Abstract
The pollen tube attractant peptide LUREs of Torenia fournieri are diffusible peptides that attract pollen tubes in vitro. Here, we report a method enabling the direct visualization of a LURE peptide without inhibiting its attraction activity by conjugating it with the Alexa Fluor 488 fluorescent dye. After purifying and refolding the recombinant LURE2 with a polyhistidine tag, its amino groups were targeted for conjugation with the Alexa Fluor dye. Labeling of LURE2 was confirmed by its fluorescence and mass spectrometry. In our in vitro assay using gelatin beads, Alexa Fluor 488-labeled LURE2 appeared to have the same activity as unlabeled LURE2. Using the labeled LURE2, the relationship between the spatiotemporal change of distribution and activity of LURE2 was examined. LURE2 attracted pollen tubes when embedded in gelatin beads, but hardly at all when in agarose beads. Direct visualization suggested that the significant difference between these conditions was the retention of LURE2 in the gelatin bead, which might delay diffusion of LURE2 from the bead. Direct visualization of LURE peptide may open the way to studying the spatiotemporal dynamics of LURE in pollen tube attraction.
Keywords: Alexa Fluor labeling, Attractants, Peptides, Pollen-tube guidance
Introduction
Plant sexual reproduction involves a complex pollen–pistil interaction between male and female cells. A pollen–pistil interaction is involved to ensure self-incompatibility, pollen tube guidance and direct gametophytic interaction after the arrival of the pollen tube at the embryo sac. Various molecules are thought to be involved in related intercellular communication. Among them, peptides (small proteins or polypeptides with <150 amino acids or <15 kDa in molecular weight; Silverstein et al. 2007) govern many processes in the pollen–pistil interaction (reviewed by Suzuki 2009, Higashiyama 2010). For example, SCR/SP11 is a peptide in the pollen coat of Brassica sp. that works as a male determinant of self-incompatibility (Schopfer et al. 1999, Takayama et al. 2000). SCR/SP11 is the ligand of the receptor kinase SRK expressed in the papilla of the female tissue (Stein et al. 1991, Takayama et al. 2001). LAT52 and LeSTIG1 of Solanum lycopersicum (tomato) pollen and stigma are peptides involved in pollen tube germination and growth (Muschietti et al. 1994, Tang et al. 2004). Chemocyanin and SCA are produced in the Lilium longiflorum stigma and style and are involved in pollen tube guidance, growth and adhesion (Jauh et al. 1997, Park et al. 2000, Kim et al. 2003, Dong et al. 2005). In the ovary, the LURE peptides of Torenia fournieri are secreted from the synergid cell and attract pollen tubes (Okuda et al. 2009). Zea mays EGG APPARATUS 4 (ZmES4) exclusively expressed in the embryo sac is thought to be involved in pollen tube discharge (Amien et al. 2010). Except for chemocyanin, most of these compounds are cysteine-rich peptides (CRPs; Higashiyama 2010). Knowledge of the dynamics of these peptides during cell–cell communication may provide helpful information regarding their function. However, the actual dynamics of these peptides during cell–cell communication remain largely unknown. The visualization of peptides would facilitate the study of intercellular signaling involving these peptides.
The spatiotemporal dynamics of attractant peptides, including diffusion, the concentration gradient and reception by target cells, are important for their function. Pollen tube guidance to the embryo sac is thought to be based on a concentration gradient of pollen tube attractants (Higashiyama and Hamamura 2008). In T. fournieri, the embryo sac protrudes from the ovule and the pollen tubes are attracted precisely to the micropylar end of the embryo sac directly, without the need to contact the surrounding female sporophytic cells (Higashiyama et al. 1998, Higashiyama et al. 2001). Defensin-like peptide LUREs have been identified as attractant peptides in the Torenia system (Okuda et al. 2009). Pollen tube attraction by LUREs can be performed in vitro using recombinant peptides expressed in Escherichia coli. If one could visualize LURE peptides, the relationship between their spatiotemporal dynamics and function might be investigated, including the role of a concentration gradient. The relationship between the attractant molecules and their concentration gradient in pollen tube guidance is controversial (Higashiyama and Inatsugi 2006). In axon guidance during neural network development, netrin proteins serve as attractant molecules (Dickson 2002). However, it remains to be elucidated whether a netrin concentration gradient actually provides spatial information during axon guidance (Dickson 2002, Kennedy et al. 2006, Mai et al. 2009). Visualization of LURE peptides may provide insights into the relationship between chemotropism and concentration gradients.
Various methods for visualizing peptides exist, including chemical labeling with fluorescent dyes and the introduction of peptide/protein tags or fluorescent proteins (Alcor et al. 2009). In this study, we examined the chemical labeling of LURE with low molecular mass florescent dyes, which would be expected to have less effect on the behavior and activity of LUREs.
