Abstract
Cellular processes are regulated by various mechanical and physical factors in their local microenvironment such as geometric confinements, cell-substrate interactions, and cell-cell contact. Systematic elucidation of these regulatory mechanisms is crucial for fundamental understanding of cell biology and for rational design of biomedical devices and regenerative medicine. Here, we report a generally applicable plasma lithography technique, which performs selective surface functionalization on large substrate areas, for achieving long-term, stable confinements with length scales from 100 nm to 1 cm toward the investigation of cell-microenvironment interactions. In particular, we applied plasma lithography for cellular confinement of neuroblastomas, myoblasts, endothelial cells, and mammary gland epithelial cells, and examined the motion of mouse embryonic fibroblasts in directionality-confined environments for studying the effect of confinements on migratory behavior. In conjunction with live cell imaging, the distance traveled, velocity, and angular motion of individual cells and collective cell migration behaviors were measured in confined environments with dimensions comparable to a cell. A critical length scale that a cell could conceivably occupy and migrate to was also identified by investigating the behaviors of cells using confined environments with subcellular length scales.
1. Introduction
In the physiological environment, cells are regulated by various mechanical and physical factors, such as geometric confinements, cell-substrate interactions, and cell-cell contacts. For instance, cell migration and adhesion, which play essential roles in numerous biological processes including wound healing, tissue morphogenesis, and embryogenesis, can be regulated mechanically [1–5], and substrate stiffness and topographical structures are known to be key factors that control cellular activities [6, 7]. Despite the fact that cells often interact with microscale and nanoscale environments such as fibrous tissues and scaffolds materials, relatively little is known about the effects of the length scale of the confinement on the migratory and adhesion behaviors of cells. Additionally, separating the effects of geometrical confinements, mechanical stiffness, topographical structures, and biochemical signals can be difficult due to their complex interactions. Another key factor that affects cellular migration is cell-cell interaction, as cells are constantly in contact and communication with neighboring cells. Recent studies have suggested that cells can migrate in cooperation with neighboring cells and the collective migration behavior is suggested to have important implications in tissue morphogenesis and cancer metastasis [8, 9]. Conventional tissue culture techniques, nevertheless, have had little emphases on investigating these physical aspects of cell-environment interactions. A systematic means to elucidate cell adhesion and migration processes in such confined environments, therefore, will have profound impacts on various biomedical applications.
As cell-environment interactions are inherently complex, it is necessary to create well-defined conditions to isolate how a cell interacts with its environment. Much knowledge has been gained by using existing cell patterning methods including microcontact printing [10], dip pen nanolithography [11], photolithography [12], inkjet printing [13], surface abrasion [14], and growth on fibers [15] for such investigations. Many existing cell patterning techniques, however, suffer from poor stability, are time and cost-intensive processes, are applicable to only small areas, or have limited resolution, or low controllability. Conventional photolithography, for example, has limited resolution for investigating the interactions between cells and submicron structures, and lithographic techniques based on self-assembled monolayers are subject to cell-dependent and other degradation [16]. To systematically investigate cell-microenvironment interactions, cell confinement needs to possess long-term stability and adjustable length scales that can span from sub-cellular to tissue-level in dimension. Additionally, physical interactions of cells with their microenvironments are dynamic and span time scales orders of magnitude apart. A system that allows for dynamic observation of cell behavior for uncovering details of cell-environment interactions, therefore, is potentially useful for shedding light upon such behavior.
To that end, we have developed a plasma lithography technique for cell confinement based on spatially selective plasma functionalization of substrate materials [17–19] to guide cellular attachment. Plasma surface treatment is simple to implement and is suitable for functionalizing polymeric materials which due to their tunability are ideally suited for biological applications. To create the confinement by plasma lithography, a 3D polymer mold is placed on a surface to form a series of holes or channels into which plasma flows to react with the surface. This produces a template of surface functionalizations to guide the attachment of cells (supplementary Fig. S1). In this work, we examined the characteristics of plasma lithography for cell confinement and explored its applications for investigating cellular adhesion and migration on directionality-confined environments with length and time scales spanning several orders of magnitude.
2. Materials and Methods
2.1 Plasma lithography
In plasma lithography, selective shielding of plasma contact to a surface is used to produce a chemical template as described above. Briefly, during this process a polydimethylsiloxane (PDMS) mold having 3D features is placed in conformal contact with a substrate and exposed to the action of atmospheric plasma. Due to physical channels formed between the 3D mold and the substrate, the substrate material is selectively, chemically modified by the plasma (Fig. S1). This selective modification forms a pattern that can serve as a means of cellular confinement. In this study, 60 mm polystyrene culture dishes (VWR) were plasma exposed for 10 minutes in a Harrick Plasma cleaner (model PDC-001) and were then UV sterilized for 10 minutes before being seeded with cells. PDMS and glass substrates can also be patterned and used to guide cellular attachment, though all results discussed pertain to using polystyrene as a substrate. Original masters used to create 3D molds via PDMS molding [20] were created by three separate methods. Patterns with line widths larger than 500 µm were created by laser engraving polyethylene terephthalate (PET) substrates using a laser etcher (VersaLASER model VLS 2.30). Patterns between 500 µm and 3 µm were created via photolithography using both positive and negative resist, and patterns with line widths from 100 nm to 2 µm were created via electron beam lithography. PDMS cast off of these structures to make shielding molds was mixed in either a 10:1 or 8:1 ratio of polymer base to curing agent and degassed before being cured for 24 hours at room temperature (See Supplementary Data S1 for details on mold creation and cell confinement).
