Abstract
Despite the importance of triacylglycerols (TAG) and steryl esters (SE) in phospholipid synthesis in cells transitioning from stationary-phase into active growth, there is no direct evidence for their requirement in synthesis of phosphatidylinositol (PI) or other membrane phospholipids in logarithmically growing yeast cells. We report that the dga1Δlro1Δare1Δare2Δ strain, which lacks the ability to synthesize both TAG and SE, is not able to sustain normal growth in the absence of inositol (Ino− phenotype) at 37 °C especially when choline is present. Unlike many other strains exhibiting an Ino− phenotype, the dga1Δlro1Δare1Δare2Δ strain does not display a defect in INO1 expression. However, the mutant exhibits slow recovery of PI content compared with wild type cells upon reintroduction of inositol into logarithmically growing cultures. The tgl3Δtgl4Δtgl5Δ strain, which is able to synthesize TAG but unable to mobilize it, also exhibits attenuated PI formation under these conditions. However, unlike dga1Δlro1Δare1Δare2Δ, the tgl3Δtgl4Δtgl5Δ strain does not display an Ino− phenotype, indicating that failure to mobilize TAG is not fully responsible for the growth defect of the dga1Δlro1Δare1Δare2Δ strain in the absence of inositol. Moreover, synthesis of phospholipids, especially PI, is dramatically reduced in the dga1Δlro1Δare1Δare2Δ strain even when it is grown continuously in the presence of inositol. The mutant also utilizes a greater proportion of newly synthesized PI than wild type for the synthesis of inositol-containing sphingolipids, especially in the absence of inositol. Thus, we conclude that storage lipid synthesis actively influences membrane phospholipid metabolism in logarithmically growing cells.
Keywords: Diacylglycerol, Inositol Phospholipid, Lipid, Lipolysis, Triacylglycerol, Inositol
Introduction
Eukaryotic organisms store excess energy as triacylglycerols (TAG)3 and steryl esters (SE) for later use during times of deprivation. TAG and SE are hydrophobic compounds separated from the aqueous cellular environment of the cytoplasm in specialized structures called lipid droplets (1). Lipid droplets are dynamic organelles that play important roles in the biosynthesis, mobilization, and trafficking of intracellular neutral lipids. They function in close apposition with other organelles, particularly the endoplasmic reticulum (ER), endosomes, mitochondria, and peroxisomes (2, 3).
In the budding yeast, Saccharomyces cerevisiae, the formation of lipid droplets is tightly linked to the synthesis of TAG and SE. The diacylglycerol acyltransferases encoded by the DGA1 and LRO1 genes (Fig. 1) are the main enzymes involved in the biosynthesis of TAG (4–6), whereas ARE1 and ARE2 (Fig. 1) encode the enzymes that primarily mediate the esterification of ergosterol and its precursors leading to SE (7, 8). These four enzymes account for all TAG and SE biosynthesis in yeast, which begins during exponential growth and reaches its peak as cells enter stationary phase (9). During times of energy scarcity or upon recovery from stationary phase when exposed to glucose, TAG degradation occurs via the activity of lipid hydrolases encoded by the TGL3, TGL4, and TGL5 genes (10–12). The products of TAG degradation, diacylglycerols (DAG) and free fatty acids, also serve as precursors for membrane lipid synthesis (13) as well as for energy production when free fatty acids are the only carbon source available in the growth medium (14). At the cellular level, TAG degradation is up-regulated by Cdc28p/Cdk1p-dependent phosphorylation of the Tgl4p lipase (12). Lipolysis contributes to bud formation, presumably by providing precursors for synthesis of lipids involved in membrane biogenesis or signaling (12). Conversely, impairment in membrane trafficking leads to a block in phospholipid synthesis and concomitant TAG accumulation (15).
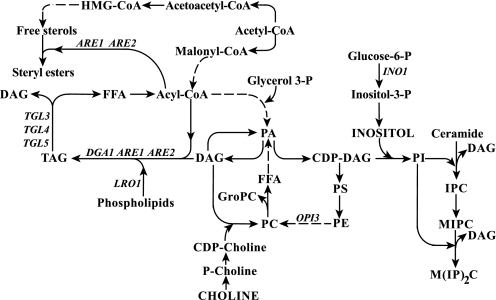
FIGURE 1.
Major pathways for the synthesis of phospholipids, inositol complex sphingolipids, and neutral lipids. The synthesis of phospholipids and neutral lipids shares DAG and PA as common precursors. In the synthesis of phospholipids, PA serves as the immediate precursor of CDP-DAG, which is in turn the precursor of both PI and PS. PS is converted sequentially into phosphatidylethanolamine (PE) and PC. PA also serves as a precursor for phosphatidylglycerol phosphate and ultimately cardiolipin (not shown). PA alternatively serves as a precursor of DAG, and PC can also be synthesized from DAG and CDP-choline. DAG also serves as the precursor for TAG and can be phosphorylated to regenerate PA. The synthesis of free sterols and fatty acids share acetyl-CoA as a common precursor. Acyl-CoA serves as fatty acid pool for the synthesis of PA, TAG, and SE. In the synthesis of inositol-containing sphingolipids, inositol phosphate derived from PI is sequentially transferred to ceramide to form IPC and to MIPC to form M(IP)2C, releasing DAG at each of the two steps. The names of the structural genes that are discussed in detail in this report are shown adjacent to the arrows of the metabolic conversions in which they are involved. FFA, free fatty acids. HMG, 3-hydroxy-3-methyl-glutacyl.
However, a yeast quadruple mutant strain (dga1Δlro1Δare1Δare2Δ) lacking the ability to store fatty acids in either TAG or SE is viable (4, 16). The sole growth phenotypes relative to wild type that have been reported for this strain are unsaturated fatty acid-induced toxicity and a prolonged lag phase after transfer to fresh YPD media (17, 18). No significant change in growth of this strain was observed during exponential or stationary phase (4, 18). These data suggest that the reserve of TAG and SE is not essential to sustain growth in yeast, at least when cells grow in rich media.
Certain mutants, defective in membrane trafficking (Sec−), were shown to exhibit increased TAG synthesis at the expense of phospholipid synthesis when shifted to a temperature that restricted membrane trafficking and cell growth (15). That study specifically showed that deleting the DAG acyltransferases, Dga1p and Lro1p, in the sec13-1 mutant, defective in membrane and protein transport from the ER, led to a lowering of the temperature at which the mutant could grow (i.e. its restrictive temperature). The lowering of the restrictive temperature in the sec13-1dga1Δlro1Δ strain, was especially pronounced when it was grown in the absence of the phospholipid precursor, inositol. In wild type cells, lack of inositol supplementation results in a substantial reduction in the synthesis of phosphatidylinositol (PI) (19, 20).
The above-summarized evidence suggests that the cell coordinates the synthesis and breakdown of storage lipids with its demand for membrane lipid synthesis. Consistent with this idea, we showed in a previous study that the fatty acids required for the rapid burst of PI synthesis after inositol supplementation to cells deprived of inositol are derived in part from both de novo fatty acid synthesis and phosphatidylcholine (PC) turnover (19, 21). However, these two sources of fatty acids do not fully account for the burst in PI synthesis (19), suggesting that additional fatty acids might be derived from hydrolysis of TAG. In the current study we tested the ability of the cells to grow in the absence of inositol and to rapidly restore PI content in response to inositol reintroduction when they are unable to mobilize TAG. We report that upon inositol reintroduction, the dga1Δlro1Δare1Δare2Δ strain, unable to synthesize TAG or SE, and the tgl3Δtgl4Δtgl5Δ strain, lacking the TAG lipases, both exhibit slow recovery of PI content in comparison to wild type cells. However, only the dga1Δlro1Δare1Δare2Δ strain failed to grow in the absence of inositol. Thus, failure to hydrolyze TAG does not fully explain the Ino− phenotype of the dga1Δlro1Δare1Δare2Δ strain. We present evidence that this mutant exhibits reduced synthesis of PI even when grown continuously in the presence of exogenous inositol. The mutant also devotes a larger percentage of newly synthesized PI to the synthesis of inositol-containing sphingolipids for which PI serves as a precursor.
EXPERIMENTAL PROCEDURES
Yeast Strains and Culture Conditions
The S. cerevisiae strains used in this study are listed in Table 1. All the strains were derived from the S288C genetic background. Cultures were maintained on 1% yeast extract, 2% peptone, 2% glucose, 2% agar media plates. All experiments were conducted using cultures grown to mid-logarithmic phase at 30 or 37 °C on a rotary shaker (New Brunswick Scientific Co., Inc.) at 200 rpm using chemically defined synthetic media as described by Jesch et al. (22). Cells were grown in 50-ml batches of complete synthetic media with (I+) or without (I−) inositol (75 μm) with (C+) or without (C−) choline (1 mm) as indicated. Solid media had the same composition plus 2% agar.
TABLE 1.
Yeast strains used in this study
Analysis of Ino− Phenotypes
For the analysis of inositol auxotrophy phenotype (Ino− phenotype) (23), yeast strains were grown overnight in I+ medium at 30 °C. After 16 h, the cultures were diluted back to A600 = 0.15 in 50 ml of the same medium and allowed to grow to mid-logarithmic phase at 30 °C. Samples were harvested and resuspended at a concentration of 0.5 A600/ml, serially diluted in 10-fold increments, spotted onto plates containing 75 μm inositol (I+) or no inositol (I−) and/or 1 mm choline (C+) or no choline (C−), and incubated at the indicated temperature for 3 days.