Results
Labeling LURE2 with Alexa Fluor 488
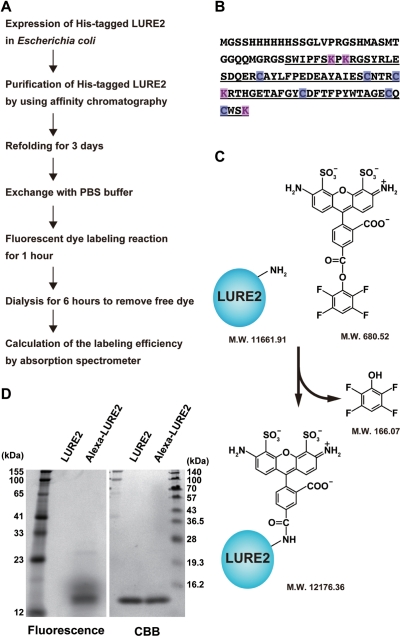
Fig. 1A–C shows a schematic of the method used for the fluorescent labeling of LURE peptide. Because the refolding step is key for the attraction activity of recombinant LURE peptides, the intramolecular disulfide bonds between six cysteine residues are thought to be important for this activity (Fig. 1B; Okuda et al. 2009). Thus, we used amine-reactive Alexa Fluor 488 dye [hydrolysis-resistant tetrafluorophenyl (TFP) ester; 680.52 Da, without triethylamine], although thiol group (cysteine)-reactive dyes can also be used for chemical labeling of proteins. Alexa Fluor 488 is a sulfonated, photostable green fluorescent dye (Panchuk-Voloshina et al. 1999). For conjugation with Alexa Fluor 488 TFP ester, we used LURE2 as a model for the chemical visualization of LURE peptides.
Fig. 1.
Labeling LURE2 peptide with Alexa Fluor 488. (A) Flow chart for Alexa 488 labeling of LURE2 peptide; see Materials and Methods. (B) Amino acid sequence of His-tagged LURE2. The LURE2 sequence following the predicted cleavage site is underlined. Lysine residues which have an amine group are labeled in magenta and cysteine residues are labeled in blue. (C) Schematic representation showing the labeling reaction of Alexa Fluor 488 TFP ester with LURE2 peptide. The molecular weights of LURE2 without the first methionine and Alexa Fluor 488 ester were calculated using the program Vector NTI (Invitrogen). (D) Purified LURE2 and Alexa–LURE2 were analyzed by SDS–PAGE. The fluorescence of peptides was visualized with a Typhoon 9400 scanner, followed by CBB staining for total peptides.
Our polyhistidine-tagged LURE2 has five primary amines that are potentially targeted by Alexa Fluor 488: an amino group at the N-terminus and the four amino groups of lysine residues in the LURE2 sequence following the predicted cleavage site (Fig. 1B). This polyhistidine-tagged LURE2 expressed in E. coli can attract pollen tubes in vitro (Okuda et al. 2009). Before labeling with Alexa Fluor 488, we investigated the molecular structure of the N-terminus of LURE2 expressed in E. coli by the Edman degradation method. The amino acid sequence of the N-terminus was GSSHH for a major population of LURE2, indicating that the methionine at the N-terminus was lacking, as frequently observed when the second amino acid of proteins in E. coli is glycine (Hirel et al. 1989). Another minor population of LURE2 contained the methionine at the N-terminus.
As shown in Fig. 1A, we purified and refolded LURE2. Then it was labeled with Alexa Fluor 488, and dialyzed to remove unbound Alexa dye. Fig. 1D shows SDS–PAGE analysis of LURE2 before and after labeling with Alexa Fluor 488. Unlabeled His-tagged LURE2 is estimated to be 11.8 kDa, but the apparent molecular mass was 14.2 kDa with Coomassie Brilliant Blue (CBB) staining (right lanes). Labeling LURE2 with Alexa Fluor 488 did not cause an apparent shift in LURE2 using 15% polyacrylamide gels. When examined using a fluorescence scanner, we detected Alexa Fluor 488 fluorescence only in the labeled LURE2 fraction (fluorescence observation, left lanes). To calculate the efficiency of LURE2 labeling by Alexa Fluor 488 dye, absorption at 280 nm for the LURE2 concentration and at 494 nm for the Alexa Fluor 488 concentration was measured using an absorption spectrometer. By applying each value to a formula (see Materials and Methods), each LURE2 molecule was estimated to be covalently bound to 1.08 ± 0.02 (mean ± SD) molecules of Alexa Fluor 488 (n = 3).
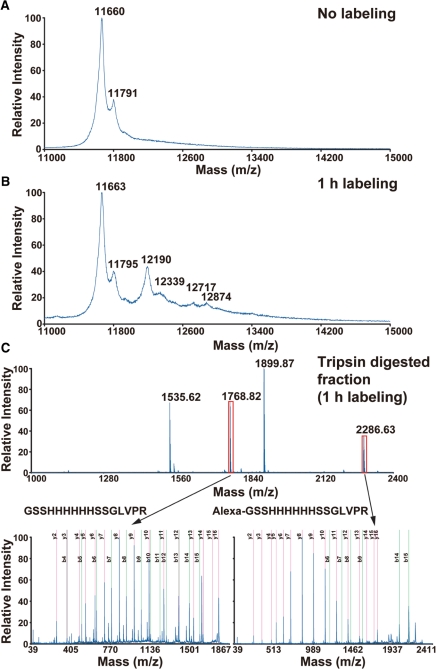
Next, the Alexa-labeled LURE2 fraction was examined using mass spectrometry (MS; Fig. 2). With matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), unlabeled LURE2 gave a major peak with an m/z of 11,660 and a minor peak with an m/z of 11,791, which corresponded to LURE2 without and with the first methionine, respectively (Fig. 2A). In contrast, Alexa-labeled LURE2 showed another four peaks (Fig. 2B). These appeared to be LURE2 molecules without and with the first methionine conjugated with a single (m/z = 12,190 and 12,339) or two Alexa Fluor 488 molecules (m/z = 12,717 and 12,874). Thus we could label the LURE2 peptide with at least one and two Alexa Fluor 488 molecules. We could not detect apparent peaks for LURE2 conjugated with more than three Alexa Fluor molecules, although conjugation of a sulfonated molecule such as Alexa Fluor might decrease the signal intensity of MS (Keough et al. 1999).