2.2 Cell culture
All cell lines used were obtained from the American Type Culture Collection (ATCC) and included: ATCC CRL-1658 mouse embryo fibroblast [3T3], ATCC HTB-26 human mammary gland adenocarcinoma MDA-MB-231 [231], ATCC CRL-1772 mouse myoblast [C2C12], ATCC CRL-1730 human umbilical vein vascular endothelial cell [HUVEC], and ATCC CRL-2266 human neuroblastoma [SH-SY5Y]. Primary cells were also studied and included embryonic chicken skeletal muscle cells and embryonic chicken cardiomyocytes. C2C12 cells were used from passage 3 to 10, HUVEC cells from passage 2 to 10, and SH-SY5Y cells from passage 2 to 10. Cells were maintained at 37°C, 100% humidity and 5% CO2 and seeded at 25–40% confluence. Growth medium and medium replacement for all cells lines was per ATCC guidelines.
2.3 Immunofluorescence
Cells were stained for F-actin with Alexa Fluor® 555 tagged phalloidin (Invitrogen), for vinculin to mark focal adhesions with monoclonal anti-vinculin−FITC antibodies (Sigma), and for nuclei with ProLong® Gold antifade reagent with 4´,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Cells were first fixed with 3.7% formaldehyde (Polysciences, inc) then permeabilized with 0.1% Triton X-100 (Astoria-Pacific). They were then stained and the antifade reagent was added before cover slips were placed over the cells and sealed with nail polish.
2.4. Live cell imaging
Sequential images of cells were captured by transferring culture dishes having patterned surfaces to a custom manufactured microscope stage mounted observation platform. The platform was comprised of a microscope slide warmer (AmScope Model TCS-100) to which was affixed a plastic enclosure that provided for proper atmosphere and humidity. The atmosphere was supplied by feeding 5% CO2 passed through a 0.3 µm in line filter at a slight overpressure to the chamber, and water trays were placed inside the enclosure to maintain humidity. Live phase contrast images of cells were then recorded with the system described below at a frequency ranging from 30 seconds to 2 minutes. After live images were collected, the cells were transferred back to a standard incubator and observed. All cells displayed normal growth for several days after being maintained on the microscope mounted incubation system.
2.5 Microscopy and image analysis
Phase contrast images of cells were captured using an inverted Nikon (model TE2000-U) microscope with either a SPOT camera (Diagnostic Instruments model 2.2.1), or, for sequential images in the microscope mounted cell chamber, an Imaging Source (model DMK41AU02) camera. Fluorescence images were captured on an inverted microscope (Leica model DMI4000B) with a Cooke SensiCam camera. Cell images were analyzed by sequential tracking of a cell’s long axes at each time point in a series of images. A custom Image J macro was used to record angle, endpoints and center point for each cell and data was then exported for further analysis.
3. Results
3.1 Characteristics of confinement via plasma lithography
The properties of the surface patterned by plasma lithography have been reported previously [17], and the technique of plasma lithography itself is stable, parallel in nature, inexpensive, involves no harsh chemicals during processing and can pattern surfaces with features from 100 nm to centimeters in size. The high resolution possible is a result of the fact that any PDMS compatible surface can be used for creation of the 3D mold, as long as the topography is suitable for the final, plasma patterning. The approach allows observation for up to several weeks of cell behavior in real time, is applicable to many kinds of materials including standard tissue culture plates, PDMS, and glass, and leaves no visual signature on the surface that is patterned.
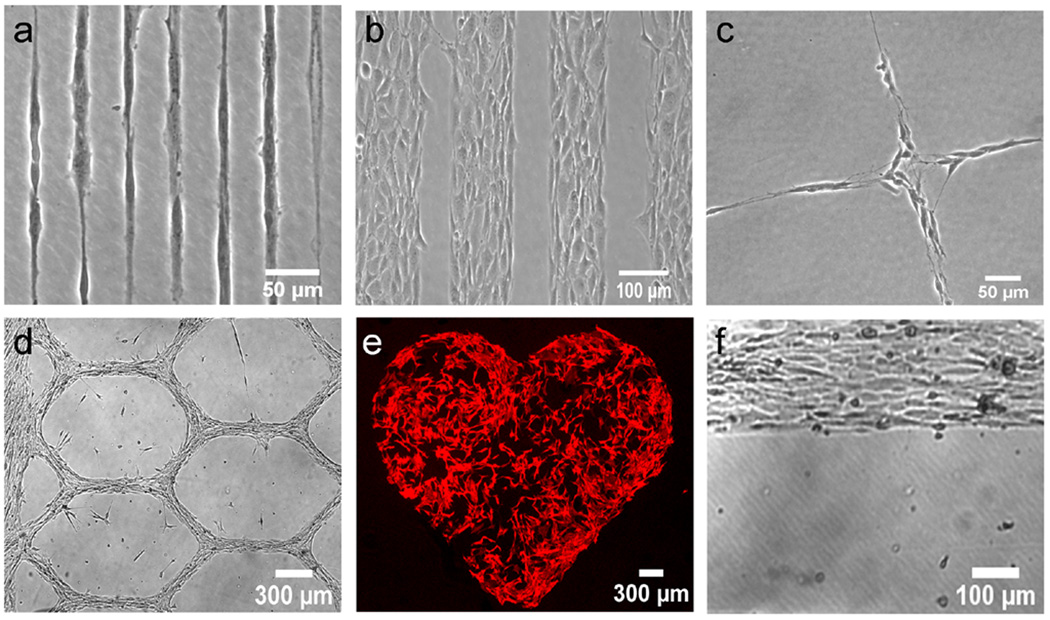