Cell Growth and Calculation of Doubling Time
To determine doubling times, cultures of wild type and dga1Δlro1Δare1Δare2Δ strains were grown overnight in I+C− medium at 30 or 37 °C and in I+C+ medium at 37 °C. After 16 h, cultures were diluted back to A600 = 0.15 in 50 ml of I+C− at 30 or 37 °C and in I+C+ medium at 37 °C and allowed to reach A600 = 0.5. At this cell density each culture was divided in half. One-half of each culture (25 ml) was filtered, washed with pre-warmed medium containing inositol, and resuspended in 25 ml of I+C− medium at its original incubation temperature of 30 or 37 °C or in I+C+ medium at 37 °C. The other half was also filtered, washed with prewarmed medium lacking inositol, and resuspended in I−C− medium at its original incubation temperature of 30 or 37 °C or in I−C+ medium at 37 °C. The samples were harvested for absorbance measurements at 1.5, 3, 4.5, and 6 h after the shift.
The A600 of the cultures was used to calculate the doubling time according to the formula Td = (t2 − t1) × log 2/log(q2/q1), in which Td is the doubling time, t2 is a time point, t1 is zero time or an earlier time point, q2 is the A600 at a given time point, q1 is the A600 at zero time. Generation times were calculated using the above formula for the intervals from 0–3 h and from 3 to 6 h after the media shift for all the growth conditions. The doubling time for yeast in complete synthetic media in the experiments reported here was found to be ∼2.5–3 h.
RNA Isolation and RT-PCR Analysis
Wild type and dga1Δlro1Δare1Δare2Δ strains were grown under the growth conditions identical to those described above for the calculation of the generation time. Total RNA was isolated using RNeasy® Mini kit including a DNA digestion with RNase-free DNase Set (both from Qiagen). 1 μg of RNA was transcribed into cDNA using oligo(dT) 12–18 primer (0.5 μg), PCR grade dNTP mix (0.5 μm), First Strand Buffer (1×), DTT (10 mm), and 100 units of SuperScript® III reverse transcriptase (Invitrogen). Real-time PCR was performed on a StepOnePlusTM Real-Time PCR System (Applied Biosystems) using TaqMan® Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems), and the following TaqMan® probes and primers: INO1, TaqMan® probe, 5′-6-carboxyfluorescein (Fam)-CTG TTG CCC ATG GTT AGC CCA AAC G-6-carboxytetramethylrhodamine (Tamra)-3′; forward primer (5′-9 GGA ATG ACG TTT ATG CTC CTT TTA A-3′) and reverse primer (5′-GTC CCA ACC AGA GAC GAC AAA-3′; OPI3, TaqMan® probe, 5′-Fam-ACG AGT CTG CAT TGC GTG AAC AGC CTA C-Tamra-3′; forward primer (5′-TCG CTA GGT ATC GTC AGA GAC ATG-3′) and reverse primer, (5′-TCG CCC GTG ATC AGA GAA C-3′); ACT1, TaqMan® probe, 5′-Fam-TGC AAA CCG CTG CTC AAT CTT CTT CAA T-Tamra-3′; forward primer (5′-CGC CTT GGA CTT CGA ACA AG-3′) and reverse primer (5′-GAC CAT CTG GAA GTT CGT AGG ATT-3′). ACT1 gene served as an internal standard for normalization.
In brief, the reaction mix in a volume of 25 μl consisted of 0.5 μm primers, 0.2 μm TaqMan® probe, 1× Master Mix, and 5 ng of cDNA. All reactions were performed in technical duplicate. Non-template control (5 ng of RNA) and non-reaction control (diethylpyrocarbonate water) were routinely performed. The thermal program for the PCR included 95 °C for 10 min (Stage 1), 95 °C for 0.5 min and 60 °C for 1 min for a total of 40 cycles (Stage 2), and hold at 4 °C (Stage 3). Relative quantitation was done using the ΔΔCt method (see StepOnePlusTM user manual of Applied Biosystems). The ΔΔCt represents the change in mRNA expression after ACT1 normalization relative to the wild type control calculated as 2−(Gene Ctx − ACT1 Ctx) − (Gene Ctcr − ACT1 Ctcr), where “gene” represents the mRNA under study (INO1 or OPI3), x refers to the strain from which the mRNA tested was derived (i.e. wild type or quadruple mutant), and “cr” refers to the control mRNA. The value cr (for control mRNA) was derived from level of mRNA in the wild type strain, pre-grown as described above in I+C− medium at 30 or 37 °C or in I+C+ medium at 37 °C but shifted to fresh medium of the same composition (i.e. containing inositol) at the same temperature for 1.5 h. The Ct (cycle threshold) is defined as the number of cycles required for the fluorescent signal to cross the threshold (i.e. to exceed background level). Each RT-PCR experiment was performed at least in triplicate.
Pulse Labeling of Phospholipids
To analyze de novo synthesis of phospholipids after the shift to fresh media, wild type and dga1Δlro1Δare1Δare2Δ cells were pre-grown to A600 = 0.5 in I+C+ medium at 37 °C, and the culture was divided in half as described above. One-half of the culture was filtered and washed in prewarmed I+C+ medium and then resuspended in a fresh aliquot of I+C+ medium, whereas the other was washed and filtered in prewarmed I−C+ medium and then resuspended in fresh, prewarmed I−C+ medium. After the shift to fresh medium with or without inositol, both cultures were incubated at 37 °C, and 5-ml samples were collected at 1.5 and 3 h and pulse-labeled for 20 min with 100 μCi/ml [32P]orthophosphate (specific activity of 32P in the medium was 13.51 mCi/mmol of phosphate). Labeled lipids were extracted as previously described (24). The individual phospholipids species were resolved by two-dimensional thin layer chromatography (25). Phospholipid identity was based on the mobility of known standards and quantified on a STORM 860 PhosphorImager (Amersham Biosciences).
Analysis of Sphingolipid Synthesis
Wild type and dga1Δlro1Δare1Δare2Δ mutant were grown and labeled under identical conditions described above for the phospholipid synthesis except that the cells were pulse-labeled for 30 min instead of 20 min to allow sufficient time for 32P label to enter through PI and be transferred to the inositol-containing sphingolipids. Growth of both strains was terminated by the addition of TCA to a final concentration of 5%. Cells were collected by centrifugation and washed once with water. Labeled lipids were extracted as previously described (24). The lipids were deacylated according to the method of Stock et al. (26). By this method, the dried lipid extracts were redissolved in 1 ml of chloroform:methanol:water (16:16:5, v/v/v). An equal volume of 0.2 n NaOH in methanol was added to each sample, and the mixture was incubated at 30 °C for 45 min. To each sample, 1.1 ml of 0.5% (w/v) EDTA was added, and the mixtures were neutralized by the addition of 0.2 ml of 1 n acetic acid. The nondeacylated lipids containing the complex sphingolipids were extracted with 0.5 ml of chloroform, dried under N2v and resuspended in 0.5 ml of chloroform:methanol:water (16:16:5, v/v/v). Labeled sphingolipids were separated by two-dimensional thin layer chromatography as described by Stock et al. (26). The amounts of the labeled sphingolipids on the chromatograms were quantified on a STORM 860 PhosphorImager.
Analysis of Total Lipid Composition Assessed by Labeling of Lipids of Logarithmically Growing Cultures to Steady State with [14C]Acetate
Cultures of wild type, dga1Δlro1Δare1Δare2Δ, and tgl3Δtgl4Δtgl5Δ were grown in I+C− medium at 30 or 37 °C and in I+C+ medium at 37 °C in the presence of 1 μCi/ml [1-14C]acetate (specific activity, 57 mCi/mmol). Yeast strains were maintained in logarithmic growth phase in the presence of label for at least seven generations. Cultures were grown from A600 = 0.1 until they reached approximately A600 = 2.0. At this cell density, the cultures were diluted in the same medium containing [1-14C]acetate at the same specific activity to A600 = 0.15 and allowed to grow until mid-logarithmic phase. At this time point, steady state labeling of all lipid classes, defined as the point at which proportional and absolute labeling relative to culture absorbance of pools of all lipids remained constant, was achieved. After taking samples for time 0 (I+ medium control), the cells were then shifted as described above to medium lacking inositol, keeping all other growth conditions and specific activity of the label unchanged (i.e. to I−C− medium at 30 or 37 °C and to I−C+ medium at 37 °C, respectively) and incubated for an additional 180 min maintaining constant specific activity of the label. After 180 min, samples were taken, and the phospholipid and neutral lipid compositions of the cultures were determined as described by Gaspar et al. (19). This represents the composition of cells grown for 180 min after the shift to inositol-free medium. After 180 min of growth in the absence of inositol, inositol was added back to the cultures to a final concentration of 75 μm, and samples were collected at 0, 5, 15, and 30 min after inositol addition. For the assessment of the phospholipid composition in the absence and presence of cerulenin, parallel cultures of wild type and tgl3Δtgl4Δtgl5Δ similarly labeled with [1-14C]acetate to steady state were shifted to I−C+ medium for 90 min. At this time point, cerulenin (final concentration of 10 μg/ml) was added to block de novo fatty acid synthesis (10), and the cultures were allowed to grow for additional 90 min. Inositol was reintroduced at 180 min after the shift, and samples were collected at 0, 5, 15, and 30 min after inositol addition. Total lipids were extracted with a mixture of chloroform:methanol 2:1 v/v (27). The chloroform phase was dried, and the residue was dissolved in chloroform-methanol 2:1 v/v. Phospholipids were separated on Whatman Silica Gel 60A HPTLC plates using the solvent system chloroform:ethyl acetate:acetone:isopropyl alcohol:ethanol:methanol:water:acetic acid (30:6:6:6:16:28:6:2, per vol) (28). The neutral lipid classes were analyzed on high performance TLC plates using hexane:diethylether:formic acid (80:20:2, per volume) (29). Metabolite identity was established based on the mobility of known standards. The amounts of labeled lipids on the chromatograms were quantified on a STORM 860 PhosphorImager.