Fig. 2.
MALDI-TOF MS analysis of Alexa–LURE2. Purified LURE2 before labeling (A) and the dialyzed reaction mixture after labeling for 1 h (B) were analyzed by MS. The peaks at m/z 11,660, 11,791 (A) and 11,663, 11,795 (B) corresponded to unlabeled LURE2 without or with the first mehionine, that at m/z 12,190, 12,339 (B) to LURE2 labeled with one molecule of Alexa Fluor 488 without or with the first mehionine, and that at m/z 12,719, 12,894 (B) to LURE2 labeled with two molecules of Alexa Fluor 488 without or with the first mehionine. (C) The trypsin-digested Alexa Fluor 488 labeling reaction mixture was analyzed by MALDI-TOF MS (upper spectrum). The peptides (GSSHHHHHHSSGLVPR) corresponding to the N-terminus of unlabeled LURE2 (m/z 1,768.82) or LURE2 labeled with one molecule of Alexa Fluor 488 (m/z 2,286.63) were further subjected to MS/MS analysis (lower spectra).
When Alexa Fluor 488-labeled LURE2 was digested by trypsin, the N-terminal fragment conjugated with Alexa Fluor 488 was found (Fig. 2C). Tandem MS (MS/MS) analysis showed that the amino acid sequence of both labeled and unlabeled fragments was GSSHHHHHHSSGLVPR. As shown in the MS/MS data in Fig. 2C, b-ions containing the N-terminal amino acids were scarcely detected in the fragment conjugated with Alexa Fluor 488. This indicated that conjugation of the Alexa Fluor dye decreased the signal intensities of b-ions as discussed above. In contrast, y-ions lacking N-terminal amino acids were detected in both peptide fragments similarly. Thus we estimated the ratio of Alexa Fluor labeling of the N-terminal amino group by comparing signal intensities of the y8-ion (HSSGLVPR), which gave the highest signal intensity and signal/noise ratio. The signal intensity of the y8-ion from the labeled N-terminal fragment contained in the peak fraction was 20,471, while that from the unlabeled N-terminal fragment was 14,557. The labeling ratio of the N-teminus of LURE2 was estimated to be 60%. At least 60% of the LURE2 was estimated to be labeled with Alexa Fluor 488. Hereafter, we call the fraction of Alexa Fluor 488-labeled LURE2 after dialysis ‘Alexa–LURE2’. Extending the labeling time from 1 to 2 h did not increase the labeling efficiency, but caused aggregation of the LURE2 peptides (data not shown).
Evaluation of the attraction activity of Alexa–LURE2
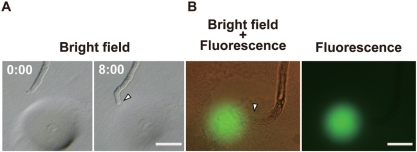
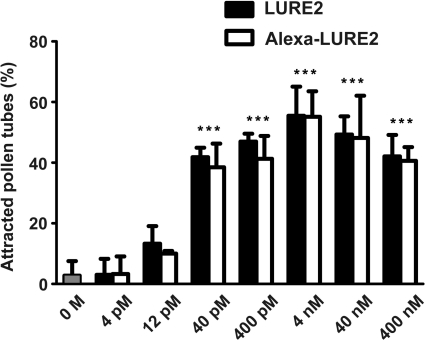
We examined whether Alexa–LURE2 could attract pollen tubes. Alexa–LURE2 was embedded in gelatin microbeads approximately 40 μm in diameter, as described by Okuda et al. (2009), at final concentrations of 4 and 400 nM. When a bead was put in front of pollen tubes using a micromanipulator, the tubes were observed to grow towards the bead (Fig. 3). We could not find any difference in the behavior of the pollen tubes between unlabeled LURE2 and Alexa–LURE2. Using fluorescence microscopy, Alexa–LURE2 molecules were visualized in the bead and diffusing from the bead (Fig. 3). Moreover, Alexa488–LURE2 showed the same activity as unlabeled LURE2 (Fig. 4). The activity of Alexa–LURE2 was highest at the same concentration as unlabeled LURE2 (4 nM); the frequency of pollen tube attraction was also the same (55.5 ± 9.6%, n = 4, for unlabeled LURE2; 55.2 ± 8.4%, n = 3, for Alexa–LURE2). Dilution of Alexa–LURE2 also resulted in no difference between labeled and unlabeled LURE2. Both Alexa–LURE2 and unlabeled LURE2 had activity above 40 pM (∼1,000 molecules in a 40 μm bead), but not below 12 pM (∼300 molecules in a 40 μm bead). Alexa–LURE2 appeared to contain at least 60% of labeled molecules, as estimated above. These results suggest that the labeling procedure did not impair the activity of LURE2.
Fig. 3.
Attraction of pollen tubes by Alexa–LURE2. A pollen tube growing through a cut style (competent pollen tube) was attracted by the Alexa–LURE2 in a gelatin bead (A, 4 nM; B, 400 nM). Numbers indicate time (minutes:seconds). Green fluorescence shows the Alexa–LURE2 remaining 12 min after positioning the gelatin bead (B). Arrowheads indicate tips of pollen tubes growing towards the beads. Scale bars: 30 μm.
Fig. 4.
