Abstract
The traditional model for transcription sees active polymerases tracking along their templates. An alternative (controversial) model has active enzymes immobilized in “factories.” Recent evidence supports the idea that the DNA moves, not the polymerase, and points to alternative explanations of how regulatory motifs like enhancers and silencers work.
Key words: transcription factories, nuclear organization, fixed RNA polymerases, chromatin loops
Although the vital processes of replication and transcription that occur within eukaryotic nuclei depend upon stochastic interactions between individual molecules, the relevant molecular machines and their templates are nonetheless highly ordered.1–3 For example, replication occurs in sub-nuclear hot-spots or “factories,”4,5 and we have also suggested that transcription does so too.6 We define such a “transcription factory‘ as a cluster of at least two RNAPs active on different templates (a typical nucleoplasmic factory in a HeLa cell contains ∼8 enzymes engaged on ∼8 templates).7 The raison d'être of all factories is the same: to enhance production by concentrating relevant machines, resources and expertise in one place. For example, HeLa nuclei contain a 1 µM pool of diffusing RNA polymerase II (RNAP II), but essentially all nascent RNA is made in nucleoplasmic factories where the concentration is 1,000-fold higher.7
This heterodox idea is controversial (reviewed in ref. 8) as it presupposes acceptance of some principles not found in our textbooks: (1) factories represent critical architectural motifs to which RNAPs and transcription factors (TFs) tether chromatin in loops, (2) active RNAPs are transiently immobilized in factories and work by reeling in their templates as they extrude their transcripts, (3) individual complexes housed in one factory carry out most (if not all) processes involved in producing a mature transcript (including RNA synthesis, processing and proofreading) and (4) different factories specialize in transcribing different sub-sets of genes.7 Here, we describe recent data supporting the idea that active polymerases are immobilized while they are active.9 Of course, movements are relative and the polymerase might be fixed to a factory, but both might be moving together through the nucleus.
Distinguishing Between Tracking and Fixed RNAPs
According to the traditional model, active RNAPs track like locomotives down their templates. As with so many received ideas, there seems to be little (if any) evidence supporting such tracking in vivo. In contrast, early experiments suggested that active polymerases were attached to the nuclear substructure, and so immobilized; most of a loop could be detached using nucleases without removing nascent RNA or transcribed templates.10 We also now know that fixed polymerases are powerful molecular motors able to reel in their templates in the required way, with many single-molecule analyses relying upon enzyme immobilization.11
We recently showed (albeit indirectly) that active RNAPs are immobile.9 For the experiment, we needed two genes that could be switched on rapidly—one to act as a reference point, while the other had to be long enough to provide sufficient spatial resolution. We stimulated human umbilical endothelial vein cells (HUVECs) by treating them with tumor necrosis factor α (TNFα); this cytokine signals through nuclear factor κB (NFκB) to activate and repress many genes and TNFAIP2 and SAMD4A—which both encode regulators of this cascade—are amongst the first to respond.12 The two lie ∼50 Mbp apart on the genetic map. TNFAIP2, a short 10 kbp gene, is turned on within ∼10 min and is then transcribed repeatedly over the next few hours. SAMD4A is 221 kbp, and although the pioneering polymerase also initiates within ∼10 min, it only terminates after another ∼75 min (as the gene is so long). We sought to monitor by chromosome conformation capture (3C; reviewed in ref. 13) how close together different parts of the two genes were at different times after stimulation.
If the conventional model for transcription applies, we would not expect the short gene to lie near enough to the long gene to give a 3C product at any time after stimulation. If, for whatever reason, the two happened to lie together (for example, before stimulation), then they would soon separate as the pioneering polymerase tracked down the long gene. But if both responding genes were transcribed by polymerases that were transiently immobilized in the same “NFκB-factory” that specialized in transcribing TNFα-responding genes, the short gene, which would repeatedly attach to (and detach from) the factory as it initiates (and terminates), should always lie close to just the part of the long gene being transcribed at that particular moment (Fig. 1). Then, we would not expect to see any 3C products before stimulation. But after 10 min (when both initiate), the short gene should lie next to the promoter of the long gene (but no other part). Then, as the polymerase reels in the long gene, introns 1, 2, 3, etc. should successively be brought into the factory to lie transiently next to the short gene. And after 85 min, when the pioneering polymerase is about to terminate, only the terminus should lie next to the short gene. 3C products appeared and disappeared exactly as predicted; moreover, they did so when three other short genes—two on different chromosomes from the long gene—were used as reference points.9
Figure 1.