RESULTS
The Absence of TAG and SE Synthesis Leads to Growth Impairment in Medium Lacking Inositol at High Temperatures
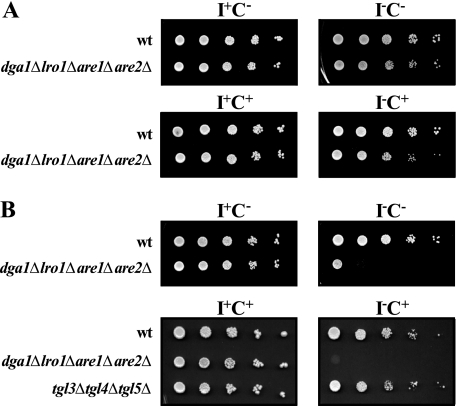
We previously observed that deletion of the DGA1 and LRO1 genes in the sec13-1 genetic background resulted in a decrease in the permissive temperature at which the triple mutant would grow in the absence of inositol compared with the sec13-1 parent (15). This finding suggested that strains blocked in TAG synthesis might display additional phenotypes under conditions limiting phospholipid synthesis. We have also previously shown that the combined effects of removing inositol while adding a second phospholipid precursor, choline, to the growth medium and increasing the incubation temperature to 37 °C has detrimental consequences for cell growth and survival of mutants defective in certain aspects of lipid metabolism and stress response signaling (30).4 Therefore, we tested the growth of the dga1Δlro1Δare1Δare2Δ mutant in four different media I+C−, I−C−, I+C+, and I−C+ on agar plates. At 30 °C the dga1Δlro1Δare1Δare2Δ mutant grew as well as wild type on these media (Fig. 2A). However, when the temperature was raised to 37 °C, growth of the mutant was significantly reduced on I−C− medium, and virtually no growth was observed on I−C+ medium (Fig. 2B). Deletion of any of the individual genes or any combinations of two or three of them did not confer this phenotype (data not shown), suggesting that the complete absence of lipid droplets is responsible for the Ino− phenotype. We also tested the growth of the tgl3Δtgl4Δtgl5Δ mutant, in which TAG lipolysis is blocked (10, 32, 33), to determine whether the failure of the dga1Δlro1Δare1Δare2Δ mutant to grow in the absence of inositol was due to inability of mobilize TAG. However, unlike the dga1Δlro1Δare1Δare2Δ mutant, growth of the tgl3Δtgl4Δtgl5Δ mutant under all of the conditions tested was similar to wild type cells (Fig. 2B).
FIGURE 2.
The dga1Δlro1Δare1Δare2Δ quadruple mutant exhibits inositol auxotrophy at 37 °C. Overnight cultures grown in I+ or I− medium at 30 °C were diluted back to A600 = 0. 15 in 50 ml of the same medium and allowed to grow to mid-logarithmic phase at 30 °C. Samples were harvested and resuspended at a concentration of 0.5 A600. Each sample was diluted in multi-well plates by 1:10 serial dilutions, and 10 μl of cells from each dilution were spotted on I+C−, I−C−, I+C+, and I−C+ plates and allowed to grow at the designated temperatures for 3 days. Panel A, wild type and dga1Δlro1Δare1Δare2Δ strains grown at 30 °C. Panel B, wild type, dga1Δlro1Δare1Δare2Δ, and tgl3Δtgl4Δtgl5Δ strains grown at 37 °C.
Mutant Strains Unable to Synthesize Storage Lipids or Mobilize TAG Exhibit Reduced Synthesis of PI upon Inositol Addition
Actively growing wild type cells deprived of inositol undergo a dramatic burst of PI synthesis when inositol is reintroduced (19–21). There are at least three different sources of fatty acids that can be utilized for PI synthesis under these conditions, namely de novo fatty acid synthesis, PC turnover, and TAG lipolysis. We had previously observed that de novo fatty acid synthesis and turnover of PC contributed to the ability of wild type cells to rapidly restore PI content (19). To determine whether TAG metabolism plays a role in the burst of PI synthesis observed in the wild type strain after addition of inositol, we compared the changes in PI content in the tgl3Δtgl4Δtgl5Δ strain as well as the dga1Δlro1Δare1Δare2Δ strain to those in the wild type strain. In the dga1Δlro1Δare1Δare2Δ mutant, TAG synthesis is disabled altogether due to the lack of the respective acyltransferases that convert DAG to TAG, whereas in the tgl3Δtgl4Δtgl5Δ mutant, TAG lipolysis is blocked. Thus, fatty acids for PI synthesis in both mutants must come largely from PC turnover or de novo synthesis.
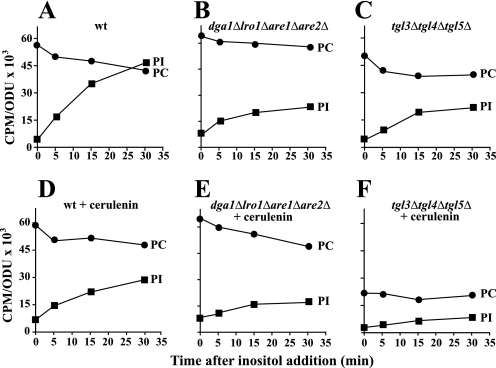
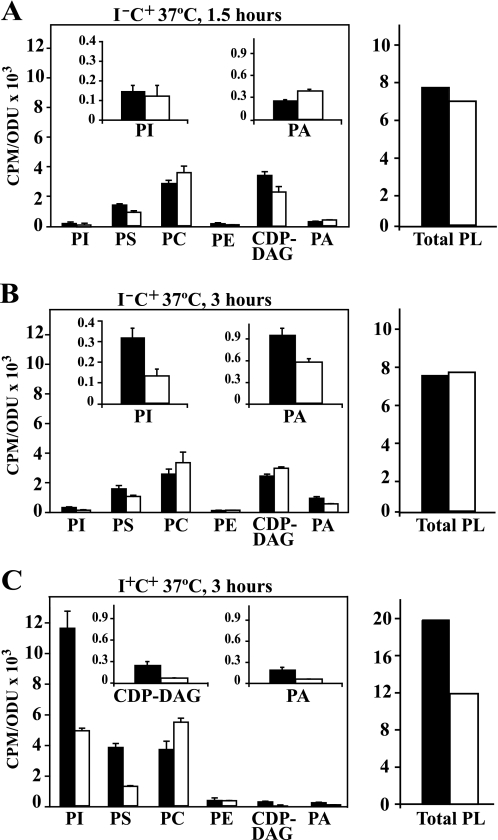
Because fatty acids generated from PC turnover are maximized in medium containing choline at 37 °C (34, 35), cells were grown under these conditions. To measure absolute phospholipid content, cells were pregrown and labeled with [14C]acetate to steady state in medium containing both inositol and choline (I+C+) at 37 °C and shifted to medium lacking inositol (I−C+) while maintaining the label constant as described under “Experimental Procedures.” After incubation for an additional 180 min, inositol was reintroduced, and samples were taken and analyzed for the rebound in PI content. The rebound in PI content under these conditions in the wild type strain was striking (Fig. 3A). In contrast, in the dga1Δlro1Δare1Δare2Δ mutant, the rate of recovery of PI content after inositol supplementation was only about 30% that of the level in the wild type strain under the same conditions (Fig. 3B). PC content in the mutant strain, however, was slightly higher than in wild type and declined somewhat less sharply upon inositol reintroduction (Fig. 3B). Similarly, in the tgl3Δtgl4Δtgl5Δ mutant, the rate of recovery of PI content after inositol supplementation was very slow (compare Fig. 3, A and C). However, in this case, whereas PC content was somewhat lower than wild type before inositol re-addition, it declined in a fashion similar to that observed in wild type cells (Fig. 3, A and C). These data overall indicate that PC turnover is not markedly altered in dga1Δlro1Δare1Δare2Δ or tgl3Δtgl4Δtgl5Δ strains in comparison to wild type. However, a lack of fatty acids derived from TAG in both strains significantly attenuates the burst in PI synthesis.
FIGURE 3.
The pattern of PI synthesis after reintroduction of inositol is reduced in mutant strains unable to mobilize or store TAG. Lipids in cultures of wild type, dga1Δlro1Δare1Δare2Δ, and tgl3Δtgl4Δtgl5Δ were labeled to steady state with [1-14C]acetate (1 μCi/ml) in I+C+ medium at 37 °C as described under “Experimental Procedures” for at least 7 generations and grown to mid-logarithmic phase and shifted to I−C+ medium at 37 °C for 180 min as described under “Experimental Procedures,” maintaining label at constant specific activity. At 180 min, inositol was reintroduced, and samples were taken at the time points depicted in the figure. Parallel cultures grown in the same fashion were treated with cerulenin at a final concentration of 10 μg/ml at 90 min after the shift to medium lacking inositol. Cells were allowed to grow for additional 90 min, and inositol was reintroduced. Samples were collected at 0, 5, 15, and 30 min after inositol addition. Lipids were extracted and analyzed as described under “Experimental Procedures”. Panel A, wild type. Panel B, dga1Δlro1Δare1Δare2Δ. Panel C, tgl3Δtgl4Δtgl5Δ. Panel D, wild type + cerulenin. Panel E, dga1Δlro1Δare1Δare2Δ + cerulenin. Panel F, tgl3Δtgl4Δtgl5Δ + cerulenin. The data represent the average of at least two independent experiments. Experimental error was less than 10% in all cases. Error bars are not shown for clarity of presentation. The lipids indicated are PI (solid squares) and PC (solid circles). ODU, optical density units.