Pollen tube attraction activity of LURE2 and Alexa–LURE2. The purified LURE2 (filled box) and Alexa–LURE2 (open box) peptides were used at different concentrations for the in vitro attraction assays. Data are the means and SD (n = 3 with >9 pollen tubes per replicate). Statistically significant differences were determined by one-way ANOVA with Bonferroni post-hoc test [n = 3, ***P < 0.0001 compared with 0 M condition (a buffer containing no LURE2 peptide)].
Dynamics of LURE2 in the bead assay
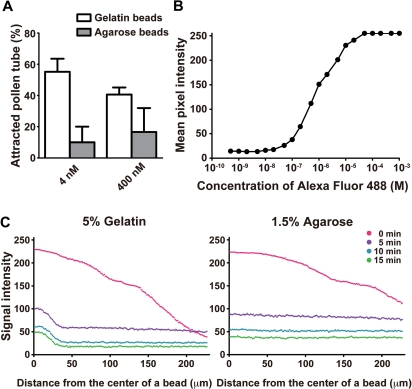
We next tried to examine the relationship between the spatiotemporal change of distribution and activity of LURE2 using Alexa–LURE2. When we originally developed the bead assay method (Okuda et al. 2009), we observed attraction using gelatin as the gellant, while attraction was unlikely to occur when we used agarose as the gellant. In this study, we confirmed this observation (Fig. 5A): agarose beads containing Alexa–LURE2 scarcely attracted pollen tubes under two different conditions (4 and 400 nM). Comparison of the spatiotemporal dynamics of Alexa–LURE2 in these two types of bead might identify the conditions necessary for pollen tube attraction in vitro by LURE.
Fig. 5.
Spatiotemporal change of distribution of Alexa-LURE2 in different conditions using gelatin and agarose beads. (A) Alexa–LURE2 peptides in gelatin beads (open box) and agarose beads (filled box) were used for the in vitro attraction assays. Data are the means and SD (n = 3 with >8 pollen tubes per replicate). (B) The fluorescence intensities of Alexa Fluor 488 hydrazide were measured in a hemocytometer for different concentrations of Alexa Fluor 488 hydrazide. The mean fluorescence intensity per pixel was plotted against the concentration of Alexa Fluor 488 hydrazide. (C) The fluorescence intensities of Alexa–LURE2 in a 5% gelatin bead (left graph) and a 1.5% agarose bead (right graph) were measured at different time points. A 40 nM concentration of Alexa–LURE2 was used. The fluorescence intensity of the lines from the center of a bead was plotted.
To analyze the spatiotemporal dynamics of Alexa–LURE2, we evaluated the quantitative capabilities of our microscope system (Fig. 5B). We measured the fluorescence intensities of various concentration of Alexa Fluor 488 using a microscopic counting chamber. In the high-sensitivity condition of our system using an electron-multiplying charge coupled device (EM-CCD) camera, linearity between the concentration of Alexa–LURE2 and the fluorescence intensity was confirmed for relative fluorescence intensities from 40 to 230. We also confirmed that Alexa Fluor 488 dye was not bleached much under our observation conditions.
We then studied the spatiotemporal change in Alexa–LURE2 in our bead assay (Fig. 5C). Alexa–LURE2 was observed to diffuse rapidly from both 5% gelatin and 1.5% agarose beads (diameter, 40 μm; Alexa–LURE2 concentration, 40 nM). A gradient of fluorescence intensity was observed over 200 μm under both conditions immediately after putting the beads on the medium (time 0 min). At 5 min after adding the beads, the apparent gradient of fluorescence intensity disappeared for both gelatin and agarose beads. However, a significant difference was observed within the beads; some Alexa–LURE2 molecules remained in the gelatin beads without diffusing (∼20 μm from the center of beads), while no such remaining fluorescence signal in the beads was observed with agarose beads. When we used 5% gelatin (Fig. 5C), the rate of decrease of Alexa–LURE2 from the bead was estimated to be 97.2% from 0 to 5 min, 63.5% from 5 to 10 min, and 29.4% from 10 to 15 min (calculated from Fig. 5B), suggesting that Alexa–LURE2 continued to diffuse from the beads at these rates.
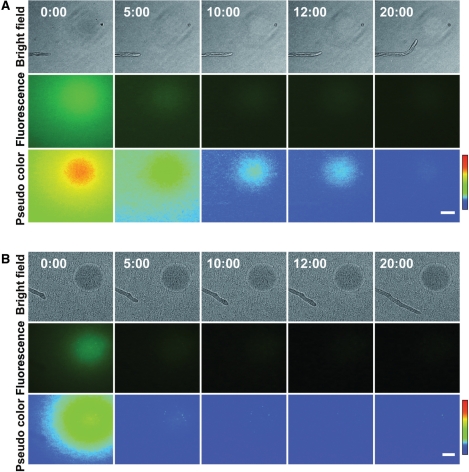
Next, the spatiotemporal change in Alexa–LURE2 was examined in the presence of pollen tubes. Fig. 6 shows a typical example of pollen tube attraction with Alexa–LURE2. The bead was made of 5% gelatin and contained 400 nM Alexa–LURE2. As seen in Fig. 5C, the Alexa–LURE2 diffused rapidly immediately after placing the bead and produced a fluorescence gradient across the tip of the pollen tube (Fig. 6, 0 min). The apparent gradient of fluorescence from the bead disappeared by 10 min. At 12 min after placing the bead, the pollen tube began to grow towards the bead. The growth rate of the pollen tube, 1.67 μm min−1, did not change during the in vitro assay, indicating that only the growth direction changed. As shown in Fig. 6, pollen tubes grew towards the bead even when no fluorescence gradient spreading from the bead was obvious. However, Alexa–LURE2 was still observed within the bead, although the amount decreased gradually, indicating that Alexa–LURE2 continued to diffuse from the gelatin bead while the pollen tube changed its direction of growth and grew towards the bead.