In order to characterize the nature of the cell confinement achieved via plasma lithography, the effects of plasma patterning on cell behavior were first examined for several different cell types. The results of cell seeding on plasma patterned surfaces are shown in Fig. 1. The diverse cell lines, including SH-SY5Y neuroblastoma, C2C12 myoblasts, and 3T3 fibroblasts, could be confined to patterns ranging in size from micrometers to millimeters (see Supplementary Data S2 for results with additional cell types). Cells could freely move on plasma treated surfaces and moved randomly within the confined areas made by the pattern. Occasionally at boundaries, a small percentage of cells (<10%) attempted to migrate over the untreated, hydrophobic areas for a short period of time. These cells, however, did not appear to be stable on the untreated areas, i.e. they would not remain off of or explore beyond the cell friendly area for more than the time a transient membrane extension was maintained, and the vast majority of cells were completely confined within treated areas. The cell types examined all responded to the patterning with similar degrees of recognition and immediately recognized the pattern as soon as adherence to the surface began (Videos V1 and V2). The 10 minute plasma treatment stated above was seen to produce repeatable, satisfactory guidance and was therefore used for all cells and substrate materials. Cell morphology, growth rate, and immunostaining of focal adhesion and cytoskeletal proteins revealed cells cultured on the plasma modified areas maintained their normal physiologies (Supplementary Data S3). In addition, we tested the stability of the confinement for long-term cell culture and it was observed that the cell guidance was stable, i.e. the patterning maintained the ability to guide cells over long time periods. In our experiments, all cell types could be confined for long time periods (~2 weeks) before experiments were stopped due to over confluence. For C2C12 myoblasts, the cells could be cultured for ~2 months after switching to differentiation media. The cells maintained the original patterns without any observable pattern degradation (Fig. 1f and Supplementary Data S4). In addition, the use of plasma lithography for cell confinement could also be integrated with microfluidics for providing multiple fluidic and biochemical stimuli simultaneously (Supplementary Data S5) [21]. These results indicate that plasma lithography is suitable for investigating the effects of confinements on the behavior of various types of cells.
Figure 1. Cell guidance.
(a) Single SH-SY5Y neuroblastomas grown on 12 µm wide lines. (b) 3T3 fibroblasts grown on 120 µm wide lines. (c) SH-SY5Y cells confined to a grid having 30 µm wide lines. (d) C2C12 myoblasts forming a grid having 100 µm wide lines. (e) 3T3 fibroblasts on large millimeter sized patterns stained for actin. (f) C2C12 myoblasts at a plasma modified polystyrene-native polystyrene interface after 53 Days in culture.
3.2 Migration of individual cells
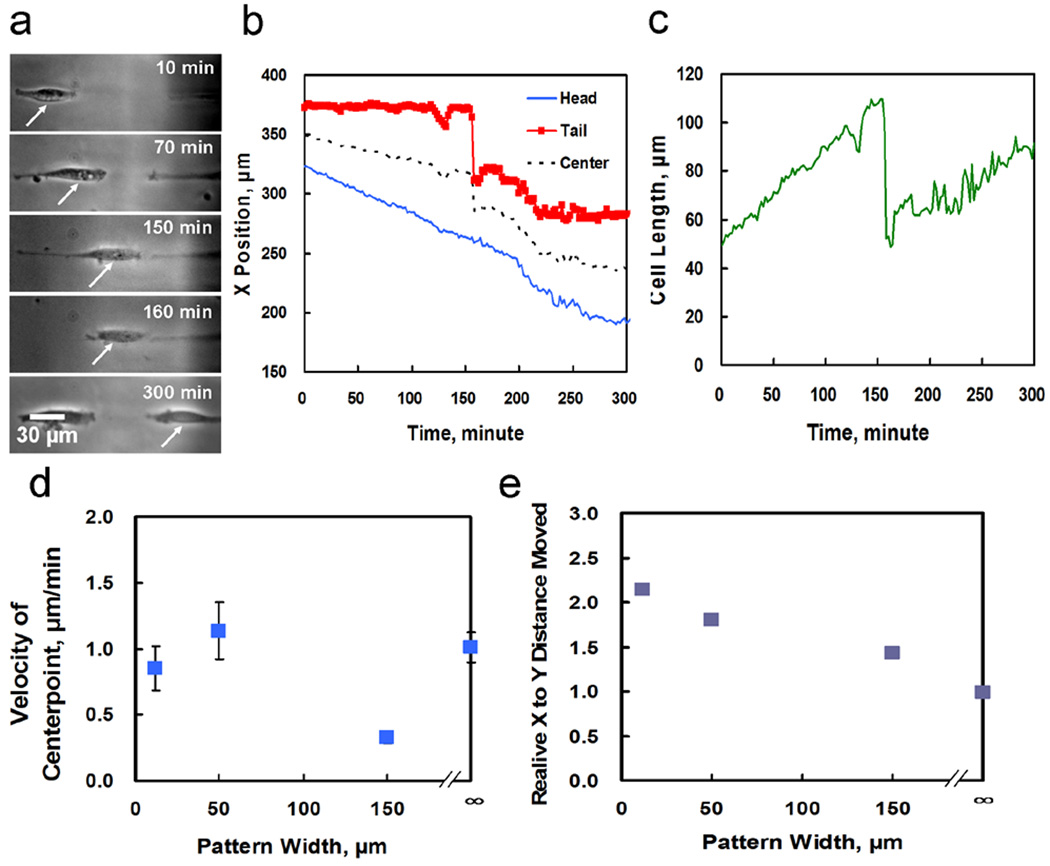
To study the effects of confinment, we applied plasma lithography in conjunction with live cell imaging to explore the dependence of cell migration on one-dimensional confined environments. The movement of 3T3 fibroblasts, an essential component of wound healing, was examined by tracking the motion of individual cells at low density. Fig. 2a shows a representative cell migrating on a 12 µm wide line for over 5 hours. Rapid migratory behaviors including tail snapping and periodic directional changes were observed [22]. As shown in Fig. 2a–c, the position of the head of the polarized cell increased linearly reflecting the constant polymerization rate at the leading side of the cell, while the movement of the tail was relatively constant initially until a rapid retraction due to breaking of focal adhesion was observed at the trailing edge of the cell. These observations support the applicability of plasma lithography for investigating the cell migration process with a high spatiotemporal resolution.