To assess the contribution of de novo fatty acid synthesis to the burst of PI synthesis, lipids in all three strains were labeled to steady state with [14C]acetate in I−C+ medium at 37 °C following the procedure described above. Cerulenin, an inhibitor of fatty acid synthesis (36), was added to the cultures at 90 min before inositol reintroduction. Wild type cells pretreated with cerulenin showed a much less dramatic rebound in PI content after inositol reintroduction than that observed in cells not treated with cerulenin (Fig. 3D), consistent with previously reported results (19). In the dga1Δlro1Δare1Δare2Δ strain treated with cerulenin, re-synthesis of PI after inositol reintroduction was also attenuated (Fig. 3E), and the mutant also experienced a greater decline in PC content than wild type under these conditions (compare Fig. 3, D to E).
The results in the tgl3Δtgl4Δtgl5Δ cultures treated with cerulenin in the same fashion were strikingly different from either wild type or the dga1Δlro1Δare1Δare2Δ cultures. The level of PI attained after inositol reintroduction in the tgl3Δtgl4Δtgl5Δ strain was lower than wild type or dga1Δlro1Δare1Δare2Δ, and PC metabolism was also dramatically altered in comparison to wild type or dga1Δlro1Δare1Δare2Δ strains (Fig. 3F). During the 90 min of treatment with cerulenin before the introduction of inositol, PC content in the tgl3Δtgl4Δtgl5Δ mutant dropped by 50% as compared with the untreated mutant cells (compare Fig. 3, C and F). This result suggests that de novo fatty acid synthesis is required in the tgl3Δtgl4Δtgl5Δ mutant to maintain PC content, unlike wild type or the dga1Δlro1Δare1Δare2Δ mutant.
These results support the notion that lack of the storage lipids, TAG and SE, and the inability to degrade TAG have major impacts on both PI and PC homeostasis in response to inositol supplementation and suggest that storage lipids may provide a source of fatty acids for these essential cellular phospholipids. The two mutant strains, however, exhibit different responses to the presence of cerulenin. These differences indicate that the inability to synthesize storage lipids has consequences independent of the ability to mobilize it. Moreover, although the reduction in the burst of PI synthesis was seen in both mutants, only the dga1Δlro1Δare1Δare2Δ mutant failed to grow in the absence of inositol. Thus, the slow rate of recovery of PI synthesis in the dga1Δlro1Δare1Δare2Δ mutant does not fully account for its Ino− phenotype.
The Kinetics of INO1 Derepression Is Altered in the dga1Δlro1Δare1Δare2Δ Mutant as Compared with Wild Type
The Ino− phenotype is frequently associated with reduced expression of INO1 and other phospholipid biosynthetic genes that are regulated by the Opi1p repressor (21, 23). INO1 expression is derepressed up to several hundred-fold in wild type cells grown in the absence of inositol as compared with expression levels in the presence of 75 μm inositol, and INO1 derepression is very rapid when cells are shifted to inositol-free medium (21, 22, 37). Because the dga1Δlro1Δare1Δare2Δ mutant, which is unable to synthesize TAG, displays an Ino− phenotype at 37 °C, we questioned whether this phenotype was due to a deficiency in INO1 expression.
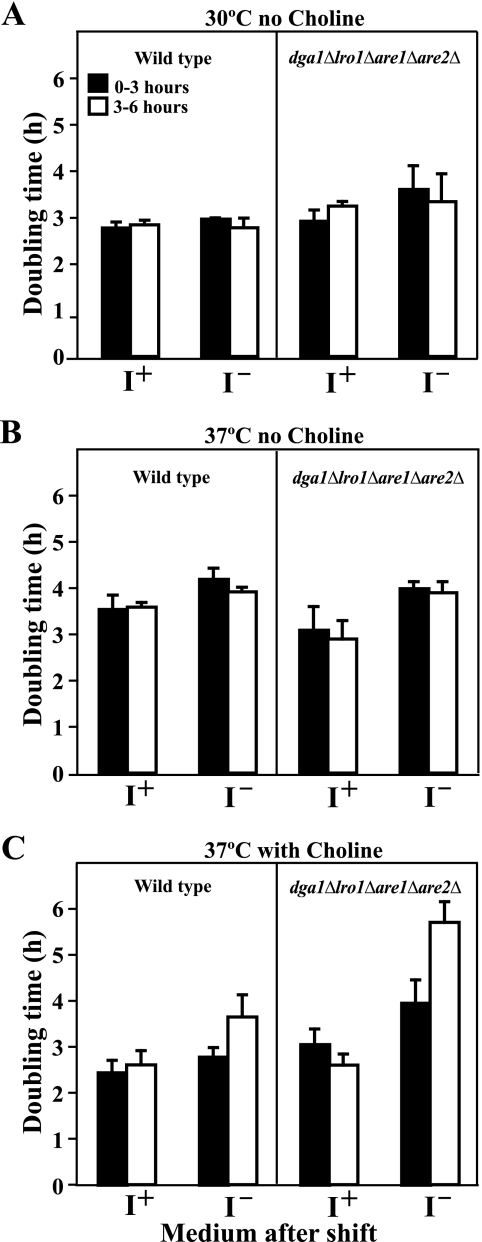
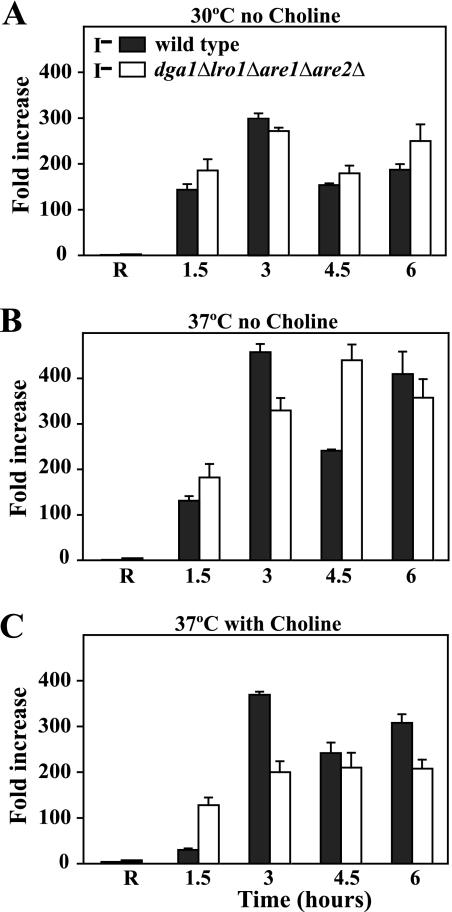
First, we compared the growth of wild type and dga1Δlro1Δare1Δare2Δ strains in liquid medium as described under “Experimental Procedures” to ensure that analyses were performed at comparable growth rates, which are known to affect INO1 expression (23). The doubling time of both strains (Fig. 4) was calculated for the time intervals corresponding to 0–3 and 3–6 h after a shift of logarithmically growing cells from I+C− medium to I−C− medium at 30 or 37 °C and from I+C+ to I−C+ at 37 °C. There was no change in the doubling time in either strain at 30 °C for the first 6 h after the shift to I−C− media (Fig. 4A). At 37 °C, between 3–6 h after the shift to I−C−, both wild type and dga1Δlro1Δare1Δare2Δ mutant exhibited somewhat longer doubling times as compared with growth at 30 °C (Fig. 4B). When shifted to I−C+ medium at 37 °C, both strains showed a significant increase in doubling time in the interval from 3 to 6 h after the shift. However, the lengthening of doubling time was significantly greater in the dga1Δlro1Δare1Δare2Δ strain (Fig. 4C). Growth of the dga1Δlro1Δare1Δare2Δ mutant for an extended period of time in I−C+ medium at 37 °C resulted in a further gradual increase in doubling time, which ultimately exceeded 8 h before the culture reached stationary phase. This reduced increase in cell mass, however, was not due to a loss of viability (data not shown).
FIGURE 4.
Growth rates of wild type and dga1Δlro1Δare1Δare2Δ immediately after a shift to medium lacking inositol. Cells were grown in I+C− medium at 30 or 37 °C and in I+C+ medium at 37 °C until mid-logarithmic growth as described under “Experimental Procedures.” Cells were harvested by filtration, washed, and resuspended into prewarmed medium identical to the original culture medium containing inositol. A second aliquot of each culture was shifted to prewarmed medium identical to the original culture medium but lacking inositol. The samples were harvested for absorbance measurements at 1.5, 3, 4.5, and 6 h after the shift, and the doubling time for each interval was calculated by using the formula described under “Experimental Procedures.” Solid bars represent the growth rate from 0 to 3 h, and open bars indicate the growth rate from 3 to 6 h. The data represent the average of three experiments.
To ensure that the INO1 gene was fully repressed at the start of the experiment, the kinetics of INO1 derepression was measured in cells pregrown at 30 or 37 °C in I+C− medium or in I+C+ medium at 37 °C, identical to the conditions used in the experiments in which doubling times were measured (Fig. 4). To determine INO1 derepression in I− medium, each culture was shifted to prewarmed medium, which lacked inositol but was otherwise identical to the original culture medium, and samples were harvested at 1.5, 3, 4.5, and 6 h after the shift.