Fig. 6.
Behavior of pollen tubes and spatiotemporal change of distribution of Alexa-LURE2 during in vitro assay using a gelatin bead (A) and an agarose bead (B). The in vitro attraction assay was performed using 400 nM Alexa–LURE2. Pseudo-colored 8-bit images correspond to the fluorescence intensities of Alexa–LURE2. Red represents the highest level, corresponding to the fluorescence intensity of 78 (arbitary unit, see color scale). Numbers indicate time (minutes:seconds). Scale bars: 50 μm.
Discussion
Visualization of active LURE2 by chemical labeling with Alexa Fluor 488
In this study, we fluorescently labeled the attractant peptide LURE2, which showed the same activity as unlabeled LURE2. The attraction activity was confirmed in an in vitro attraction assay. For the chemical labeling of peptides, the thiol groups of cysteines, amino groups and non-natural amino acids are generally used to target a complementary reactive group appended to the dye (Ferreon et al. 2010). We used the amine-reactive TFP ester of Alexa Fluor, which involved a simple procedure and was unlikely to affect the intramolecular disulfide bonds between the six cysteines of LURE2; the activity of LURE2 depends on the refolding of the recombinant peptide (Okuda et al. 2009). We used the Alexa Fluor dye because it is photostable and emits bright fluorescence. Dyes such as Cy and TAMRA could be used similarly. Alternatively, the fusion of peptide/protein tags is possible (Chen and Ting 2005), including Halo tag, SNAP tag, Lumio tag and ACP tag. Compared with the fusion of these tags, direct chemical labeling causes less change in the molecular mass and consequently does not alter the behavior or molecular structure of the peptides, which are critical for their activity. With these tags, it is possible to label peptides in a strict one-to-one ratio. Thus, fusion tags can also be considered for labeling LURE, depending on the purpose of the experiments. Fluorescent proteins, including green fluorescent protein (GFP), are much bigger than peptides, which might affect the behavior and activity of the peptides.
While at least 60% of the LURE2 was estimated to be labeled with Alexa Fluor 488 and the labeled molecules appeared to have the ability to attract pollen tubes, the Alexa–LURE2 fraction still contains various molecules, as shown by MS (Fig. 2), including unlabeled LURE2, LURE2 with different numbers of Alexa Fluor molecules and possibly LURE2 in which different amino groups are labeled. Because LUREs tend to be absorbed by the column, the fractionation of the molecules using gel filtration, ion exchange or reverse phase chromatography is still being studied. Alternatively, the substitution of the four lysine residues by silent mutations may be a way to target the N-terminal amino group specifically. We showed that the N-terminal amino group of the recombinant LURE2 could be labeled by Alexa Fluor 488 (Fig. 2). Labeling of the N-terminal amino group is less likely to inhibit the activity of LUREs than that of lysine residues of LUREs because fusion of the polyhistidine tag of 34 amino acids did not impair the activity of pollen tube attraction (Okuda et al. 2009).
The visualization of various ligand peptides has been reported in animals. Studies include the chemical labeling of insulin with fluorescent dyes (Schlessinger et al. 1978). Epidermal growth factor (EGF) was visualized by labeling it with amine-reactive Cy3 or Cy5 dyes to investigate EGF receptor (EGFR) signaling at the surface of living cells by single-molecule imaging (Sako et al. 2000). Conversely, there are few reports on the visualization of peptides/ligands in plants, despite the increasing number of studies of plant peptide hormones (Matsubayashi 2011). An elicitor, 22 kDa fungal protein ethylene-inducing xylanase, has been visualized to examine its binding to its receptor (Ron and Avni 2004). LURE peptides can induce a quick response by pollen tube cells in vitro, and a small number of LURE molecules can attract pollen tubes. As shown in Fig. 4 and previously (Okuda et al. 2009), as few as 1,000 molecules in a bead can attract pollen tubes. Based on the fluorescent visualization in this study, direct imaging analyses of LURE peptides might serve as a model for plant studies.
Dynamics of LURE2 peptide during in vitro pollen tube attraction
This is the first reported study to visualize the spatiotemporal dynamics of LURE2 during an in vitro pollen tube attraction assay. Comparison of spatiotemporal change in the distribution of Alexa–LURE2 between gelatin and agarose bead assays provided insight into the factors responsible for pollen tube attraction. The former resulted in high pollen tube attraction, while the latter did not (Fig. 5A). A gradient of fluorescence intensity was observed immediately after adding both types of bead. We found that most of the LURE2 peptide molecules diffused rapidly from both types of bead (Figs. 5, 6). When we used gelatin beads, >95% of the LURE2 peptide diffused from the beads within 5 min. After 5 min, a small amount of Alexa–LURE2 was retained in the gelatin beads, but not in the agarose beads. In the gelatin beads, Alexa–LURE2 was suggested to diffuse from the bead with a longer duration, even after 5 min. This sustained flux of a small amount of LURE2 might resemble the secretion from the synergid cell and might be critical for pollen tube attraction.