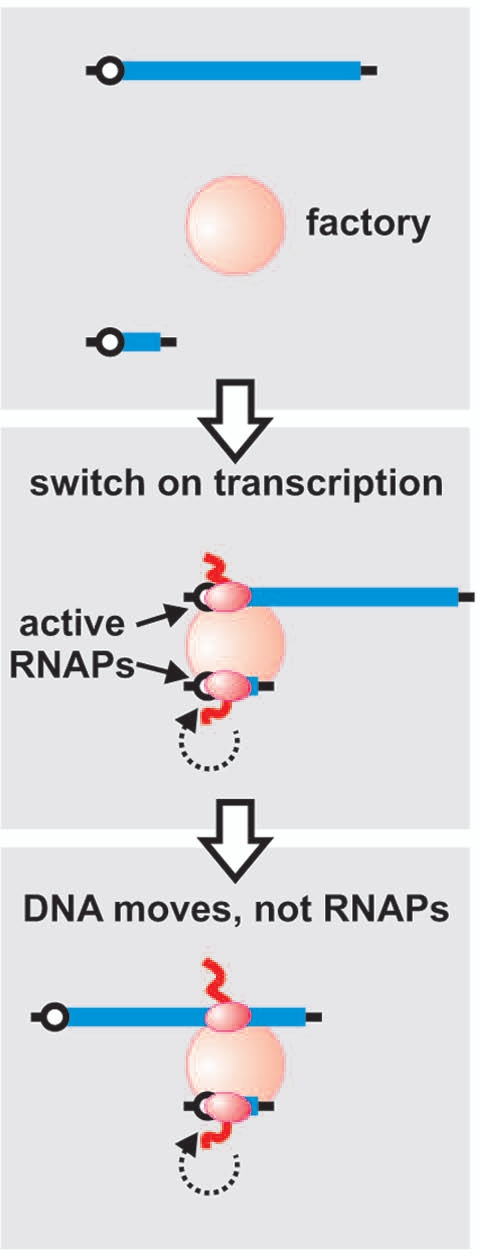
Cartoon depicting the relative movements of two genes after switching on their transcription. On adding TNFα, the two genes (blue) diffuse to a factory (pink) and collide with RNAPs (red) bound there; after initiation, the immobilized RNAPs reel in their templates as they extrude their transcripts (red wavy lines). The short gene is soon transcribed; it detaches and is then transcribed again repeatedly (dotted arrow). It takes much longer to transcribe the long gene. Initially (middle part), its promoter lies next to the short gene (proximity detected by 3C). When the pioneering polymerase has transcribed two-thirds into the long gene (bottom part), the transcribed region now lies next to the short gene. As a result, the short gene lies next to just that segment of the long gene that is being transcribed at that particular moment. This is the result obtained, indicating that the DNA moves (and not the polymerase).
These 3C experiments showed that just the transcribed parts of the long and short genes were together. Use of an independent method (i.e., RNA FISH with probes targeting intronic sequences) confirmed that the relevant nascent RNAs lay very close together at the appropriate times. “Super-resolution” microscopy also demonstrated that the pairs of nascent transcripts colocalized to the degree expected if they were randomly distributed within a 35-nm shell around an 87 nm factory—the known average dimensions of a nucleoplasmic factory.9,14 All these results are difficult to reconcile with the idea that RNAPs track. Consider, for example, some alternatives. If polymerases do track, what is the nature of the corral or “force field” that confines them within this shell? Even if the promoter and terminator of a gene are juxtaposed—and there is evidence for this in mammalian cells15—why should a pioneering polymerase transcribing two-thirds of the way into the gene now lie so close to a polymerase transcribing a different gene? In both cases, fixing the active polymerases in a factory provides a simple solution, with loops appearing and disappearing as polymerases initiate and terminate.