After the shift to I−C− medium either at 30 or 37 °C, no striking difference in the level of expression of INO1 was observed in the dga1Δlro1Δare1Δare2Δ mutant as compared with wild type (Fig. 5, A and B). In general, the extent of derepression of INO1 observed in both strains at 37 °C upon a shift to I−C− medium was higher than that observed at 30 °C in the same medium (Fig. 5, A and B). Moreover, there was oscillation in the level of INO1 mRNA over the time course of 6 h after the shift to I−C− medium at 37 °C in the wild type strain, indicative of an auto-regulatory circuit controlling INO1 expression.5 At 1.5 h after the shift to I−C+ medium at 37 °C, the dga1Δlro1Δare1Δare2Δ mutant actually exhibited a higher level of INO1 expression than the wild type strain (Fig. 5C), whereas at 3 h in I−C+ medium at 37 °C, the wild type strain showed a spike in expression that was not seen in the dga1Δlro1Δare1Δare2Δ mutant (Fig. 5C). However, by 4.5 h after the shift, INO1 expression leveled off in both strains at fairly similar values (Fig. 5C).
FIGURE 5.
Derepression of the INO1 gene in the wild type and quadruple mutant under various growth conditions. Cells were grown in I+C− medium at 30 or 37 °C and in I+C+ medium at 37 °C until mid-logarithmic growth as described under “Experimental Procedures.” Cells were harvested by filtration, washed, and resuspended into prewarmed medium identical to the original culture medium containing inositol. A second aliquot of each culture was shifted to prewarmed medium identical to the original culture medium except that it did not contain inositol. The samples were harvested at 1.5, 3, 4.5, and 6 h after the shift, and total RNA was isolated and analyzed by RT-PCR as described under “Experimental Procedures.” R refers to the INO1 expression under repressing conditions defined as the level of INO1mRNA in each strain at the stated temperature and choline supplementation condition and shifted to fresh medium containing inositol and maintaining temperature and choline supplementation. All other values in each of the three panels represent the increase in INO1mRNA over the time course of growth relative to the wild type repressed level after the shift to medium lacking inositol but maintaining temperature and choline supplementation level. Data are the averages from three independent experiments. Solid bars represent wild type cells, and open bars indicate the dga1Δlro1Δare1Δare2Δ strain.
Similar measurements were made of the patterns of derepression of OPI3, encoding a phospholipid methyltransferase involved in PC biosynthesis, which like INO1 is regulated by the Opi1p repressor but shows a much less dramatic derepression ratio in wild type cells shifted to I− medium (38). Derepression of OPI3 in the dga1Δlro1Δare1Δare2Δ mutant was also similar to wild type (data not shown). Thus, the observed Ino− phenotype of the dga1Δlro1Δare1Δare2Δ mutant grown in the presence of choline at 37 °C is not due to an inability to derepress INO1 or coregulated lipid biosynthetic genes.
The Rate of Phospholipid Synthesis in the dga1Δlro1Δare1Δare2Δ mutant Is Significantly Decreased in Comparison to Wild Type at 37 °C during Active Growth Especially in the Presence of Inositol
Because the growth defect of the dga1Δlro1Δare1Δare2Δ mutant on I−C+ medium at 37 °C cannot be explained simply by a defect in INO1 expression, we analyzed phospholipid synthesis by pulse-labeling with [32P]orthophosphate. The wild type and dga1Δlro1Δare1Δare2Δ strains were grown in the presence of inositol (I+C+ medium) at 37 °C to mid-logarithmic phase and shifted by filtration to prewarmed I−C+ or I+C+ medium as described under “Experimental Procedures” and previous experiments involving derepression of INO1. At 1.5 and 3 h after the shift, aliquots were taken and pulse-labeled for 20 min followed by lipid extraction and analysis.
Wild type and dga1Δlro1Δare1Δare2Δ strains initially showed fairly similar patterns of incorporation of 32P into PI at 1.5 h after the shift to I−C+ medium at 37 °C (Fig. 6A). At this initial time point, however, labeling of phosphatidic acid (PA) in mutant cells was 30% higher than that observed in wild type cells (Fig. 6A), correlating well with the relative expression levels of INO1 in these two strains (Fig. 5C). These results are consistent with the model of regulation of INO1 proposed by Loewen et al. (21). It is plausible that the higher level of labeling of PA in the dga1Δlro1Δare1Δare2Δ mutant at this initial time point is due to its inability to synthesize TAG from DAG, thus, slowing the utilization of newly synthesized PA during the period immediately after removal of inositol. However, in cultures grown in I−C+ medium for 3 h, the level of PA in the wild type strain had increased by more than 3-fold over the levels observed in the same medium at 1.5 h, whereas labeling of PA in the mutant increased only modestly (Fig. 6B), again correlating to the lower level of INO1 expression observed in the mutant compared with wild type at this time point after the shift (Fig. 5). At 3 h in I−C+ medium, labeling of PI had also increased substantially in the wild type strain over the levels observed in the same medium at 1.5 h (Fig. 6B). However, labeling of PI in the mutant was less than half that of wild type at 3 h and, unlike wild type, showed little change between 1.5 and 3 h (compare Fig. 6, A and B). Moreover, incorporation of 32P into total phospholipids was similar in both strains at both time points in I−C+ medium (Fig. 6, Total PL).
FIGURE 6.
Pulse labeling of wild type and dga1Δlro1Δare1Δare2Δ with 32[P]orthophosphate. Cultures were grown in I+C+ medium at 37 °C until mid-logarithmic phase of growth (A600 = 0. 5) and then divided in half. One half of the culture was filtered, washed in pre-warmed I+C+, and resuspended in the same medium, whereas the remaining half was filtered-washed with prewarmed I−C+ medium and resuspended in I−C+. Both cultures were incubated at 37 °C, and samples were collected at 1.5 and 3 h and pulse-labeled for 20 min with 32P. Lipids were extracted and analyzed as described under “Experimental Procedures.” Data are expressed as counts of radiolabel 32P incorporated into total and individual phospholipids per absorbance units in the cell culture. PE, phosphatidylethanolamine; Total PL, total phospholipids. The inset on each of the panels is an expansion of the data that accounts for PI, PA, and CDP-DAG. Panel A, shown are cultures harvested at 1.5 h after the shift to I−C+ at 37 °C. Panel B, shown are samples harvested at 3 h after the shift to I−C+ at 37 °C. Panel C, shown are samples taken at 3 h after the shift to I+C+ at 37 °C. Solid bars represent wild type cells, and open bars indicate dga1Δlro1Δare1Δare2Δ.
When wild type cells acclimated to grow in I+C+ medium were transferred to fresh I+C+ medium for 3 h at 37 °C, overall labeling of phospholipids (Fig. 6, Total PL) was more than double in comparison to that observed in cells transferred for the same period of time from I+C+ medium to medium lacking inositol (I−C+) (Fig. 6C). Although the greatest proportion of the relative increase in total phospholipid synthesis in the wild type strain in the presence of inositol as opposed to its absence was accounted for by an increase in PI synthesis, labeling of PA, CDP-DAG, and PS were also increased (Fig. 6C). Labeling of total phospholipids and PI were also higher in the mutant in the presence of inositol, but the increase was less dramatic than in wild type cells (Fig. 6C). Thus, the lower rate of synthesis of PI in the mutant in comparison to wild type in I+C+ medium is clearly independent of INO1 expression, as INO1 is not expressed by either strain in the presence of inositol (Fig. 5). Also, the presence of exogenous inositol did not correct the relative reduction in the synthesis of PI in the mutant in comparison to wild type (Fig. 6). Because inositol is not limiting for PI production in inositol-containing medium, other factors must limit PI synthesis in the mutant relative to wild type. Because other phospholipids that derive label directly from PA all show reduced labeling in the mutant in I+C+ medium, it is likely that PA synthesis is among the limiting factors.
However, the reduced labeling of PA and PA-derived phospholipids in the dga1Δlro1Δare1Δare2Δ mutant could be due to either reduced PA synthesis and/or increased turnover of newly synthesized PA to produce DAG. In the presence of exogenous choline, PC is made largely via the CDP-choline pathway by the reaction of CDP-choline with DAG, which is derived from PA (Fig. 1). However, in the formation of DAG, the phosphate label from PA is lost, and the labeled phosphate in PC under these conditions is derived from CDP-choline. Consistent with the idea that a greater fraction of PA synthesis is being diverted to DAG, labeling of PC in the mutant strain was higher than in wild type after 3 h following the shift to either I+C+ or I−C+ medium (Fig. 6, B and C). However, the increase in synthesis of PC in the mutant is not sufficient to explain the extent of the reduction in the mutant in comparison to wild type in total labeling of all the lipids that derive phosphate from PA, including PS and PI. Thus, the dga1Δlro1Δare1Δare2Δ strain, which is completely unable to synthesize storage lipids, also experiences decreased synthesis of membrane phospholipids relative to wild type even when growing continuously in the presence of inositol.
A Higher Proportion of Newly Synthesized PI Is Used to Support Inositol-containing Sphingolipid Synthesis in the dga1Δlro1Δare1Δare2Δ Mutant
Rapid use of PI as a precursor in the synthesis of inositol-containing sphingolipids (Fig. 1) could also contribute to the reduced labeling of PI observed in the dga1Δlro1Δare1Δare2Δ mutant as compared with the wild type strain (Fig. 6). During the synthesis of these lipids, inositol phosphate derived from PI is sequentially transferred to ceramide to form inositol-phosphorylceramide (IPC) and to mannosyl-inositol-phosphorylceramide (MIPC) to form mannosyl-diinositol-phosphorylceramide (M(IP)2C), releasing DAG at each of the two steps that could also contribute to PC synthesis (Figs. 1 and 6). We analyzed the synthesis of inositol-containing sphingolipids by pulse labeling with [32P]orthophosphate following the growth conditions described above for phospholipid labeling (Fig. 6), except that the cells were pulse-labeled for 30 min instead of 20 min to allow sufficient time for 32P label to enter through PI and be transferred to the inositol-containing sphingolipids.