The timing of the directional change of pollen tube growth ranged from a few minutes to >10 min. As shown in Fig. 6, pollen tube attraction still occurred after the gradient of fluorescence intensity disappeared. Pollen tubes might sense a very low LURE2 concentration gradient below the range of sensitivity of our imaging system, although we cannot determine exactly when the pollen tube senses the LURE2. Single-molecule imaging of the LURE peptide on the surface of pollen tubes would be useful for further study of the dynamics of LURE.
Single-molecule imaging is a powerful technique for studying the interaction between ligands and receptors. For example, chemotaxis of the slime mold Dictyostelium toward Cy3-labeled cAMP has been used to show gradient sensing by receptors (Ueda et al. 2001). Dimerization of EGFR during the binding of EGF peptide has also been demonstrated (Sako et al. 2000). These single-molecule analyses have been performed on cell surfaces using total internal reflection fluorescence (TIRF) microscopy. Recently, a highly inclined thin illumination method has been developed by modifying TIRF microscopy (Tokunaga et al. 2008, Konopka and Bednarek 2008), enabling us to perform single-molecule analysis in cells several microns thick. Single-molecule imaging analysis of LURE peptides might be possible for examining the relationship between the number and position of LURE molecules bound on the pollen tube surface and the directional change of the pollen tube. The development of an in vitro attraction method without agarose medium, as well as the search for LURE receptors, is now in progress for single-molecule imaging of LURE peptides.
Pollen tubes can follow the synergid cells of ovules moved by a micromanipulator (Higashiyama and Hamamura 2008). This indicates that the pollen tube can sense dynamic changes in the distribution of attractant molecules around it and respond precisely to the attraction signal. LURE1 and LURE2 are involved in the attraction signal and there are other possible attractant CRPs expressed in the synergid cells of T. fournieri (Okuda et al. 2009). Visualization of each LURE may provide insight into how a precise attraction signal is generated by mixture of different attractant peptides.
Materials and Methods
Plant materials
Torenia fournieri L. cv ‘Blue and White’ was grown on soil in a growth chamber at 25°C under long-day conditions (16 h light/8 h dark), as described by Higashiyama et al. (2006).
Purification and refolding of recombinant LURE2
The expression vector was cloned and LURE2 lacking the putative N-terminal peptide (70 amino acids) was purified as described previously (Okuda et al. 2009). A His-tagged recombinant LURE2 fusion peptide was expressed in E. coli strain BL21 CodonPlus (DE3)-RIL (Stratagene). Cells were grown to an OD600 of 0.6 at 37°C in 1.5 liters of LB medium containing 2% glucose and incubated for an additional 20 h at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation, resuspended in 20 mM Tris–HCl buffer (pH 8.0) containing 0.5 M NaCl, Complete Protease Inhibitor Cocktail (Roche), 1 mg ml−1 lysozyme (Wako) and 2.5 μg ml−1 DNase I (Sigma), and disrupted by incubation for 1 h at 4°C, followed by sonication on ice. After centrifugation (14,400 × g, 30 min, 4°C), the pellet was washed three times with 0.5% (v/v) Triton X-100, containing 1 mM EDTA, and Complete Protease Inhibitor Cocktail by sonication, as described above. Inclusion bodies were resuspended in 50 mM Tris–HCl (pH 8.0) containing 6 M guanidine-HCl, 0.5 M NaCl, 5 mM imidazole, 1 mM 2-mercaptoethanol, and Complete Protease Inhibitor Cocktail for 1 h at room temperature. After centrifugation (14,400 × g, 30 min, 4°C), the supernatant was filtered through a 0.22 μm filter (Millipore).
The supernatant was applied to a 1 ml HisTrap HP column (GE Healthcare) and washed with 10 ml of buffer. The bound protein was refolded by washing with a linear 6→0 M urea gradient of refolding buffer (50 mM Tris–HCl, pH 8.0, 5 mM imidazole, 1 mM 2-mercaptoethanol and Complete Protease Inhibitor Cocktail) at a flow rate of 0.5 ml min−1 for 13 h using the ÄKTA purifier system (GE Healthcare). His-tagged LURE2 was eluted with 50 mM Tris–HCl (pH 8.0) containing 0.5 M NaCl, 0.5 M imidazole, 1 mM 2-mercaptoethanol and Complete Protease Inhibitor Cocktail. After 2-fold concentration using an Amicon Ultra 3 K (Millipore), the peptide was dialyzed (Spectra/Por3 MWCO:3,500; Spectrum Laboratories) and refolded for 3 d at 4°C in 50 mM Tris–HCl (pH 8.0) containing 1 mM oxidized glutathione, 10 mM reduced glutathione (Wako), 10 mM l-arginine ethyl ester dihydrochloride (Sigma) and Complete Protease Inhibitor Cocktail.
Protein sequencing
The N-terminal amino acid sequence of purified recombinant LURE2 peptide was determined by Edman degradation using an ABI Procise 491 HT protein sequencer (Applied Biosystems).
Alexa Fluor 488 labeling and biochemical evaluation
LURE2 was labeled with Alexa Fluor 488 using an Alexa Fluor 488 protein labeling kit (Invitrogen), according to the manufacturer's instructions. After dialysis with phosphate-buffered saline (PBS; pH 7.4) for 6 h at 4°C, purified LURE2 peptide was diluted to 1 mg ml−1 in PBS. After adding 1 M sodium bicarbonate to 0.2 ml of LURE2 solution, the sample was allowed to react with one vial of reactive dye. The reaction mixture was stirred for 1 h at room temperature in the dark, and then ultrafiltered using a Microcon Ultracel YM-3 (Millipore) to remove unincorporated dye from the labeled peptide. Finally, the Alexa–LURE2 was dialyzed twice with 1 M Tris–HCl (pH 8.0) for 3 h at 4°C.