Figure 2. Cell motility.
(a) Images of a 3T3 fibroblast confined to a 12 µm line tracked over time. (b) Position data gathered for the cell in (a) moving over time. The sharp jumps at ~160 minutes correspond to rapid tail retractions after tail detachment. (c) Cell length over time for the cell shown in (a). (d) Velocity of cells on different size patterns, the smallest pattern is 12 µm wide. Error bars are standard deviation. (e) Relative horizontal and vertical distances moved showing spatial confinement by patterning for different sized patterns. ∞ corresponds to a homogeneously plasma modified substrate.
Cellular motion was also investigated on line patterns of different widths and on homogeneous surfaces. The adhesion and migratory behaviors of the cells including the size, distance traveled, angular movements, cell velocity, and the ratio of distances moved in the horizontal and vertical directions relative to the direction of line patterns were measured. When the movement of cells on line widths ranging from 12 µm to 150 µm was examined, it was observed that the cells retained their overall velocities in confined environments as compared to their movement when migrating on homogeneous surfaces (Fig. 2d). In addition, the cells were not affected in terms of their average length or total distance moved (data not shown). The principle difference from the homogeneous surface was the effect of the cellular confinement on the lines of the pattern which changed the directionality of the movement. As can be seen in Fig. 2e which shows the ratio of cellular movement between the X and Y directions, the patterns served to confine the cellular motion, with smaller line widths able to confine cells more than large line widths. For the smallest line widths (i.e., 12 µm), the direction of movement becomes essentially one dimensional as the cells are confined on a pattern that is narrower than a normal cell width and hence cannot move perpendicular to the direction of the line. Normal width is considered to be the cell width measured on a homogeneous plasma modified surface where a cell has large room to spread. Therefore, the principle characteristics of cell migration are maintained in directionality-confined environments.
3.3 Cell-cell Interactions
To explore the role of cell-cell contact on migratory behavior, 3T3 fibroblasts were confined to a one-dimensional line pattern. In the experiment, the cell seeding density was adjusted to manipulate the chance of cell-cell contact occurring. Fig. 3 shows the behavior of fibroblasts migrating on a pattern of plasma modified lines 12 µm wide separated by 35 µm wide native polystyrene areas with data collection commencing from directly after cells were plated. The cell motion and cell-cell contact on the pattern were observed continuously for over 12 hours (Video V1). The area examined was near the end of a pattern of lines which meant that immediately at the edge of the pattern there existed a large hydrophilic area with a large number of cells (see Supplementary Data S6). Fig. 3a shows a video time series of a cell migrating leftward and contacting a neighboring cell and Fig. 3b shows selected traces of cell motions along the direction of the line pattern.
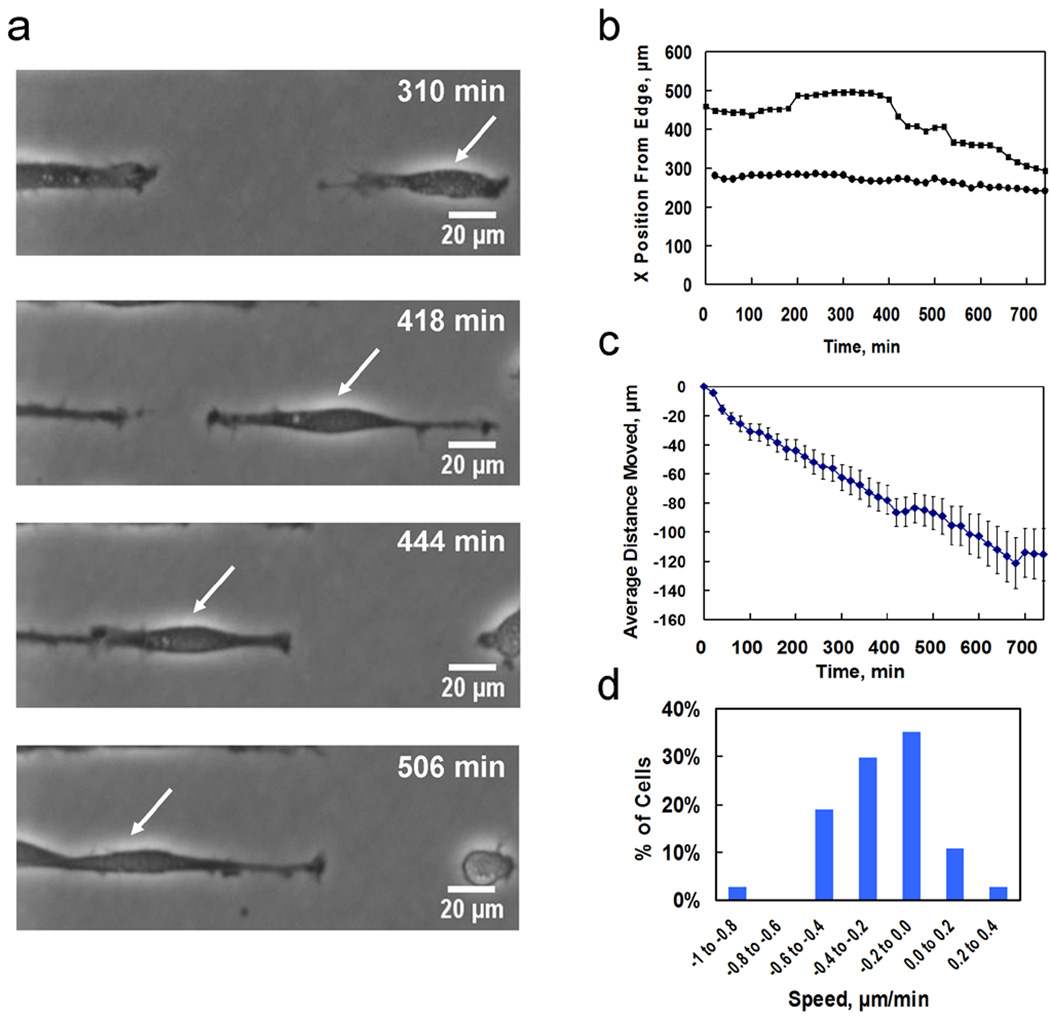
Figure 3. Cell-to-cell interactions.