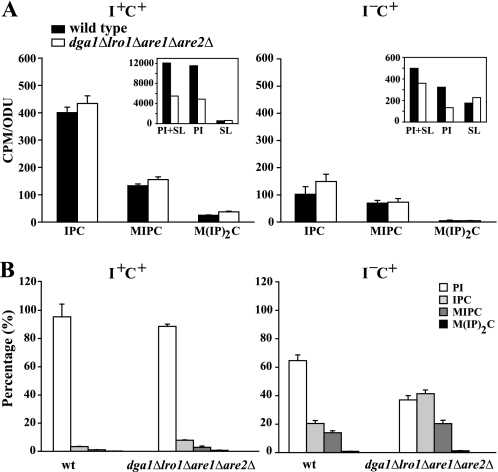
Because 32P label appearing in the inositol-containing sphingolipids must enter through PI (Fig. 1), the total amount of PI synthesized in a 30-min pulse is actually greater than the amount of label remaining in PI. This is reflected in the sum of the label retained in PI plus the label transferred to the three inositol-containing sphingolipids (see PI + SL, Fig. 7A inset). When inositol was present, the label appearing in PI was far greater in both strains, and the same was true of the label accounted for by the sum of PI plus the inositol-containing sphingolipids (Fig. 7A inset, compare PI + SL, I+C+ to I−C+). In both media, however, the mutant incorporated less label into either PI or PI + SL than the wild type strain. In I−C+ medium, labeling of PI alone in the mutant represented only 41% that in the wild type strain (Fig. 7A inset), whereas labeling of PI plus the inositol-containing sphingolipids (PI + SL) in the mutant represented 72% of that observed in the wild type (Fig. 7A inset). The cause of this discrepancy in labeling of PI versus PI + SL in the mutant versus wild type became apparent when labeling of the sphingolipids was examined. In the absence of inositol, the label incorporated into inositol-containing sphingolipids was reduced by 3-fold in both strains (Fig. 7A inset). However, in contrast to the wild type strain, the label incorporated into the sum of the three inositol-containing shingolipids in I−C+ medium in the mutant actually exceeded the label retained in PI in I−C+ medium (Fig. 7A inset). Moreover, the rate of synthesis of the individual inositol-containing sphingolipid species in the mutant was comparable or even slightly higher than in wild type in both I+C+ and I−C+ media (Fig. 7A) despite the lower rate of PI synthesis in the mutant. In Fig. 7B, label incorporated into each individual lipid (i.e. PI, IPC, MIPC, and M(IP)2C) is presented as a percentage of the total label incorporated into PI + SL. When the data are displayed in this fashion it is evident that of the total label incorporated into PI + SL in the mutant, almost 60% was used in producing inositol-containing sphingolipids in I−C+ medium in a 30-min pulse. In contrast, less than 40% of the total label in PI + SL was recovered in inositol-containing sphingolipids in the wild type under the same conditions (Fig. 7B). Indeed, the mutant incorporated a higher proportion of the total label entering PI (i.e. PI + SL) into sphingolipids, even in I+C+ medium (Fig. 7B).
FIGURE 7.
A higher proportion of newly synthesized PI is used to support inositol-containing sphingolipid synthesis in the dga1Δlro1Δare1Δare1Δ mutant. Cells were grown and labeled as described in Fig. 6 except that the cells were pulse-labeled for 30 min instead of 20 min to allow sufficient time for 32P label to enter through PI and be transferred to the inositol-containing sphingolipids. The total lipids were extracted and deacylated. The nondeacylated fraction contained the inositol-containing sphingolipids. Panel A, absolute 32P incorporation into IPC, MIPC, and M(IP)2C is shown. The inset shows the 32P incorporated into the total of inositol containing sphingolipids plus PI. The rate of 32P incorporation into PI was measured from lipid extracts before the deacylation step that were prepared from pulse-labeled parallel cultures as describe under “Experimental Procedures.” Panel B, data from the panel A inset is plotted as a percentage of total 32P label derived from PI (i.e. PI + SL) to illustrate the synthesis of each lipid relative to PI + SL. SL, total inositol-containing sphingolipids. ODU, optical density units.
Thus, the dga1Δlro1Δare1Δare2Δ mutant utilizes a higher proportion of newly synthesized PI to synthesize sphingolipids than the wild type strain. The net result is that the two strains synthesize similar amounts of inositol-containing sphingolipids (Fig. 7A) despite the reduced rate of synthesis of PI in the mutant. These data suggest that yeast cells have a mechanism(s) for ensuring adequate sphingolipid synthesis at the expense of PI. Furthermore, increased synthesis of inositol-containing sphingolipids relative to PI synthesis is clearly a factor in the reduction in labeling of PI in the mutant in comparison to wild type in I−C+ medium (Fig. 6). However, these considerations do not explain the fact that synthesis of both PI and PI + SL is greatly reduced in the mutant even in the presence of inositol. In fact, the proportionate decrease in PI + SL in the mutant versus wild type is actually greater in the presence of inositol. Nevertheless, reduced synthesis of PI coupled with increased demand for sphingolipid synthesis relative to total PI synthesis are likely contributing factors in the poor growth of the dga1Δlro1Δare1Δare2Δ mutant in the absence of inositol.
Exogenous Inositol Affects the Metabolism of Neutral Lipids in Wild Type and the dga1Δlro1Δare1Δare2Δ Mutant
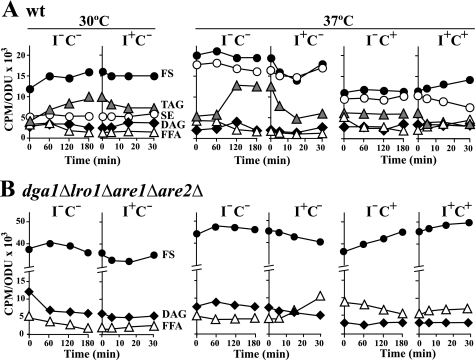
We also assessed changes in the abundance of neutral lipid classes in wild type and the dga1Δlro1Δare1Δare2Δ strains after the shift from I+ to I− medium. Cultures were allowed to reach mid-logarithmic phase in I+ medium (with or without choline at 30 or 37 °C) in the presence of [14C]acetate as described under “Experimental Procedures.” Cells were washed and transferred into medium lacking inositol at the same temperature and choline concentration, maintaining constant specific activity of the label. After incubation at 30 or 37 °C, samples were taken at 0, 60, 120, and 180 min. After 180 min in I− medium, inositol was reintroduced, and samples were collected at 5, 15, and 30 min (Fig. 8). Among the different neutral lipid classes, the most dramatic change in wild type cells after a shift from inositol-containing medium to I−C− medium at either 30 or 37 °C was an increase in the abundance of TAG (Fig. 8A). At both temperatures, the increase in TAG levels was accompanied by an almost “mirror image” decrease in free fatty acid content (Fig. 8A). When inositol was added back to the wild type cultures, TAG content decreased with a concomitant increase in the level of DAG that is presumably derived from lipolysis (Fig. 8A). A similar but more accentuated decline in TAG levels in response to inositol re-addition occurred in wild type cells incubated at 37 °C in the absence of choline (Fig. 8A). In contrast, when wild type cells were shifted to I−C+ at 37 °C, only a marginal change was observed in the abundance of TAG (Fig. 8A), but free fatty acids decreased significantly after the media shift (Fig. 8A). However, when inositol was reintroduced, TAG levels declined by about 50% within 30 min, whereas fatty acids increased (Fig. 8A). SE levels also decreased by about 30% after inositol re-addition, and free sterols increased (Fig. 8A). These data further demonstrate that TAG and SE degradation is stimulated upon the addition of inositol to actively growing cultures, increasing the availability of fatty acids for subsequent channeling into synthesis of PI and other membrane phospholipids derived from PA in wild type cells.
FIGURE 8.
The availability of inositol affects the abundance of neutral lipid classes in wild type. Lipids in all strains were labeled to steady state with [1-14C]acetate (1 μCi/ml) in I+C− medium at 30 or 37 °C and in I+C+ medium at 37 °C as described under “Experimental Procedures” for at least seven generations and grown to mid-logarithmic phase. They were shifted by filtration either in I−C− medium at 30 and 37 °C or to I−C+ medium at 37 °C as described under “Experimental Procedures” maintaining label at constant specific activity and cultured for 180 min. At this time point, inositol was reintroduced into the cultures, and aliquots were collected at 0, 5, 15, and 30 min. Lipids were extracted and analyzed as described under “Experimental Procedures.” Each panel of this figure contains two graphs. The graph on the left indicates the neutral lipid composition of both strains shifted to inositol free media for 180 min. The graph on the right represents the neutral lipid composition of both strains when inositol was added back to cultures deprived of inositol. Panel A, wild type. Panel B, dga1Δlro1Δare1Δare2Δ. The data represent the average of three independent experiments. The magnitude of the error is similar to the data in Fig. 3, but error bars were omitted for clarity of the presentation. The lipids indicated are free fatty acids (open triangles), TAG (gray triangles), free sterols (solid circles), DAG (solid diamonds), and SE (open circles). ODU, optical density units.