Specialized Transcription Factories
The results described above imply that these TNFα-responsive genes are being transcribed in dedicated “NFκB” factories; indeed, they all have NFκB bound to their promoters.9 There is now excellent evidence for the specialization of factories in such a manner, and the nucleolus provides the prototypic example. Simply put, it is a “mega-factory” where RNAP I transcribes ribosomal DNA to produce the ribosomal RNA that is then assembled into ribosomes.16 Active RNAP II and III are also each concentrated in distinct nucleoplasmic factories.17 Moreover, different RNAP II factories specialize in transcribing intron-less and intron-containing genes.18 In other examples, transcription units encoding factors involved in globin production (e.g., Hbb-b1, its locus control region or LCR, Eraf) are often co-transcribed in dedicated “EKLF-factories”,19 and genes regulated by estrogen receptor α (ERα) appear to co-associate20 (presumably in “ERα-factories”).
Polymerases Fixed in Factories: Some Implications
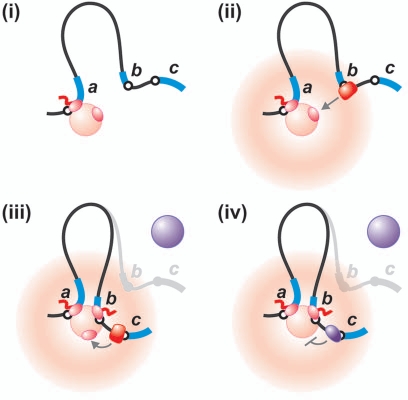
The model illustrated in Figure 1 has various implications, not only on how we perceive polymerases work, but also on the way related processes are arranged and executed. For example, it has been difficult to explain how regulatory motifs like enhancers, silencers, barriers and insulators all work. But if transcription only occurs in factories, it becomes immediately obvious that tethering a promoter more or less closely to a factory will determine (to a significant extent) how often that promoter will be transcribed; promoters tethered close to a factory (e.g., those in the “hot” halo in Fig. 2) are much more likely than others lying further away to diffuse (randomly) and collide with RNAPs concentrated in the factory. As a result, the position of a promoter in a loop relative to a factory is one critical determinant of initiation frequency. Then, we suggest that an enhancer acts by bringing its target promoter closer to the relevant factory containing the appropriate TFs—and it could do so if it first attached to a factory and was itself transcribed (Fig. 2). Conversely, transcription of a silencer element might tether its target promoter close to a factory containing the “wrong” kind of TFs. In both cases, the regulatory motifs are transcription units, and their activity depends upon them being transcribed (Fig. 2).7 Consistent with this, old studies showed that canonical enhancers/silencers were transcribed, and recent genome-wide ones confirm that most are and that they bear activation-related chromatin marks.21,22 We also now know that a large number of tightly-regulated genes have RNAPs on their promoters before they are “turned on”—and this ensures a prompt transcriptional response.23 For example, the promoter of the uPA gene is “poised” by attachment to a factory, looping the adjacent chromatin to organize the genome locally.24
Figure 2.
A parsimonious model for transcriptional regulation. (i) Gene a is being transcribed by a polymerase in a factory (pink) and, as a result, genes b and c are tethered close to the factory. (ii) Intuition suggests (and computer simulations support; see ref. 7) the idea that genes tethered close to the pink factory are more likely to be transcribed (i.e., those in the “hot” halo around the pink factory) especially if bound by the “right” TFs (in this case red). (iii) Gene b has attached to the factory and is now being transcribed; this has brought c into the “hot” zone (which makes it more likely to be transcribed). In other words, b acts as an enhancer of c. Another factory (purple) is also shown. (iv) The structure is the same as in (iii) but we are at a different stage during development. Now, different transcription factors have bound to c (purple), enhancing its affinity for a different transcription factory (also purple). Even though c is in the “hot” zone around the pink factory, it remains unlikely to be transcribed there. In this case, b has silenced c by distancing it from its favored (purple) factory.
