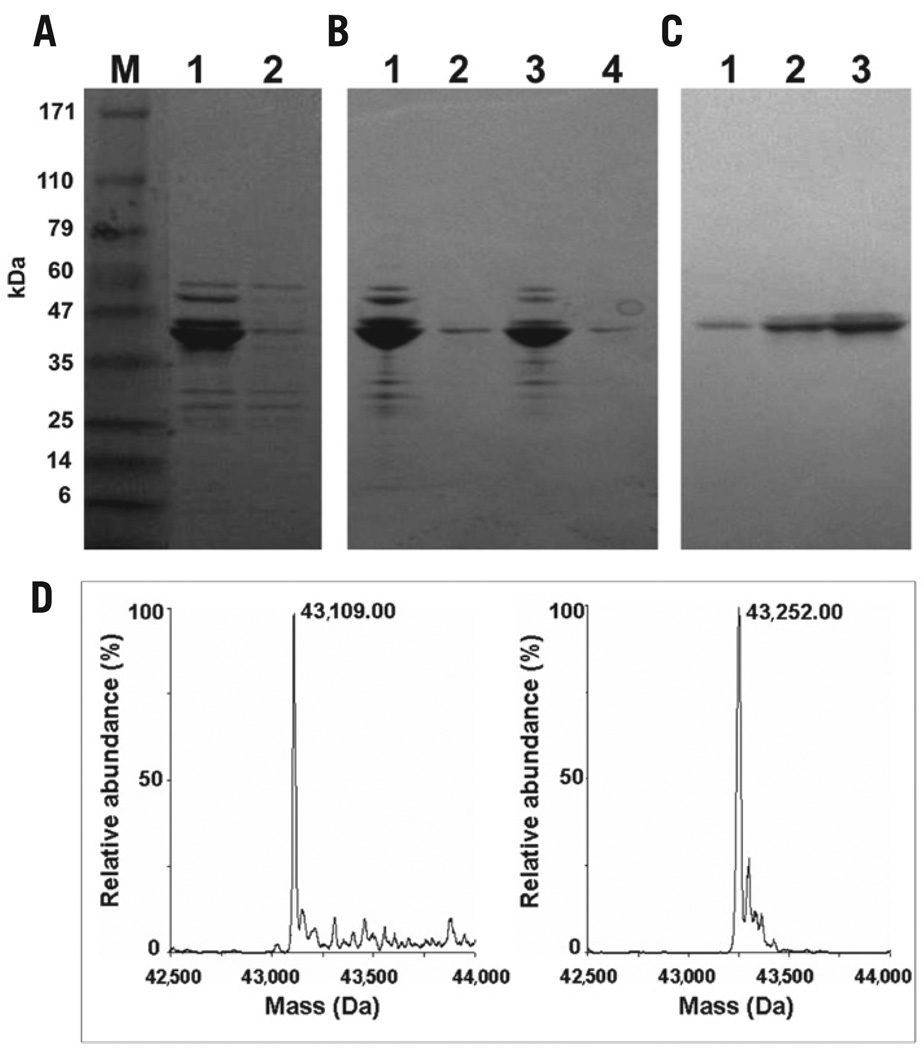
MBP was produced in high yield by the glyphosate-containing auto-induction medium but not when lactate was absent from the medium (noninducing medium) (Figure 2A). The amount of protein produced in the presence of glyphosate was only somewhat lower than in its absence (Figure 2B). 6F-Trp-MBP was purified by amylose resin affinity chromatography with elution by maltose and yielded a single major band on SDS-PAGE (Figure 2C). The average yield of purified 6F-Trp–labeled MBP, obtained after proteolysis of the fusion (MBP-lacZ-α) protein, was ~20–30 mg/L (50-mL working volume) for three expression trials (average OD600 ≈ 6.5 at cell harvest; wet cell mass 8.7 g/L).
Figure 2. Expression and purification of MBP produced from E. coli [BL21-Codon Plus(DE3)-RIL] cells carrying pMAL-p2E.
(A) SDS-PAGE analysis of extracts (lane 1) from cells grown in the auto-induction medium and (lane 2) from cells grown in the same medium without lactate [noninduction medium MDAG (13)]. Lane M contains a mixture of molecular weight markers with masses indicated. (B) SDS-PAGE analysis of cell extracts: (lane 1) soluble fraction; (lane 2) insoluble fraction from cells grown on the auto-induction medium with glyphosate (1 g/L), amino acid mixture I (all 20 amino acids except Cys and Trp), and Trp (200 µg/L); (lane 3) soluble fraction; (lane 4) insoluble fraction from cells grown on the auto-induction medium with glyphosate (1 g/L), amino acid mixture I, 6F-Trp (316 mg/L), and Trp (50 µg/L). (C) SDS-PAGE of 6F-Trp-labeled MBP purified by affinity chromatography. Lanes 1, 2, and 3 show three sequential fractions eluted with 10 mM maltose. (D) Deconvoluted ESI-mass spectra of purified MBP obtained from cultures grown in a chemically defined auto-induction medium for aromatic unusual amino acid incorporation: (left) unlabeled MBP and (right) 6F-Trp-MBP.