In comparison to wild type, the dga1Δlro1Δare1Δare2Δ mutant, which does not synthesize TAG or SE, contained 2–3-fold higher levels of free sterols under all the growth conditions tested (Fig. 8B). DAG and free fatty acid content were also higher under all three-growth conditions in the mutant compared with wild type, consistent with the metabolic block in the mutant preventing acylation of DAG to form TAG. After the shift to I−C− medium at 30 °C, the DAG content in the dga1Δlro1Δare1Δare2Δ mutant declined by 50% within the first 60 min and remained constant at this lower level over the remaining time course. A decrease in the content of free sterols was also observed starting at about 60 min after the shift to I−C− at 30 °C, and a gradual decline in free fatty acid content occurred over the entire 180 min (Fig. 8B). However, after the shift to I−C− medium at 37 °C, the levels of free sterols, free fatty acids, and DAG remained essentially unchanged (Fig. 8B). When the dga1Δlro1Δare1Δare2Δ mutant was grown in I+C+ medium at 37 °C, initial levels of free sterols were similar to those observed in I+C− medium at 30 or 37 °C (Fig. 8B, time 0 before the media shift). However, in contrast to the two other growth conditions, upon shift to I−C+ medium at 37 °C, a continuous rise in free sterol content was observed in the mutant strain together with a decrease in free fatty acids.
The addition of inositol to cultures of the dga1Δlro1Δare1Δare2Δ mutant after 180 min growth in I−C−at 30 °C or in I−C+ medium at 37 °C had little impact on the content of any of the neutral lipids (Fig. 8B). However, at 37 °C in I−C− medium, free sterols in the dga1Δlro1Δare1Δare2Δ mutant declined slightly in response to inositol addition, and free fatty acid content doubled when inositol was reintroduced (Fig. 8B).
DISCUSSION
The Ino− phenotype is classically associated with a deficiency in expression of INO1, the structural gene for the inositol-3-phosphate synthase (23, 39). However, we report that the dga1Δlro1Δare1Δare2Δ mutant, which totally lacks lipid droplets (4, 16), exhibited no significant defect in INO1 expression at 37 °C in I−C− medium compared with wild type. Nevertheless, its ability to grow on agar plates under these conditions was greatly reduced (Fig. 2). Immediately after a shift of actively growing cells to I−C+ medium at 37 °C, the mutant actually exhibited higher INO1 expression than the wild type strain (Fig. 5), but within a few hours after the shift to I−C+ medium at 37 °C, INO1 expression was similar to that in the wild type strain. The dga1Δlro1Δare1Δare2Δ strain is not the first mutant reported to exhibit an Ino− phenotype and yet to be able to express INO1. The sac1Δ mutant (40) and mutants defective in the protein kinase C (PKC) stress response pathway also have Ino− phenotypes and yet express INO1 (30). Moreover, PKC signaling is triggered both by growth in the absence of inositol and by growth at high temperature (30, 41, 42). The Ino− phenotypes of mutants defective in the PKC pathway, like that of the dga1Δlro1Δare1Δare2Δ mutant, are strengthened in the presence of choline at 37 °C (30). Growth in the absence of inositol activates a number of stress response pathways in addition to PKC (43). The phenotype of the dga1Δlro1Δare1Δare2Δ strain suggests that the absence of storage lipid synthesis adds to the stress-associated with growth in I−C+ medium at 37 °C. The fact that the mutant devotes a higher proportion of newly synthesized PI to sphingolipid synthesis than the wild type strain under these conditions is consistent with this hypothesis, as sphingolipids have been implicated in several stress response pathways including PKC signaling (44).
However, the mutant also displayed alterations in phospholipid biosynthesis including reduced synthesis of PI even when grown continuously in I+C+ medium (Fig. 6), a growth condition under which inositol was completely derived from the medium. Labeling of PA, CDP-DAG, and PS were also lower in the mutant in comparison to wild type in the presence of inositol, suggesting that the mutant synthesizes PA, the immediate precursor of CDP-DAG, PI, and PS at a reduced rate. Because the INO1 gene was completely repressed in the mutant as well as in the wild type strain in medium containing inositol (Fig. 5), the decrease in PI synthesis in the mutant under these conditions (Fig. 6) is clearly independent from its ability to derepress the INO1 gene or its gene product, inositol-3-phosphate synthase. Moreover, in the mutant, in comparison to wild type, newly synthesized PI was also utilized to a greater extent in the synthesis of inositol-containing sphingolipids, especially when it was growing in the absence of inositol, a topic that will be discussed below. Based on these observations, we conclude that the storage lipid synthesis actively influences the metabolism of membrane lipids, both phospholipids and sphingolipids, in logarithmically growing cells.
Lack of TAG Hydrolysis in Logarithmically Growing Cells Attenuates the Burst in PI Synthesis in Response to Inositol Reintroduction
In studies on stationary-phase cells reentering active growth, Taylor and Parks in 1979 (13) proposed that fatty acids derived from TAG are utilized for the synthesis of phospholipids upon resumption of growth. Most recently, the lipid hydrolases that catalyze TAG degradation were identified (10, 12, 32, 33). In this study we report that both the dga1Δlro1Δare1Δare2Δ mutant, which lacks the ability to produce TAG (4, 16) and the tgl3Δtgl4Δtgl5Δ mutant, which is unable to mobilize TAG (10, 12, 33), are compromised in rebuilding PI content to the levels seen in wild type cells immediately after the addition of inositol. Consistent with these results, TAG is consumed in proliferating wild type cells upon inositol reintroduction coincident with the rebuilding of PI content. We propose a new metabolic role for TAG as donor of fatty acids for the synthesis of PI in logarithmically growing yeast cells, particularly when inositol is reintroduced to cultures previously lacking this lipid precursor.
However, the dga1Δlro1Δare1Δare2Δ strain also exhibits impaired growth in medium lacking inositol. This growth defect is not simply due to the inability of the dga1Δlro1Δare1Δare2Δ mutant to mobilize TAG for PI synthesis, as the tgl3Δtgl4Δtgl5Δ strain is able to sustain growth in I− medium at 37 °C in the presence of choline. As we discuss below, the ability to synthesize TAG was shown to be protective under conditions of impaired membrane trafficking and declining phospholipid synthesis (15). In addition, the reduced recovery of PI content immediately after inositol reintroduction displayed by both mutants in comparison to wild type suggests that a pool of fatty acids derived from TAG degradation may be used for rapid restoration of PI after inositol reintroduction. The data presented here also confirm the combined contribution of de novo fatty acid synthesis and PC turnover for the burst of PI content after inositol reintroduction, as previously reported (19). Thus, we conclude that there are at least three different metabolic pathways capable of providing fatty acids for the dramatic increase in PI upon inositol reintroduction in wild type cells.
The Absence of Lipid Droplets Affects Synthesis of Both PI and Inositol-containing Sphingolipids
The Ino− phenotype of the dga1Δlro1Δare1Δare2Δ strain is clearly not due simply to its inability to mobilize TAG, and thus, the inability to store fatty acids in TAG and SE must contribute to its phenotype. Despite the fact that the dga1Δlro1Δare1Δare2Δ mutant is not able to store fatty acids in either TAG or SE, it is viable under the culture conditions employed in the studies by Oelkers et al. (4) and Sandager et al. (16). These two studies showed that storage lipids are not essential to sustain active logarithmic growth in yeast. However, TAG hydrolysis does play an important role in the recovery of cells from stationary phase into active growth (12, 13). Moreover, active TAG biosynthesis was shown to be protective under conditions of secretory stress (15). The sec13-1 strain, which has a defect in COPII vesicle trafficking from the ER when shifted to its restrictive temperature of 37 °C, experienced a dramatic decrease in phospholipid synthesis relative to the wild type control, whereas synthesis of TAG increased. Thus, it appears that when membrane trafficking pathways are compromised, synthesis of phospholipids required for membrane proliferation declines in a fashion that is coordinated with the slowing of membrane trafficking. Under these conditions, excess fatty acids are channeled into TAG (15). Moreover, when the genes encoding the DAG acyltransferases, Dga1p and Lro1p, were deleted in the sec13-1 mutant, the restrictive temperature of the sec13-1 dga1Δ lro1Δ strain decreased by two degrees compared with the parental strain (sec13-1 DGA1 LRO1) in media lacking inositol (15). Thus, the increased synthesis of TAG appears to serve under these conditions to absorb the excess fatty acids not being used in membrane proliferation, providing a degree of protection under conditions of secretory stress (15).
The above evidence suggests that both TAG synthesis and breakdown are interdependent with ongoing membrane lipid synthesis. In agreement with these reports, we found that the lack of storage lipids in the dga1Δlro1Δare1Δare2Δ mutant has a profound impact on the synthesis of membrane lipids. The inability of the mutant to synthesize TAG and SE affects the synthesis of all the phospholipids derived directly from PA (Fig. 6), and this effect is not specific to PI synthesis alone. These results indicate that the storage lipids in lipid droplets are required to maintain normal rates of phospholipid synthesis in proliferating cells. Furthermore, the dga1Δlro1Δare1Δare2Δ mutant utilizes more of its newly synthesized PI in the formation of inositol-containing sphingolipids than the wild type strain. In contrast, the tgl3Δtgl4Δtgl5Δ mutant was reported to exhibit decreased formation of sphingolipids in comparison to wild type (45).
Sphingolipids together with sterols are highly enriched in the plasma membrane (46–49), where they are believed to form microdomains within a phospholipid environment, referred to as lipid rafts (50, 51). Thus, the maintenance of sphingolipid synthesis in the dga1Δlro1Δare1Δare2Δ mutant despite lower PI synthesis may be correlated with the elevated synthesis of free sterols seen in the mutant under all growth conditions examined (Fig. 8). It has also been proposed that lipid droplets are in part involved in the transport of free sterols from the ER to the plasma membrane in wild type cells (52). Because this mode of transport is not possible in the dga1Δlro1Δare1Δare2Δ mutant, the delivery of free sterols must occur by routes such as membrane transport or non-vesicular transport pathways. There is also evidence indicating a role for sterols and sphingolipids in the generation of lipid gradients along the secretory pathway (53). Thus, the active production of complex sphingolipids in the dga1Δlro1Δare1Δare2Δ mutant in the absence of lipid droplets might also be required to facilitate the transport of the excess free sterols produced in the mutant from the ER to the plasma membrane by means of the membrane transport pathways.