(a) Image sequence showing a 3T3 fibroblast migrating leftward and contacting a neighboring cell then continuing to migrate leftward at an increased speed. (b) Migration data for selected cells shown in video V1, tracked over 12 hours. Black squares depict the cell in (a) which changes migration behavior (change in slope) upon contact with a neighboring cell. Black circles show migration behavior of a cell which does not change migration behavior upon contact with a neighboring cell. (c) Average distance moved for all cells over the observation period, showing the leftward bias, error bars are SEM. (d) Average speed distribution of cells during 740 minutes. (e) Number of cells displaying types of migration behavioral changes.
Several unique aspects of cellular movement could be discerned from the experiment. Firstly, the majority of cells have a bias to move leftwards towards the large mass of cells on the hydrophilic area connected to the pattern of lines. This can be seen in Fig. 3c which shows that the average distance moved displays a clear bias motion that is distinct from the random motion observed for isolated cells. Additionally, Fig. 3d shows the average speed distribution which indicates that 86% of the cells moved leftwards towards the mass of cells over the time period examined. The cells, secondly, displayed a strong randomness to their movement even though they are moving on identical lines under identical conditions. This includes random directional changes, movement of cells against the larger trend of a leftward bias, and a wide distribution of migration speeds. This is indicative of the complex and stochastic behavior of multicellular migration and aggregation. Furthermore, the directionality-confined environment created by plasma lithography allowed us to observe the effect of cell-cell contacts on the speed of cell migration. Interestingly, 65% of cells which experienced cell-cell contact showed an increase in speed immediately after contact. The remaining 35%, however, did not show any noticeable change in migration upon contact with a neighboring cell. This highlights the complexity of the global behavior of the cell migration process, which can not be explained by considering only an individual factor.
3.4 Cell-subcellular pattern interactions
In many physiological and biomedical scenarios, cellular interaction with subcellular scale structures including various fibrous materials and geometric cues is key to determining cell behavior. To study subcellular confinements to which cellular migration responds, lines from 100 nm to 12 µm in width spaced 35 µm apart were produced as described above. Cells living on large (4 – 12 µm), cell friendly, plasma modified areas were seen to migrate or extend membrane segments onto thin lines (see Supplementary Data S7). This resulted in the cell becoming highly elongated and such cells were observed to maintain this extended morphology over long time periods (days to weeks). This implied that a pattern at this length scale was not a significant hindrance to cellular movement or viability and that a cell could effectively interact with such a stimulus. A principle indication of this was the fact that cells migrate normally, i.e., at speeds and total distances equal to that on totally plasma modified areas. Additionally, cells would adhere stably for long time periods on such lines even when the cell did not spread out and span multiple lines [23].