The labeling efficiency of Alexa–LURE2 was estimated using an absorption spectrometer according to the manufacturer's instructions as follows.
| (1) |
| (2) |
where 24,350 cm−1 M−1 is a calculated molar extinction coefficient of LURE2 at 280 nm, 0.11 is a correction factor for absorption of the dye at 280 nm, and 71,000 cm−1 M−1 is the approximate molar extinction coefficient of the Alexa Fluor 488 dye at 494 nm.
The purified LURE2 and Alexa–LURE2 were subjected to 15% SDS–PAGE, and visualized with a Typhoon 9400 scanner (GE Healthcare). Total proteins were stained with CBB.
Mass spectrometry
To desalt the peptides, 5 μg of purified LURE2 and Alexa LURE2 were absorbed to ZipTip C4 pipet tips (Millipore), and then washed with 0.1% trifluoroacetic acid (TFA) and eluted with 5 μl of 70% acetonitrile containing 0.1% TFA. The samples (0.5 μl) were deposited on a MALDI target plate, followed by the deposition of 0.5 μl of matrix (4 mg ml−1 α-cyano-4-hydroxycinnamic acid in 70% acetonitrile containing 0.1% TFA). MALDI-TOF MS was performed on a 4700 Proteomics Analyzer with version 3.6 software (ABSciex). MS spectra were acquired in linear negative ion mode. Bovine insulin (average molecular mass 5,733.51), equine cytochorme c (average molecular mass 12,360.96) and equine apomyoglobin (average molecular mass 16,951.27) were used as standard proteins (ProteoMass™ Protein MALDI-MS Calibration Kit, Sigma).
Alexa Fluor 488 labeling reaction mixture containing unlabeled and Alexa Fluor 488-labeled LURE2 was digested by trypsin in the presence of 0.01% Protease MAX surfactant (Promega). Resultant peptides were fractionated by a Dina nanoHPLC system with reverse phase chromatography (KYA Technologies) and each fraction was directly spotted on a MALDI plate with α-cyano-4-hydroxycinnamic acid by the DiNa Map system (KYA Technologies). MALDI-TOF MS and MS/MS were performed on a 4700 Proteomics Analyzer. MS spectra were acquired in reflector-positive ion mode and MS/MS spectra were acquired in positive ion mode with CID off. MS/MS data were analyzed by MASCOT.
In vitro attraction assays and observation
For in vitro assays using gelatin and agarose beads, 2 μl of peptide were mixed with 2 μl of 10% (w/v) gelatin (Nacalai) or 1.5% (w/v) agarose (Sigma) pre-melted at 50°C. After adding 150 μl of hydrated silicone oil, they were mixed by vortexing and cooled on ice to allow the formation of gelatin beads. Single beads (approximately 40 μm in diameter) were picked up using a glass needle and placed in front of the pollen tubes. We used the criteria described by Okuda et al. (2009) for judging ‘attracted’ and ‘non-attracted’ pollen tubes. The behavior of pollen tubes was recorded on the inverted platform of a fluorescence microscope (IX-71; Olympus) equipped with a time-lapse digital video system (Sigma Koki) and an EM-CCD camera (KP-DE500; Hitachi). Images were acquired with a ×10 objective lens (UPlanSApo, NA 0.40) or a ×20 objective lens (LUCPlanFLN, NA 0.45).
One-way analysis of variance (ANOVA) with a Bonferroni post-hoc test was performed using GraphPad Prism version 5.01 for Windows (GraphPad Software).
To examine the relationship between the amount of Alexa–LURE2 and the signal intensity in our camera system, 2 μl of Alexa Fluor 488 hydrazide (Invitrogen) at different concentrations were applied to a Thoma-type counting chamber (depth 0.10 mm; Hirschmann). The fluorescence intensities were measured and analyzed with the MBF ImageJ software. Pseudo-colored 8-bit images of the fluorescence intensities were obtained using the rainbow spectrum of Look-up Tables (LUT) of ImageJ.
Funding
This study was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan [Grant Nos. 18075004 and 19370017 to T.H.; No. 18GS0314-01 to N.S.]; the Japan Society for the Promotion of Science for Young Scientists [to S.O., A.M. and D.K.]; the Japan Science and Technology Agency [PRESTO project to T.H.]; the Yamada Science Foundation [to T.H.]; the Mitsubishi Foundation [to T.H.].
Acknowledgments
We thank Dr. Masahiro Kanaoka, Mr. Hidenori Takeuchi, Dr. Hidefumi Shinohara and Dr. Yoshikatsu Matsubayashi at Nagoya University, and Dr. Minako Ueda at Nara Institute of Science and Technology (NAIST) for helpful suggestions and discussion.
Glossary
Abbreviations
- ANOVA
analysis of variance
- CBB
Coomassie Brilliant Blue
- CRP
cysteine-rich peptide
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EM-CCD
electron-multiplying charge-coupled device
- MALDI-TOF MS
matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- TIRF
total internal reflection fluorescence
- TFA
trifluoroacetic acid
- TFP
tetrafluorophenyl.