In summary, we have found that the dga1Δlro1Δare1Δare2Δ strain is not able to sustain growth in the absence of inositol at 37 °C. This is not due to a defect in INO1 expression but may be due in part to the absence of a pool of storage lipids from which to draw fatty acids as precursors to sustain optimal synthesis of PI and other membrane phospholipids in logarithmically growing yeast cells. The need for a higher proportion of newly synthesized PI in producing sphingolipids may also be a factor contributing to the growth defect experienced by the mutant in the absence of inositol. Together, these results suggest that synthesis of storage lipids and membrane-forming lipids, in particular PI and inositol-containing sphingolipids, are interdependent and operate in a coordinated fashion to maintain lipid homeostasis in actively growing yeast cells.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-19629 (to S. A. H.). This work was also supported by grants from the Austrian Science Fund FWF (Project Lipotox F3005) and the Austrian Federal Ministry for Science and Research (Project GOLD (Genomics of Lipid-associated Disorders)) in the framework of the Austrian Genomics Program (GEN-AU) (to S. D. K.).
M. Villa-Garcia, M. S. Choi, F. I. Hinz, M. L. Gaspar, S. A. Jesch, and S. A. Henry, submitted for publication.
H. F. Hofbauer, unpublished observation.
- TAG
- triacylglycerol(s)
- PI
- phosphatidylinositol
- PA
- phosphatidic acid
- ER
- endoplasmic reticulum
- CDP-DAG
- CDP-diacylglycerol
- DAG
- diacylglycerol
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- SE
- steryl ester(s)
- SL
- total sphingolipids
- IPC
- inositolphosphorylceramide
- MIPC
- mannosyl-inositol-phosphorylceramide
- M(IP)2C
- mannosyl-diinositol-phosphorylceramide
- I
- inositol
- C
- choline.
REFERENCES
- 1. Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 2. Goodman J. M. (2008) J. Biol. Chem. 283, 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murphy S., Martin S., Parton R. G. (2009) Biochim. Biophys. Acta. 1791, 441–447 [DOI] [PubMed] [Google Scholar]
- 4. Oelkers P., Cromley D., Padamsee M., Billheimer J. T., Sturley S. L. (2002) J. Biol. Chem. 277, 8877–8881 [DOI] [PubMed] [Google Scholar]
- 5. Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., Sturley S. L. (2000) J. Biol. Chem. 275, 15609–15612 [DOI] [PubMed] [Google Scholar]
- 6. Sorger D., Daum G. (2002) J. Bacteriol. 184, 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jensen-Pergakes K., Guo Z., Giattina M., Sturley S. L., Bard M. (2001) J. Bacteriol. 183, 4950–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sandager L., Dahlqvist A., Banaś A., Ståhl U., Lenman M., Gustavsson M., Stymne S. (2000) Biochem. Soc. Trans. 28, 700–702 [PubMed] [Google Scholar]
- 9. Müllner H., Daum G. (2004) Acta Biochim. Pol. 51, 323–347 [PubMed] [Google Scholar]
- 10. Athenstaedt K., Daum G. (2005) J. Biol. Chem. 280, 37301–37309 [DOI] [PubMed] [Google Scholar]
- 11. Kohlwein S. D. (2010) J. Biol. Chem. 285, 15663–15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., Kohlwein S. D. (2009) Mol. Cell 33, 53–63 [DOI] [PubMed] [Google Scholar]
- 13. Taylor F. R., Parks L. W. (1979) Biochim. Biophys. Acta 575, 204–214 [DOI] [PubMed] [Google Scholar]
- 14. Rajakumari S., Grillitsch K., Daum G. (2008) Prog. Lipid Res. 47, 157–171 [DOI] [PubMed] [Google Scholar]
- 15. Gaspar M. L., Jesch S. A., Viswanatha R., Antosh A. L., Brown W. J., Kohlwein S. D., Henry S. A. (2008) J. Biol. Chem. 283, 25735–25751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sandager L., Gustavsson M. H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. (2002) J. Biol. Chem. 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- 17. Garbarino J., Padamsee M., Wilcox L., Oelkers P. M., D'Ambrosio D., Ruggles K. V., Ramsey N., Jabado O., Turkish A., Sturley S. L. (2009) J. Biol. Chem. 284, 30994–31005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., Natter K., Kohlwein S. D. (2009) J. Biol. Chem. 284, 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaspar M. L., Aregullin M. A., Jesch S. A., Henry S. A. (2006) J. Biol. Chem. 281, 22773–22785 [DOI] [PubMed] [Google Scholar]
- 20. Kelley M. J., Bailis A. M., Henry S. A., Carman G. M. (1988) J. Biol. Chem. 263, 18078–18085 [PubMed] [Google Scholar]
- 21. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 22. Jesch S. A., Zhao X., Wells M. T., Henry S. A. (2005) J. Biol. Chem. 280, 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carman G. M., Henry S. A. (1999) Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 24. Atkinson K., Fogel S., Henry S. A. (1980) J. Biol. Chem. 255, 6653–6661 [PubMed] [Google Scholar]
- 25. Steiner M. R., Lester R. L. (1972) Biochim. Biophys. Acta. 260, 222–243 [DOI] [PubMed] [Google Scholar]
- 26. Stock S. D., Hama H., Radding J. A., Young D. A., Takemoto J. Y. (2000) Antimicrob. Agents Chemother. 44, 1174–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 28. Weerheim A. M., Kolb A. M., Sturk A., Nieuwland R. (2002) Anal. Biochem. 302, 191–198 [DOI] [PubMed] [Google Scholar]
- 29. Christie W. (2003) in Lipid Analysis: Isolation, Separation, Identification, and Structural Analysis of Lipids (Christie W. ed.) pp. 105–112, The Oily Press, Bridgwater, UK [Google Scholar]
- 30. Nunez L. R., Jesch S. A., Gaspar M. L., Almaguer C., Villa-Garcia M., Ruiz-Noriega M., Patton-Vogt J., Henry S. A. (2008) J. Biol. Chem. 283, 34204–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 32. Athenstaedt K., Daum G. (2003) J. Biol. Chem. 278, 23317–23323 [DOI] [PubMed] [Google Scholar]
- 33. Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S. D. (2006) J. Biol. Chem. 281, 491–500 [DOI] [PubMed] [Google Scholar]
- 34. Dowd S. R., Bier M. E., Patton-Vogt J. L. (2001) J. Biol. Chem. 276, 3756–3763 [DOI] [PubMed] [Google Scholar]
- 35. Zaccheo O., Dinsdale D., Meacock P. A., Glynn P. (2004) J. Biol. Chem. 279, 24024–24033 [DOI] [PubMed] [Google Scholar]
- 36. Nomura S., Horiuchi T., Omura S., Hata T. (1972) J. Biochem. 71, 783–796 [DOI] [PubMed] [Google Scholar]
- 37. Hirsch J. P., Henry S. A. (1986) Mol. Cell. Biol. 6, 3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gaynor P. M., Gill T., Toutenhoofd S., Summers E. F., McGraw P., Homann M. J., Henry S. A., Carman G. M. (1991) Biochim. Biophys. Acta 1090, 326–332 [DOI] [PubMed] [Google Scholar]
- 39. Greenberg M. L., Lopes J. M. (1996) Microbiol. Rev. 60, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rivas M. P., Kearns B. G., Xie Z., Guo S., Sekar M. C., Hosaka K., Kagiwada S., York J. D., Bankaitis V. A. (1999) Mol. Biol. Cell 10, 2235–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levin D. E. (2005) Microbiol. Mol. Biol. Rev. 69, 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levin D. E., Bartlett-Heubusch E. (1992) J. Cell Biol. 116, 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jesch S. A., Liu P., Zhao X., Wells M. T., Henry S. A. (2006) J. Biol. Chem. 281, 24070–24083 [DOI] [PubMed] [Google Scholar]
- 44. Dickson R. C. (2008) J. Lipid Res. 49, 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajakumari S., Rajasekharan R., Daum G. (2010) Biochim. Biophys. Acta 1801, 1314–1322 [DOI] [PubMed] [Google Scholar]
- 46. Hechtberger P., Zinser E., Saf R., Hummel K., Paltauf F., Daum G. (1994) Eur. J. Biochem. 225, 641–649 [DOI] [PubMed] [Google Scholar]
- 47. Lange Y., Swaisgood M. H., Ramos B. V., Steck T. L. (1989) J. Biol. Chem. 264, 3786–3793 [PubMed] [Google Scholar]
- 48. Patton J. L., Lester R. L. (1991) J. Bacteriol. 173, 3101–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schneiter R., Brügger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G., Paltauf F., Wieland F. T., Kohlwein S. D. (1999) J. Cell Biol. 146, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simons K., Ikonen E. (1997) Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 51. Simons K., van Meer G. (1988) Biochemistry 27, 6197–6202 [DOI] [PubMed] [Google Scholar]
- 52. Sorger D., Athenstaedt K., Hrastnik C., Daum G. (2004) J. Biol. Chem. 279, 31190–31196 [DOI] [PubMed] [Google Scholar]
- 53. Klemm R. W., Ejsing C. S., Surma M. A., Kaiser H. J., Gerl M. J., Sampaio J. L., de Robillard Q., Ferguson C., Proszynski T. J., Shevchenko A., Simons K. (2009) J. Cell Biol. 185, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]