Analysis by liquid chromatography-electrospray ionization mass spectrometry (LC-ESI MS), interpreted with the deconvolution algorithm MaxEnt (Figure 2D), gave 43,109.0 Da (versus calculated value of 43,100.4 Da) for the purified unlabeled MBP, and 43,252.0 Da (versus calculated value of 43,244.38 Da) for the purified labeled MBP (grown on 50 µg/L Trp and 316 mg/L 6F-Trp). The difference of 143.0 Da—in comparison to the predicted molecular weight difference of 143.98 Da for 100% labeling of the eight Trp residues—indicated that the incorporation rate was 99.3%.
The approach described herein should be applicable to the incorporation of a variety of aromatic amino acids. Its advantages are simplicity (no need for auxotrophs or induction), high protein yields, and high-level incorporation of the amino acid analog.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH; grant nos. RR02301, P50 GM64598, and 1U54 GM074901). This paper is subject to the NIH Public Access Policy.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/113491.
Competing interests
The authors declare no competing interests.
References
- 1.Mohammadi F, Prentice GA, Merrill AR. Protein-protein interaction using tryptophan analogues: novel spectroscopic probes for toxin-elongation factor-2 interactions. Biochemistry. 2001;40:10273–10283. doi: 10.1021/bi011035u. [DOI] [PubMed] [Google Scholar]
- 2.Ross JB, Szabo AG, Hogue CW. Enhancement of protein spectra with tryptophan analogs: fluorescence spectroscopy of protein-protein and protein-nucleic acid interactions. Methods Enzymol. 1997;278:151–190. doi: 10.1016/s0076-6879(97)78010-8. [DOI] [PubMed] [Google Scholar]
- 3.Steward LE, Chamberlin AR. Protein expression by expansion of the genetic code. In: Meyers RA, editor. Encyclopedia of Molecular Biology and Molecular Medicine. New York: VCH Publishers; 1996. [Google Scholar]
- 4.Senear DF, Mendelson RA, Stone DB, Luck LA, Rusinova E, Ross JB. Quantitative analysis of tryptophan analogue incorporation in recombinant proteins. Anal. Biochem. 2002;300:77–86. doi: 10.1006/abio.2001.5441. [DOI] [PubMed] [Google Scholar]
- 5.Kim HW, Perez JA, Ferguson SJ, Campbell ID. The specific incorporation of labelled aromatic amino acids into proteins through growth of bacteria in the presence of glyphosate. Application to fluorotryptophan labelling to the H(+)-ATPase of Escherichia coli and NMR studies. FEBS Lett. 1990;272:34–36. doi: 10.1016/0014-5793(90)80442-l. [DOI] [PubMed] [Google Scholar]
- 6.Neerathilingam M, Greene LH, Colebrooke SA, Campbell ID, Staunton D. Quantitation of protein expression in a cell-free system: efficient detection of yields and 19F NMR to identify folded protein. J. Biomol. NMR. 2005;31:11–19. doi: 10.1007/s10858-004-5357-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, Cellitti SE, Geierstanger BH, Schultz PG. Efficient expression of tyrosine-sulfated proteins in E. coli using an expanded genetic code. Nat. Protocols. 2009;4:1784–1789. doi: 10.1038/nprot.2009.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones DH, Cellitti SE, Hao X, Zhang Q, Jahnz M, Summerer D, Schultz PG, Uno T, Geierstanger BH. Site-specific labeling of proteins with NMR-active unnatural amino acids. J. Biomol. NMR. 2010;46:89–100. doi: 10.1007/s10858-009-9365-4. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki M, Roy R, Zheng H, Woychik N, Inouye M. Bacterial bioreactors for high yield production of recombinant protein. J. Biol. Chem. 2006;281:37559–37565. doi: 10.1074/jbc.M608806200. [DOI] [PubMed] [Google Scholar]
- 10.Comai L, Sen LC, Stalker DM. An altered aroA gene product confers resistance to the herbicide glyphosate. Science. 1983;221:370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- 11.Qing G, Ma LC, Khorchid A, Swapna GV, Mal TK, Takayama MM, Xia B, Phadtare S, et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- 12.Sivashanmugam A, Murray V, Cui C, Zhang Y, Wang J, Li Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009;18:936–948. doi: 10.1002/pro.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, et al. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr. Purif. 2005;40:256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Tyler RC, Sreenath H, Aceti DJ, Bingman CA, Singh S, Markley JL, Fox BG. Auto-induction medium for the production of [U-15N]- and [U-13C, U-15N]-labeled proteins for NMR screening and structure determination. Protein Expr. Purif. 2005;40:268–278. doi: 10.1016/j.pep.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 16.Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol. Prog. 2007;23:585–598. doi: 10.1021/bp070011x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.