References
- Alcor D., Gouzer G., Triller A. Single-particle tracking methods for the study of membrane receptors dynamics. Eur. J. Neurosci. 2009;30:987–997. doi: 10.1111/j.1460-9568.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- Amien S., Kliwer I., Márton M.L., Debener T., Geiger D., Becker D., et al. Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 2010;8:e1000388. doi: 10.1371/journal.pbio.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I., Ting A.Y. Site-specific labeling of proteins with small molecules in live cells. Curr. Opin. Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Dickson B.J. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Dong J., Kim S.T., Lord E.M. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 2005;138:778–789. doi: 10.1104/pp.105.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon A.C., Moran C.R., Gambin Y., Deniz A.A. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- Higashiyama T. Peptide signaling in pollen–pistil interactions. Plant Cell Physiol. 2010;51:177–189. doi: 10.1093/pcp/pcq008. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Hamamura Y. Gametophytic pollen tube guidance. Sex. Plant Reprod. 2008;21:17–26. [Google Scholar]
- Higashiyama T., Inatsugi R. Comparative analysis of biological models used in the study of pollen tube growth. Plant Cell Monogr. 2006;3:265–286. [Google Scholar]
- Higashiyama T., Inatsugi R., Sakamoto S., Sasaki N., Mori T., Kuroiwa H., et al. Species preferentiality of the pollen tube attractant derived from the synergid cell of Torenia fournieri. Plant Physiol. 2006;142:481–491. doi: 10.1104/pp.106.083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Kuroiwa H., Kawano S., Kuroiwa T. Guidance in vitro of the pollen tube to the naked embryo sac of Torenia fournieri. Plant Cell. 1998;10:2019–2031. doi: 10.1105/tpc.10.12.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Yabe S., Sasaki N., Nishimura Y., Miyagishima S., Kuroiwa H., et al. Pollen tube attraction by the synergid cell. Science. 2001;293:1480–1483. doi: 10.1126/science.1062429. [DOI] [PubMed] [Google Scholar]
- Hirel P.H., Schmitter M.J., Dessen P., Fayat G., Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl Acad. Sci. USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh G.Y., Eckard K.J., Nothnagel E.A., Lord E.M. Adhesion of lily pollen tubes on an artificial matrix. Sex. Plant Reprod. 1997;10:173–180. [Google Scholar]
- Kennedy T.E., Wang H., Marshall W., Tessier-Lavigne M. Axon guidance by diffusible chemoattractants: a gradient of netrin protein in the developing spinal cord. J Neurosci. 2006;26:8866–8874. doi: 10.1523/JNEUROSCI.5191-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keough T., Youngquist R.S., Lacey M.P. A method for high-sensitivity peptide sequencing using postsource decay matrix-assisted laser desorption ionization mass spectrometry. Proc. Natl Acad. Sci. USA. 1999;96:7131–7136. doi: 10.1073/pnas.96.13.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Mollet J.C., Dong J., Zhang K., Park S.Y., Lord E.M. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl Acad. Sci. USA. 2003;100:16125–16130. doi: 10.1073/pnas.2533800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- Mai J., Fok L., Gao H., Zhang X., Poo M.M. Axon initiation and growth cone turning on bound protein gradients. J. Neurosci. 2009;29:7450–7458. doi: 10.1523/JNEUROSCI.1121-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 2011;52:5–13. doi: 10.1093/pcp/pcq169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti J., Dircks L., Vancanneyt G., McCormick S. LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994;6:321–338. doi: 10.1046/j.1365-313x.1994.06030321.x. [DOI] [PubMed] [Google Scholar]
- Okuda S., Tsutsui H., Shiina K., Sprunck S., Takeuchi H., Yui R., et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458:357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- Panchuk-Voloshina N., Haugland R.P., Bishop-Stewart J., Bhalgat M.K., Millard P.J., Mao F., et al. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J. Histochem. Cytochem. 1999;47:1179–1188. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Jauh G.Y., Mollet J.C., Eckard K.J., Nothnagel E.A., Walling L.L., et al. A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell. 2000;12:151–163. doi: 10.1105/tpc.12.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Avni A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell. 2004;16:1604–1615. doi: 10.1105/tpc.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y., Minoghchi S., Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- Schlessinger J., Shechter Y., Willingham M.C., Pastan I. Direct visualization of binding, aggregation, and internalization of insulin and epidermal growth factor on living fibroblastic cells. Proc. Natl Acad. Sci. USA. 1978;75:2659–2663. doi: 10.1073/pnas.75.6.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer C.R., Nasrallah M.E., Nasrallah J.B. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- Silverstein K.A., Moskal W.A., Jr, Wu H.C., ., Underwood B.A., Graham M.A., Town C.D., et al. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007;51:262–280. doi: 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- Stein J.C., Howlett B., Boyes D.C., Nasrallah M.E., Nasrallah J.B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl Acad. Sci. USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G. Recent progress in plant reproduction research: the story of the male gametophyte through to successful fertilization. Plant Cell Physiol. 2009;50:1857–1864. doi: 10.1093/pcp/pcp142. [DOI] [PubMed] [Google Scholar]
- Takayama S., Shiba H., Iwano M., Shimosato H., Che F.S., Kai N., et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl Acad. Sci. USA. 2000;97:1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.S., Watanabe M., et al. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature. 2001;413:534–538. doi: 10.1038/35097104. [DOI] [PubMed] [Google Scholar]
- Tang W., Kelley D., Ezcurra I., Cotter R., McCormick S. LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J. 2004;39:343–353. doi: 10.1111/j.1365-313X.2004.02139.x. [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- Ueda M., Sako Y., Tanaka T., Devreotes P., Yanagida T. Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science. 2001;294:864–867. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]