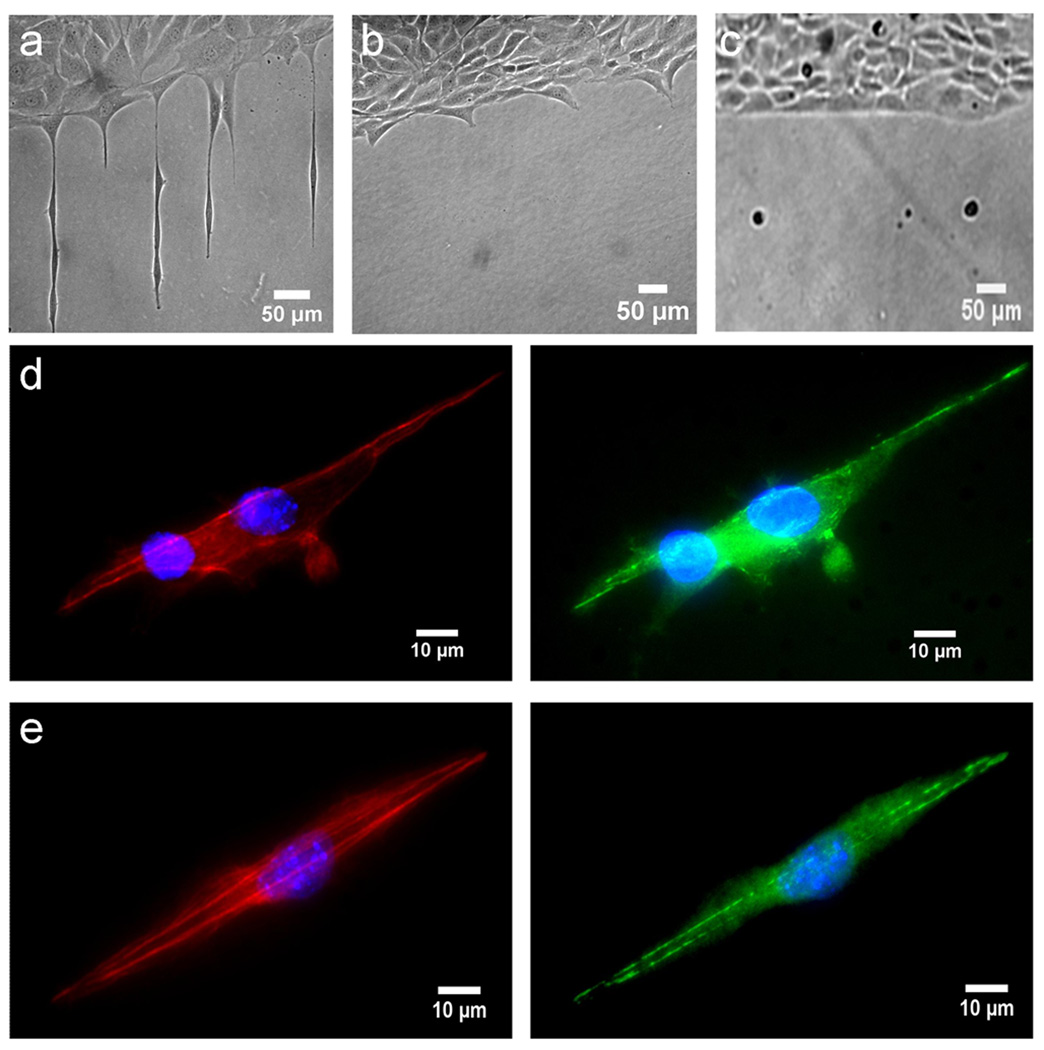
We therefore further investigated the behaviors of cells on line widths from 100 nm – 2 µm (see Supplementary Data S8). The growth of cells on these smaller patterns can be seen in Fig. 4 which shows the cell growth on subcellular size patterns. At lines with larger widths, i.e. the 1 or 2 µm lines, cells could live or extend membrane segments onto the lines. For these two larger patterns it was observed that cells would extend a membrane segment onto the pattern if spreading next to it, and that cells that settled out of solution far from a pattern edge could grow on such patterns even when isolated. For the two smaller size patterns, i.e., 100 nm and 500 nm, however, different cellular behavior was seen and cells were not able to occupy the lines. For the 500 nm wide lines, cells were no longer able to migrate or live on the lines though they did attempt to occupy the lines. The attempt to occupy a line can be seen in Fig. 4b which shows cells growing on a homogenously plasma treated area that have extended membrane segments onto the lines. Confirmation that these extensions are related to the patterning and not random probings of the surface was seen by comparing the spacing of the membrane extrusions and observing that they matched with the pattern of the confinement. The cells growing on the 100 nm wide lines also did not occupy the submicron patterns and furthermore did not extend any membrane segment to investigate the area. Based on these results, a critical length scale may exist for a cell to occupy and migrate in a confined environment.
Figure 4. Cell interaction with subcellular stimuli.
(a) Cells on a 2 µm wide pattern of plasma treated polystyrene. (b) Cells on a 500 nm pattern with cells attempting to extend membrane segments onto the pattern. (c) Cells on a 100 nm wide pattern. (d) 3T3 Cells on a 1 µm wide line pattern stained for F-actin (red), vinculin (green), and nucleus (blue). (e) Cells on a 2 µm wide line pattern stained for F-actin (red), vinculin (green), and nucleus (blue). Images are representative of three experiments using identical line sizes. Cells are 3T3 fibroblasts.
4. Discussion
In this study, plasma lithography is demonstrated as a useful tool to study cell migration and adhesion processes in confined environments. The basis for the cell confinement is the chemical functionalization by plasma on the surface of polystyrene. Areas that are plasma modified possess altered, hydrophilic functional groups compared to regions not exposed to plasma which are hydrophobic and naturally possess phenyl functional groups composed of the native polystyrene. The resulting chemical and topographical nature of the surface has been described previously [17] and is composed of a mix of oxygen based functional groups having a roughness of 3.8 ± 0.7 nm. The pattern has a topographical component that is below that which cells typically sense [7, 24]. This suggests the confinement is primarily chemical, instead of mechanical or topological, in nature. Though the cells can attach on unmodified areas, they generally have an abnormal morphology, are less healthy, and displayed disrupted focal adhesion and actin configurations [25] (see also supplementary S3 and Video V3). Additionally, live observation of cells has shown that when encountering a plasma modified area during migration, cells have a strong tendency to stay in the modified region. These observations have also shown that cells seeded initially on plasma modified areas probe their surroundings and when encountering unmodified areas retract their filopodia with a preference to remain on a modified area. Collectively, the results support that plasma lithography can create chemically functionalized confinements for guiding the adhesion and migration of cells. The nature of the differing affinity of surface attachments is believed to be related to both the hydrophilicity and the composition of the altered functional groups present after exposure to plasma. The preference for treated areas has been studied before [25, 26], and likely results from a higher affinity of both the cells own attachment mechanisms and attachment of other proteins in favorable configurations on the treated surfaces.