We now also know that transcripts initiate not only from classical promoters, but from many other points on one or other strand.25 Therefore, we imagine that the average rate of production of any transcript (whether sense or anti-sense, genic or non-genic) will depend on how closely the template is tethered to a factory. Of course, other factors like the underlying DNA sequence, histone modifications and chromatin compaction will play important roles. Note also that where overlapping sense and anti-sense transcripts are seen, they must be produced sequentially, as complementary bases in a template cannot be transcribed simultaneously by either tracking or fixed polymerases.
Finally, trans-splicing—a regulated process that leads to the formation of chimeric transcripts encoded by distant genomic regions26,27—is another phenomenon that has been difficult to explain using the conventional model. Although rare in mammals, it is seen more frequently in the protein-coding transcripts of various metazoans. We expect the splicing machinery acting on two nascent transcripts in one factory to occasionally (mis-)splice one transcript to another and, as factories specialize, we would also predict that trans-splicing should mainly occur between transcripts generated from promoters that bind the same transcription factors.
Outstanding Questions
Obviously, fixing active polymerases begs many questions. For example, how many factories are “dedicated” to transcribing TNFα-responsive genes, do other signaling pathways adopt similar strategies and act through analogous specialized factories, how many types of such specialized factories might there be, and how rapidly can one be converted into another? Fortunately, the techniques for answering these questions are now at hand.
Acknowledgements
We thank the BBSRC and Wellcome Trust for support; A.P. is the Kemp Junior Research Fellow of Lincoln College, Oxford and P.R.C. holds the EP Abraham Chair of Cell Biology.
Abbreviations
- 3C
chromosome conformation capture
- HUVECs
human umbilical vein endothelial cells
- NFκB
nuclear factor κB
- RNAP
RNA polymerase
- TF
transcription factor
- TNFα
tumor necrosis factor α
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/14275
References
- 1.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: balancing stability and plasticity. Biochim Biophys Acta. 2008;1783:2061–2079. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papantonis A, Cook PR. Genome architecture and the role of transcription. Curr Opin Cell Biol. 2010;22:271–276. doi: 10.1016/j.ceb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hozák P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie PJ, Blow JJ. Clusters, factories and domains: The complex structure of S-phase comes into focus. Cell Cycle. 2010;9:3218–3226. doi: 10.4161/cc.9.16.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook PR. A model for all genomes: the role of transcription factories. J Mol Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 9.Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, et al. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol. 2010;8:100041. doi: 10.1371/journal.pbio.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson DA, McCready SJ, Cook PR. RNA is synthesized at the nuclear cage. Nature. 1981;292:552–555. doi: 10.1038/292552a0. [DOI] [PubMed] [Google Scholar]
- 11.Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu Rev Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by chromosome conformation capture. Curr Protoc Mol Biol. 2006;21:21. doi: 10.1002/0471142727.mb2111s74. [DOI] [PubMed] [Google Scholar]
- 14.Eskiw CH, Rapp A, Carter DR, Cook PR. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J Cell Sci. 2008;121:1999–2007. doi: 10.1242/jcs.027250. [DOI] [PubMed] [Google Scholar]
- 15.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci USA. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 17.Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raab JR, Kamakaka RT. Insulators and promoters: closer than we think. Nat Rev Genet. 2010;11:439–446. doi: 10.1038/nrg2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price DH. Poised polymerases: on your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Ferrai C, Xie SQ, Luraghi P, Munari D, Ramirez F, Branco MR, et al. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol. 2010;8:1000270. doi: 10.1371/journal.pbio.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 26.Douris V, Telford MJ, Averof M. Evidence for multiple independent origins of trans-splicing in Metazoa. Mol Biol Evol. 2010;27:684–693. doi: 10.1093/molbev/msp286. [DOI] [PubMed] [Google Scholar]
- 27.Gingeras TR. Implications of chimaeric non-colinear transcripts. Nature. 2009;461:206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]