A unique advantage of plasma lithography for cell study is the long-term stability that can be achieved. This is conceivably due to the direct surface functionalization of the substrate materials in plasma lithography. It has been previously observed that functionalization created by a plasma on polymeric materials can be stable over long time periods in liquids that avoid direct exposure to atmospheric air [27, 28]. This is advantageous as loss of guidance observed with other types of patterning such as by cellular degradation of the pattern [16] or physical desorption are not factors with plasma lithography, since the pattern is created directly as part of the surface. This facilitates long term investigations of cell behaviors, especially those with time periods longer than a few days where pattern degradation may occur (e.g., cell differentiation and tissue morphogenesis). The patterning process also does not involve any cytotoxic compounds and solvents during any process step. This is useful to both eliminate any accidental or residual contamination and for in vivo uses such as for patterning tissues for therapeutic usage or for functionalizing the surfaces of implants.
Using plasma lithography, our data indicate that the principle characteristics of cell motion are not affected by the directionality-confined environment. This reflects the independence of the local actin-based migration machinery within the cell [29]. This finding also enables cell migration studies to be performed in one-dimensional spaces. This presents several advantages over conventional techniques for studying cell migration. With cells being spatially confined in one dimension, the uncertainty in cell motion observed in conventional migration assays can be eliminated and the single cell migration dynamics can be observed with high spatiotemporal resolution. Furthermore, the well-defined migratory path potentially facilitates quantitative comparison of cell motion in response to chemotactic, pharmacological, and other biochemical stimuli. We also observed that cells at a higher density displayed biased random motion collectively in the confined environments. The cause of the collective migration bias is uncertain though it may be a result of the high density of cells just at the end of the pattern. The large number of cells could create attractive signals via cell-cell contacts or by release of diffusible factors that induce the cells to aggregate [30]. Observation of the complex collective migration behaviors in unconfined environments is more challenging due to the cell-cell interactions in multiple directions with contact areas changing dynamically. The ability to confine cells one-dimensionally using plasma lithography, on the other hand, simplifies cell motion and can facilitate quantification of cell behaviors, such as the probability of velocity change upon cell-cell contact. The quantitative information obtained will potentially be useful for computational analyses and systems biology studies of collective cell behaviors.
The observations of cells growing on subcellular patterns indicate a length scale dependence of cellular adhesion and migration in confined environments. This ranges from the larger patterns where the cells can migrate in a stable manner to the 500 nm wide line pattern that appears to be at a transition zone where the interaction with the surface is sufficient to induce the cell to partially adhere, but not sufficient for the cell to migrate there or to occupy the line. Smaller patterns appeared to be inadequate to induce cellular occupancy or even membrane extrusion. These responses could ultimately be related to a critical length scale that is suitable for cell viability or investigation by membrane extension. Related minimum adhesion area requirements have been investigated before, though previous studies usually have cells spreading across multiple adhesion areas, which represents different conditions mechanobiologically [1, 2, 31]. In the case of the current experiments, the confinements are spaced farther apart than a single cell can span and the total adhesion area is not limited. Line widths of 100 and 500 nm are in fact larger than the 58–100 nm stated as being the dimension needed for integrin attachment [31, 32]. As can be seen, the cells could not fully occupy the area at a width of 500 nm and did not even probe the area for a signal of 100 nm. The result provides insights into the length scale dependence for cellular adhesion and migration. The course of this critical length scale may be related to the insufficient space for cellular processes to proceed in a normal manner, the local stress in such a highly elongated configuration, or insufficient adhesion sites to initiate the signaling pathways needed to induce the cell to extend in a given direction. Further experiments are required to determine the conclusive molecular limiting factor for cell migration and adhesion in confined environments.
5. Conclusions
In this study, we have developed an easy-to-use, cost effective, and stable plasma lithography approach for long term investigation of cell-environment interactions across multiple length scales for a diverse range of cell types. With the simplicity and effectiveness of plasma lithography, the effects of complex cell-environment interactions on cell migration and other cellular behaviors can be investigated systematically toward applications in mechanoregulation and regenerative medicine.
Supplementary Material
Acknowledgements
We thank Dr Brook Beam from the University of Arizona’s Keck Facility for assistance in SEM, ASU Center for Solid State Electronics Research for technical assistance in nanofabrication, and Samantha Whitman for work done on primary cells. M. J. is supported by the NIH Cardiovascular Training Grant, the Arizona Technology Research Initiative Fund (TRIF), and Achievement Rewards for College Scientists (ARCS). This work is supported by the NIH Director’s New Innovator Award (1DP2OD007161-01), James S. McDonnell Foundation, and NSF Nanomanufacturing (0855890).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ingber DE. Tensegrity-based mechanosensing from macro to micro. Prog Biophys Mol Biol. 2008;97(2–3):163–179. doi: 10.1016/j.pbiomolbio.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D-H, Wong PK, Park J, Levchenko A, Sun Y. Microengineered platforms for cell mechanobiology. Annu Rev Biomed Eng. 2009;11(1):203–233. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. Directing cell migration with asymmetric micropatterns. Proc Natl Acad Sci U S A. 2005;102(4):975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 5.Kaji H, Kawashima T, Nishizawa M. Patterning cellular motility using an electrochemical technique and a geometrically confined environment. Langmuir. 2006;22(25):10784–10787. doi: 10.1021/la0610654. [DOI] [PubMed] [Google Scholar]
- 6.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 7.Yim EKF, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26(26):5405–5413. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rørth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25(1):407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 9.Friedl P, Hegerfeldt Y, M. T. Collective cell migration in morphogenesis and cancer. Int J Dev Biol. 2004;48(5–6):441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- 10.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20(23–24):2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 11.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295(5560):1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 12.He W, Halberstadt CR, Gonsalves KE. Lithography application of a novel photoresist for patterning of cells. Biomaterials. 2004;25(11):2055–2063. doi: 10.1016/j.biomaterials.2003.08.055. [DOI] [PubMed] [Google Scholar]
- 13.Kim JD, Choi JS, Kim BS, Chan Choi Y, Cho YW. Piezoelectric inkjet printing of polymers: stem cell patterning on polymer substrates. Polymer. 2010;51(10):2147–2154. [Google Scholar]
- 14.Diener A, Nebe B, Lüthen F, Becker P, Beck U, Neumann HG, et al. Control of focal adhesion dynamics by material surface characteristics. Biomaterials. 2005;26(4):383–392. doi: 10.1016/j.biomaterials.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Bashur CA, Dahlgren LA, Goldstein AS. Effect of fiber diameter and orientation on fibroblast morphology and proliferation on electrospun poly(d,l-lactic-co-glycolic acid) meshes. Biomaterials. 2006;27(33):5681–5688. doi: 10.1016/j.biomaterials.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Nelson CM, Raghavan S, Tan JL, Chen CS. Degradation of micropatterned surfaces by cell-dependent and -independent processes. Langmuir. 2002;19(5):1493–1499. [Google Scholar]
- 17.Junkin M, Watson J, Vande Geest JP, Wong PK. Template-guided self-assembly of colloidal quantum dots using plasma lithography. Adv Mater. 2009;21(12):1247–1251. [Google Scholar]
- 18.Keyes J, Junkin M, Cappello J, Wu X, Wong PK. Evaporation-induced assembly of biomimetic polypeptides. Appl Phys Lett. 2008;93(2):023120–023123. [Google Scholar]
- 19.Song M, Uhrich KE. Optimal micropattern dimensions enhance neurite outgrowth rates, lengths, and orientations. Ann Biomed Eng. 2007;35(10):1812–1820. doi: 10.1007/s10439-007-9348-0. [DOI] [PubMed] [Google Scholar]
- 20.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography - a convenient and versatile tool for micropatterning bifunctional synthetic surfaces for applications in biosensing and tissue engineering. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 21.Wong PK, Yu F, Shahangian A, Cheng G, Sun R, Ho C-M. Closed-loop control of cellular functions using combinatory drugs guided by a stochastic search algorithm. Proc Natl Acad Sci USA. 2008;105(13):5105–5110. doi: 10.1073/pnas.0800823105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttenlocher A, Sandborg RR, Horwitz AF. Adhesion in cell migration. Curr Opin Cell Biol. 1995;7(5):697–706. doi: 10.1016/0955-0674(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 24.Loesberg WA, te Riet J, van Delft FCMJM, Schön P, Figdor CG, Speller S, et al. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials. 2007;28(27):3944–3951. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 25.van Kooten TG, Spijker HT, Busscher HJ. Plasma-treated polystyrene surfaces: model surfaces for studying cell-biomaterial interactions. Biomaterials. 2004;25(10):1735–1747. doi: 10.1016/j.biomaterials.2003.08.071. [DOI] [PubMed] [Google Scholar]
- 26.Webb K, Hlady V, Tresco PA. Relative importance of surface wettability and charged functional groups on NIH 3T3 fibroblast attachment, spreading, and cytoskeletal organization. J Biomed Mater Res. 1998;41(3):422–430. doi: 10.1002/(sici)1097-4636(19980905)41:3<422::aid-jbm12>3.0.co;2-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami T, Kuroda S-i, Osawa Z. Dynamics of polymeric solid surfaces treated with oxygen plasma: effect of aging media after plasma treatment. J Colloid Interface Sci. 1998;202(1):37–44. [Google Scholar]
- 28.Thurston RM, Clay JD, Schulte MD. Effect of atmospheric plasma treatment on polymer surface energy and adhesion. J Plast Film Sheet. 2007;23(1):63–78. [Google Scholar]
- 29.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 30.Adams CL, Chen Y-T, Smith SJ, James Nelson W. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin- green fluorescent protein. J Cell Biol. 1998;142(4):1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehnert D, Wehrle-Haller B, David C, Weiland U, Ballestrem C, Imhof BA, et al. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J Cell Sci. 2004;117(1):41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 32.Girard PP, Cavalcanti-Adam EA, Kemkemer R, Spatz JP. Cellular chemomechanics at interfaces: sensing, integration and response. Soft Matter. 2007;3:307–326. doi: 10.1039/b614008d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.